Abstract
Since the discovery of the phosphorylation of the 40S ribosomal protein S6 (rpS6) about four decades ago, much effort has been made to uncover the molecular mechanisms underlying the regulation of this post-translational modification. In the field of neuroscience, rpS6 phosphorylation is commonly used as a readout of the mammalian target of rapamycin complex 1 signaling activation or as a marker for neuronal activity. Nevertheless, its biological role in neurons still remains puzzling. Here we review the pharmacological and physiological stimuli regulating this modification in the nervous system as well as the pathways that transduce these signals into rpS6 phosphorylation. Altered rpS6 phosphorylation observed in various genetic and pathophysiological mouse models is also discussed. Finally, we examine the current state of knowledge on the physiological role of this post-translational modification and highlight the questions that remain to be addressed.
Keywords: rpS6 phosphorylation, mRNA translation, ribosome, mTOR, S6K, PP-1, brain, signaling cascades
Introduction
The eukaryotic ribosome is composed of the small 40S and the large 60S subunits, comprising together 4 ribosomal RNA species and 79 ribosomal proteins (Kressler et al., 2010). In many organisms, ribosomal proteins undergo various post-translational modifications, including phosphorylation, acetylation, methylation, O-linked β-N-acetylglucosaminylation, and ubiquitylation (Xue and Barna, 2012). Historically, the phosphorylation of the 40S ribosomal protein S6 (rpS6) was the first post-translational modification described (Gressner and Wool, 1974). The presence of phospho-rpS6 (p-rpS6) at different levels in a 2D gel provided the first evidence that rpS6 phosphorylation could occur at several residues (Lastick et al., 1977). Ensuing studies identified five evolutionary conserved and clustered carboxy-terminal phospho-sites, which undergo phosphorylation in an ordered manner, beginning with Ser236 and followed sequentially by Ser235, Ser240, Ser244, and Ser247 (Martin-Pérez and Thomas, 1983; Wettenhall et al., 1992; Meyuhas, 2008, 2015). Intriguingly, the exact function of the post-translational modification of this indispensable ribosomal protein remains enigmatic. Despite the large debate regarding its physiological role, rpS6 phosphorylation is commonly used as a marker for neuronal activity and a readout of mammalian target of rapamycin complex 1 (mTORC1) activity (Meyuhas, 2008, 2015; Mahoney et al., 2009; Knight et al., 2012). This review summarizes our current knowledge regarding the molecular mechanisms as well as the variety of stimuli modulating rpS6 phosphorylation in the nervous system.
Regulation of rpS6 phosphorylation
The p70/p85 S6 kinase 1 (S6K1), which is able to catalyze the phosphorylation of rpS6 at all sites, was the first kinase identified (Krieg et al., 1988; Ferrari et al., 1991; Bandi et al., 1993; Meyuhas, 2008, 2015). Further studies described additional protein kinases targeting selectively the Ser235 and Ser236 residues. These include p90 Ribosomal S6 Kinases (RSK1-4) (Roux et al., 2007), Protein Kinase C (House et al., 1987), Protein Kinase A (PKA) (Moore et al., 2009; Valjent et al., 2011; Yano et al., 2014; Biever et al., 2015), Protein Kinase G (Yano et al., 2014), and Death-Associated Protein Kinase (DAPK) (Schumacher et al., 2006) (Figure 1). Although less studied, the residue Ser247 has been identified as a target of Casein Kinase 1 (Hutchinson et al., 2011). Contrasting with the diversity of kinases regulating rpS6 phosphorylation, the dephosphorylation of the five residues is achieved by a single phosphatase: the Protein Phosphatase-1 (PP-1) (Belandia et al., 1994; Hutchinson et al., 2011) (Figure 1). Since the molecular mechanisms regulating rpS6 phosphorylation have been recently extensively reviewed (Meyuhas, 2015), we focus on the contribution of S6K1/2 kinases and the PKA/PP-1 pathway, being the main upstream mechanisms described to regulate rpS6 phosphorylation in the nervous system.
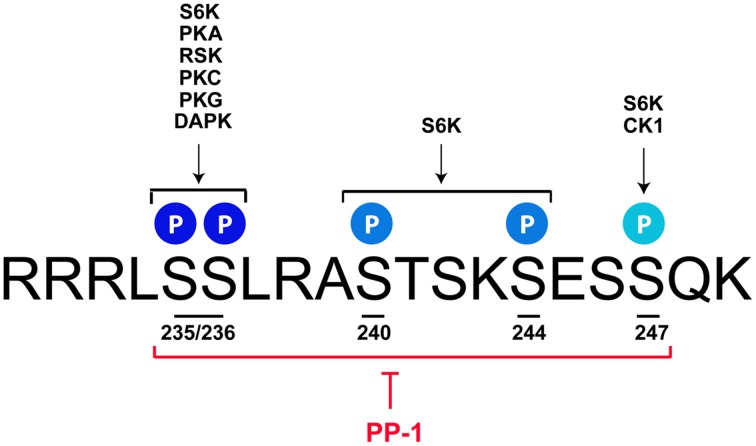
Figure 1.
rpS6 phosphorylatable residues are targeted by multiple kinases and dephosphorylated by PP-1. Mus musculus sequence of the C-terminal domain of rpS6 depicting the 5 phosphorylatable sites and their respective kinases. S6K catalyzes the phosphorylation of all the residues, while PKA, RSK, PKC, PKG, and DAPK target the S235 and S236 sites. CK1 selectively phosphorylates the S247 residue. All phospho-sites are dephosphorylated by PP-1. See text for details.
S6K1/2 pathway
In mammalian cells, two different genes encode two isoforms of the S6 Kinase, S6K1, and S6K2. S6K1 has cytosolic and nuclear isoforms (p70 S6K1 and p85 S6K1, respectively), whereas both S6K2 isoforms (p54 S6K2 and p56 S6K2) are primarily nuclear (Martin et al., 2001). As demonstrated by the use of S6K1/S6K2 double knockout mice, both isoforms contribute to the regulation of basal and inducible rpS6 phosphorylation at S235/236 and S240/244 sites (Pende et al., 2004; Kroczynska et al., 2009; Chauvin et al., 2014). Different observations were made in single S6K knockout mice. Indeed, while the S6K2 knockout mice display a reduction of rpS6 phosphorylation only at S235/236 sites in the hippocampus (Antion et al., 2008a), S6K1-deficient mice show no alterations (Antion et al., 2008a; Bhattacharya et al., 2012). Compensatory mechanisms taking place in the single knockout mice could explain these latter observations since viral-mediated overexpression of a constitutive-active S6K1 (S6K1 CA) or kinase-inactive S6K1 (S6K1 KI) in the medial prefrontal cortex increases or decreases basal rpS6 phosphorylation, respectively (Dwyer et al., 2015). Pharmacological evidences also support the critical role of S6K in the regulation of rpS6 phosphorylation. S6K1/2 undergo phosphorylation at 8 Ser/Thr phospho-sites, including 4 serine residues in the C-terminal autoinhibitory domain. Phosphorylation of the autoinhibitory domain was originally proposed to trigger a more relaxed conformation of the protein allowing its phosphorylation at T389 by mTORC1 leading to S6K activation (Dennis et al., 1998). Thus, the blockade of canonical mTORC1/S6K signaling by the mTORC1 inhibitor rapamycin suppresses both basal and stimuli-induced rpS6 phosphorylation in various brain areas (Kelleher et al., 2004; Takei et al., 2004; Cota et al., 2006; Antion et al., 2008a; Gobert et al., 2008; Géranton et al., 2009; Santini et al., 2009; Zeng et al., 2009; Huang et al., 2010; Cao et al., 2011; Troca-Marín et al., 2011; Wu et al., 2011; Bailey et al., 2012; Bertran-Gonzalez et al., 2012; Meffre et al., 2012; Brewster et al., 2013; Macias et al., 2013; Bowling et al., 2014; Biever et al., 2015).
Although the activation of S6K and the subsequent phosphorylation of rpS6 are commonly used as a readout of mTORC1 activation, several evidences point out the existence of a synergistic crosstalk between mTORC1 and the extracellular signal-regulated kinase (ERK) signaling to control rpS6 phosphorylation. Thus, ERK can promote S6K activation by enhancing its phosphorylation at T421/S424 sites (Mukhopadhyay et al., 1992). When phosphorylated, these sites located in the autoinhibitory domain of the S6K1/2 are thought to prime the activation of S6K, thereby facilitating the subsequent phosphorylation of the other sites of S6K by the upstream kinases (Dennis et al., 1998). Alternatively, ERK can also modulate the mTORC1/S6K cascade upstream of S6K. On the one hand, ERK-mediated inhibitory phosphorylation of Tuberous Sclerosis Complex 2 (TSC2) stimulates the Ras Homolog Enriched in Brain (Rheb) protein, which in turn activates mTORC1 (Roux and Blenis, 2004; Long et al., 2005; Ma et al., 2005). On the other hand, ERK can enhance mTORC1 activation through RSK-mediated phosphorylation of Raptor (Wettenhall et al., 1992). Examples of this synergistic interaction between ERK and mTORC1 in the regulation of rpS6 phosphorylation have been reported in several models and in various brain areas (Kelleher et al., 2004; Antion et al., 2008a; Gobert et al., 2008; Santini et al., 2009; Gangarossa et al., 2014) Interestingly, ERK can also regulate rpS6 phosphorylation at S235/236 through RSK independently of mTORC1/S6K signaling (Roux et al., 2007). Indeed, the increase in pS235/236-rpS6 promoted by tetraethylammonium in cultured cortical neurons is prevented by RSK3 inhibition (Gu et al., 2015). Finally, a recent study performed in hippocampal neurons demonstrated that the cdk5-dependent phosphorylation of S6K at S411 site is also critical in the regulation of S6K activation and the subsequent rpS6 phosphorylation at S235/236 sites (Lai et al., 2015).
cAMP/PKA pathway
The enhanced rpS6 phosphorylation in the cerebral cortex following the administration of N6O-2′dibutyryl cAMP was one of the first demonstrations that cAMP could modulate in vivo the state of phosphorylation of rpS6 (Roberts and Morelos, 1979). Despite this evidence, the contribution of the cAMP/PKA pathway in the regulation of rpS6 phosphorylation in the nervous system has been largely neglected. However, recent studies highlighted the importance of cAMP/PKA signaling in the regulation of rpS6 phosphorylation at S235/236 sites. Thus, the direct stimulation of the adenylate cyclase by forskolin increases pS235/236-rpS6 in the striatum and the hippocampus (Gobert et al., 2008; Biever et al., 2015) (Table 1). Similar results are obtained when the degradation of cAMP is prevented by the administration of papaverine, a potent inhibitor of type 10A phosphodiesterase (Biever et al., 2015). Although the demonstration that PKA directly catalyzes rpS6 phosphorylation in the brain is still lacking, several indirect evidences support its contribution in the control of pS235/236-rpS6 following cAMP elevation. Indeed, forskolin-induced rpS6 phosphorylation in striatal slices is reduced in the presence of a PKA inhibitor (Biever et al., 2015). Moreover, stimulation of PKA activity with the cAMP analog cBIMPS increases pS235/236-rpS6 in striatal culture (Valjent et al., 2011) (Table 1). Finally, the administration of pharmacological agents promoting PKA activation triggers rpS6 phosphorylation in several brain areas (Gobert et al., 2008; Valjent et al., 2011; Knight et al., 2012; Bonito-Oliva et al., 2013; Rapanelli et al., 2014; Biever et al., 2015; Sutton and Caron, 2015).
Table 1.
Pharmacological stimuli modulating rpS6 phosphorylation ex vivo.
| Brain areas | Model | Treatment | S235/236 | S240/244 | References |
|---|---|---|---|---|---|
| WHOLE BRAIN | |||||
| Culture | Insulin | ↑ (NS) | ↑ (NS) | Heidenreich and Toledo, 1989 | |
| HIPPOCAMPUS | |||||
| Culture | BDNF | ↑ | ND | Kelleher et al., 2004; Troca-Marín et al., 2011 | |
| Bicuculline | ↑ | ND | Kelleher et al., 2004 | ||
| Slice | DHPG | ↑ | ↑ | Antion et al., 2008a | |
| Forskolin | ↑ | ND | Gobert et al., 2008 | ||
| STRIATUM | |||||
| Culture | cBIMPS | ↑ | ND | Valjent et al., 2011 | |
| Haloperidol | ND | ↑ | Bowling et al., 2014 | ||
| Slice | Forskolin | ↑ | ND | Biever et al., 2015 | |
| 6-OHDA/SKF81297 | ↑ | ↑ | Santini et al., 2009 | ||
| CORTEX | |||||
| Synaptoneurosomes | BDNF | ND | ↑ | Takei et al., 2004 | |
| Culture | Leucine | ND | ↑ | Ishizuka et al., 2008 | |
| Bicuculline/glycine | ↑ | ND | Lai et al., 2012 | ||
| Tetraethylammonium | ↑ | ND | Gu et al., 2015 | ||
| BDNF | ND | ↑ | Lenz and Avruch, 2005 | ||
| GNA | ND | ↑ | Lenz and Avruch, 2005 | ||
| Glutamate/NMDA | ND | ↓ | Lenz and Avruch, 2005 | ||
| Bicuculline/4-AP | ND | ↑ | Lenz and Avruch, 2005 | ||
NS, not specified; ND, not determined; GNA, Glutamate + NMDA followed 5 s later by the NMDA antagonist APV; 4-AP, 4-aminopyridine.
Although PKA targets selectively S235/236 residues (Moore et al., 2009), recent evidences suggest that PKA also contributes indirectly to rpS6 phosphorylation through a protein phosphatase cascade. This mechanism has been particularly well-studied in the striatum, where the inhibition of PP-1, controlled by the PKA-dependent phosphorylation of dopamine- and cAMP-regulated phosphoprotein, Mr 32,000 (DARPP-32) at T34 (Hemmings et al., 1984; Greengard, 2001), promotes pS235/236-rpS6 induced by d-amphetamine or haloperidol (Valjent et al., 2011; Bonito-Oliva et al., 2013; Biever et al., 2015). This mechanism also contributes to the regulation of rpS6 phosphorylation at S240/244 sites (Bonito-Oliva et al., 2013). These findings highlight the importance of PKA/DARPP-32/PP-1 signaling in the regulation of rpS6 phosphorylation in the striatum and raise the intriguing hypothesis that similar mechanisms could take place in other brain areas.
Stimuli modulating rpS6 phosphorylation in the brain
Recently an increasing number of studies have used rpS6 phosphorylation as a marker for neuronal activation in the context of synaptic plasticity or in response to variety of therapeutic agents in physiological and pathophysiological contexts.
Synaptic plasticity
Increased rpS6 phosphorylation during synaptic plasticity was reported for the first time by Klann and colleagues in 1991 using a synthetic peptide of rpS6 containing the residues 222–249. Since then, enhanced rpS6 phosphorylation has been observed in several electrical or chemical models of synaptic plasticity. Thus, pS235/236-rpS6 increases in the CA1 subfield of the hippocampus during long-term potentiation following high frequency stimulation (Antion et al., 2008b) or forskolin application (Kelleher et al., 2004; Antion et al., 2008b; Gobert et al., 2008). Similarly, mGluR-dependent long-term depression induced by application of the mGluR1 agonist [3,5-RS] dihydroxyphenylhydrazine (DHPG) is associated with marked increases in pS235/236- and pS240/244-rpS6 in hippocampal slices (Antion et al., 2008a). Interestingly, a recent study reported that the state of phosphorylation of rpS6 at S240/244 sites could be used to estimate the neuronal activity state of striatal cholinergic interneurons (Bertran-Gonzalez et al., 2012).
Pharmacological stimuli
A large number of pharmacological stimuli have been described to promote rpS6 phosphorylation in neurons (Tables 1, 2). Indeed, several ex vivo studies performed in slices or neuronal cultures showed that rpS6 phosphorylation is enhanced by stimuli triggering multiple forms of neuronal activity (Table 1). In vivo, the phosphorylation of rpS6 has been assessed following a single or repeated administration of a large variety of pharmacological agents in various brain areas (Table 2). Thus, proconvulsant drugs such as kainate, pilocarpine, pentylenetetrazol (PTZ), or the dopamine D1R agonist SKF81297 lead to a massive increase in pS235/236- and pS240/244-rpS6 in principal cells in the hippocampus and in various cortical areas (Table 2). Moreover, the administration of drugs of abuse (cocaine, d-amphetamine, methamphetamine, morphine, and tetrahydrocannabinol) as well as antipsychotics (haloperidol, clozapine, and olanzapine) also regulates rpS6 phosphorylation at multiple sites in several brain areas including the striatum, the nucleus accumbens, the cortex, and the hippocampus (Table 2). Finally, several hormones involved in the regulation of energy balance enhance rpS6 phosphorylation in hypothalamic nuclei (Table 2).
Table 2.
Pharmacological stimuli modulating rpS6 phosphorylation in vivo.
| Brain areas | Model | Treatment | Doses (mg/kg) | S235/236 | S240/244 | Cell-Type | References |
|---|---|---|---|---|---|---|---|
| HIPPOCAMPUS | |||||||
| Mouse | SKF81297 | 5 | ↑ | = | Granule cells DG | Gangarossa et al., 2011; Gangarossa and Valjent, 2012 | |
| 5* | ↑ | ↑ | Granule cells DG | Gangarossa et al., 2014 | |||
| Kainate | 12.5 | ND | ↑ | Principal cells | Knight et al., 2012 | ||
| THC | 10 | ↑ | ND | Pyramidal cells | Puighermanal et al., 2009 | ||
| Rat | Kainate | 10 | ↑ | ND | Pyramidal cells | Macias et al., 2013 | |
| 12 | ↑ (NS) | ↑ (NS) | ND | Zeng et al., 2009 | |||
| ND | ↑ | ND | Chen et al., 2012 | ||||
| Pilocarpine | 300 | ↑ (NS) | ↑ (NS) | ND | Huang et al., 2010 | ||
| PTZ | 75 | ↑ | ↑ | ND | Zhang and Wong, 2012 | ||
| STRIATUM | |||||||
| Mouse | Cocaine | 30 | ND | ↑ | MSNs | Knight et al., 2012 | |
| D-amphetamine | 5 | ↑ | = | ND | Rapanelli et al., 2014 | ||
| 10 | ↑ | = | D1-MSNs | Biever et al., 2015 | |||
| 10* | ↑ | = | ND | Biever et al., 2015 | |||
| Haloperidol | 0.5 | ↑ | ↑ | D2-MSNs | Valjent et al., 2011; Bonito-Oliva et al., 2013 | ||
| Clozapine | 5 | ↑ | ↑ | D2-MSNs | Valjent et al., 2011 | ||
| Papaverine | 30 | ↑ | ↑ | D1- and D2-MSNs | Biever et al., 2015 | ||
| SKF81297 | 5 | ↑ | ND | D1-MSNs | Gangarossa et al., 2013 | ||
| Quinpirole | 1 | ↓ | = | D1-MSNs | Gangarossa et al., 2013 | ||
| Apomorphine | 3 | ↑ | ND | D1-MSNs | Gangarossa et al., 2013 | ||
| 6-OHDA/L-DOPA | 20 | ↑ | ↑ | D1-MSNs | Santini et al., 2009 | ||
| 20* | ↑ | ↑ | D1-MSNs | Santini et al., 2009 | |||
| 10 | ↑ | ↑ | D1-MSNs | Santini et al., 2012 | |||
| Rat | Quinelorane | 0.16 | = | ↑ | ND | Salles et al., 2013 | |
| Monkey | MPTP/L-DOPA | 20 | ↑ | ND | ND | Santini et al., 2010 | |
| NUCLEUS ACCUMBENS | |||||||
| Mouse | Quinelorane | 0.63 | = | ↑ | ND | Salles et al., 2013 | |
| Cocaine | 15* | ↑ | ND | ND | Sutton and Caron, 2015 | ||
| Rat | Quinelorane | 0.16 | = | ↑ | ND | Salles et al., 2013 | |
| Ketamine | 5 | ↑ | ND | ND | Tedesco et al., 2013 | ||
| Cocaine | 15 | ↑ (NS) | ↑ (NS) | ND | Wu et al., 2011 | ||
| NMDA | 250 (ng) icv | ↑ (NS) | ↑ (NS) | ND | Wang et al., 2010 | ||
| CORTEX | |||||||
| Prefrontal | Mouse | WAY181187 | 10 | ND | ↑ | ND | Meffre et al., 2012 |
| Rat | NMDA | 500 (ng) icv | ↑ (NS) | ↑ (NS) | ND | Yu et al., 2013 | |
| WAY181187 | 10 | ND | ↑ | ND | Meffre et al., 2012 | ||
| MK801 | 2 | ND | ↓ | ND | Yoon et al., 2008 | ||
| 1* | ND | ↑ | ND | Yoon et al., 2008 | |||
| 1* | ND | ↓ | Principal cells | Kim et al., 2010 | |||
| Oubain | 1 (mM) icv | ↑ | ↑ | Principal cells | Kim et al., 2013 | ||
| Cingulate | MK801 | 1* | ND | ↓ | Principal cells | Kim et al., 2010 | |
| Oubain | 1 (mM) icv | ↑ | ↑ | Principal cells | Kim et al., 2013 | ||
| Insular | MK801 | 1* | ND | ↓ | Principal cells | Kim et al., 2010 | |
| Prelimbic | Ketamine | 5 | ↑ | ND | ND | Tedesco et al., 2013 | |
| 10 | ↑ | ND | ND | Tedesco et al., 2013 | |||
| Infralimbic | 5 | ↑ | ND | ND | Tedesco et al., 2013 | ||
| 10 | ↑ | ND | ND | Tedesco et al., 2013 | |||
| Somatosensory | Kainate | 10 | ↑ | ND | pyramidal cells | Macias et al., 2013 | |
| Piriform | ↑ | ND | ND | Macias et al., 2013 | |||
| NS | 12 | ND | ↑ | ND | Chen et al., 2012 | ||
| ↑ (NS) | ↑ (NS) | ND | Zeng et al., 2009 | ||||
| Pilocarpine | 300 | ↑ (NS) | ↑ (NS) | ND | Huang et al., 2010 | ||
| PTZ | 75 | ↑ | ↑ | ND | Zhang and Wong, 2012 | ||
| Cocaine | 15 | ↑ (NS) | ↑ (NS) | ND | Wu et al., 2011 | ||
| MK801 | 0.5 | ↓ | ↓ | ND | Kim et al., 2010 | ||
| 1 | ↓ | ↓ | ND | Kim et al., 2010 | |||
| L-phenylalanine | 2 (mg/g) | ↓ (NS) | ↓ (NS) | ND | Roberts and Morelos, 1979 | ||
| VENTRAL TEGMENTAL AREA | |||||||
| Mouse | Morphine | 25 (pellet)* | ↑ (NS) | ↑ (NS) | TH+ cells | Mazei-Robison et al., 2011 | |
| Rat | Cocaine | 15 | ↑ (NS) | ↑ (NS) | ND | Wu et al., 2011 | |
| AMYGDALA | |||||||
| Basolateral | Rat | Ketamine | 10 | ↑ | ND | ND | Tedesco et al., 2013 |
| NS | Kainate | 10 | ↑ | ND | ND | Macias et al., 2013 | |
| HYPOTHALAMUS | |||||||
| Arcuate | Mouse | Olanzapine | 20 | ↑ | ND | ND | Knight et al., 2012 |
| Ghrelin | 66 (mg) | ↑ | ↑ | NPY+ cells | Knight et al., 2012 | ||
| 6 (mg) icv | ND | ↑ | ND | Villanueva et al., 2009 | |||
| Leptin | 5 (mg) icv | ND | ↑ | ND | Gong et al., 2007 | ||
| Insulin | 300 (mU/ml) icv | ND | ↑ | ND | Villanueva et al., 2009 | ||
| 400 (mU/ml) | ND | ↑ | ND | Villanueva et al., 2009 | |||
| Ventromedial | 300 (mU/ml) icv | ND | ↑ | ND | Villanueva et al., 2009 | ||
| Paraventricular | Clozapine | 10 | ↑ | ND | ND | Knight et al., 2012 | |
| Rat | Leptin | 10 (mg) icv | ND | ↑ | ND | Cota et al., 2006, 2008 | |
| NS | CNTFAx15 | 1.5 (mg) icv | ND | ↑ | ND | Cota et al., 2008 | |
| C75 | 30 (mg) icv | ND | ↑ | ND | Proulx et al., 2008 | ||
| Cerulenin | 90 (mg) icv | ND | ↑ | ND | Proulx et al., 2008 | ||
When not specified, a single dose of drug was administered;
repeated doses of drug were administered. NS, not specified; ND, not determined; DG, Dentate Gyrus; MSNs, Medium-sized Spiny Neurons; THC, tetrahydrocannabinol; PTZ, Pentylenetetrazol; 6-OHDA, 6-hydroxydopamine; L-DOPA, levodopa; MPTP, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine.
Physiological and pathophysiological conditions
Since the pioneering report demonstrating that rpS6 phosphorylation was enhanced in the CA1 subfield of the hippocampus in mice trained to contextual fear conditioning (Kelleher et al., 2004), rpS6 phosphorylation has been used as a marker of neuronal and circuit activation following physiological conditions (Table 3). Thus, phospho-rpS6 levels oscillate in the hippocampus and the suprachiasmatic nucleus of the hypothalamus along the circadian cycle (Table 3). rpS6 phosphorylation is also strongly modulated in the amygdala and the hypothalamus when defensive behaviors (freezing, escape or attacks) are engaged or in the hypothalamus in response to nutritional perturbations (Table 3). Finally, altered rpS6 phosphorylation has been reported in rodents following spontaneous seizures and traumatic brain injury and in humans in several neurodevelopmental disorders including Down syndrome, Tuberous sclerosis, Autism, and Rett syndrome (Table 3).
Table 3.
rpS6 phosphorylation regulation under physiological and pathophysiological conditions.
| Brain areas | Species | Model | S235/236 | S240/244 | References |
|---|---|---|---|---|---|
| HIPPOCAMPUS | |||||
| Mouse | Contextual FC | ↑ | ND | Kelleher et al., 2004; Saraf et al., 2014 | |
| Circadian cycle | ↑ | ND | Saraf et al., 2014 | ||
| Rat | Pilocarpine-induced spontaneous seizure | ↑ (NS) | ↑ (NS) | Huang et al., 2010; Saraf et al., 2014 | |
| Kainate-induced spontaneous seizure | ↑ | ↑ | Zeng et al., 2009 | ||
| Kainate-induced early life seizure | = | ↑ | Bernard et al., 2013 | ||
| Traumatic brain injury | ↑ | ND | Chen et al., 2007 | ||
| Human | Down syndrome | ↑ (NS) | ↑ (NS) | Iyer et al., 2014 | |
| Alzheimer's disease (severe-stage) | ↑ | ND | Sun et al., 2014 | ||
| STRIATUM | |||||
| Mouse | Stroke | ND | ↑ | Xiong et al., 2014 | |
| NUCLEUS ACCUMBENS | |||||
| Mouse | Highly palatable isocaloric food | ↑ | ND | Guegan et al., 2013 | |
| Rat | Cue-induced cocaine reinstatement | ↑ | ND | Wang et al., 2010 | |
| CORTEX | |||||
| Prefrontal | Mouse | Highly palatable isocaloric food | ↑ | ND | Guegan et al., 2013 |
| NS | Middle cerebral artery occlusion | ND | ↑ | Xiong et al., 2014 | |
| Prefrontal | Rat | Focal cerebral ischemia | ↓ | ND | Koh, 2013 |
| Neonatal phencyclidine | ND | ↑ | Meffre et al., 2012 | ||
| Social isolation | ND | ↑ | Meffre et al., 2012 | ||
| Prelimbic | Rat | Cue-induced alcohol reinstatement | ↑ | ND | Barak et al., 2013 |
| Extinction after retrieval of FC | ↑ | ND | Tedesco et al., 2014 | ||
| Infralimbic | Extinction after retrieval of FC | ↑ | ND | Tedesco et al., 2014 | |
| Orbitofrontal | Cue-induced alcohol reinstatement | ↑ | ND | Barak et al., 2013 | |
| Parietal | Traumatic brain injury | ↑ | ND | Chen et al., 2007 | |
| NS | Pilocarpine-induced spontaneous seizure | ↑ (NS) | ↑ (NS) | Huang et al., 2010 | |
| Kainate-induced spontaneous seizure | ↑ (NS) | ↑ (NS) | Zeng et al., 2009 | ||
| Cortex | Human | Focal cortical dysplasia | ↑ | = | Baybis et al., 2004 |
| Focal cortical dysplasia | ↑ | ND | Jansen et al., 2015 | ||
| Hemimegalencephaly | ↑ | ND | Jansen et al., 2015 | ||
| Cortical Tubers | Tuberous sclerosis (unspecified) | ↑ | = | Baybis et al., 2004 | |
| Tuberous sclerosis (specific mutations) | ↑ | ND | Parker et al., 2011 | ||
| Tuberous sclerosis (TSC1 mutation) | ↑ (NS) | ↑ (NS) | Jansen et al., 2004 | ||
| Frontal cortex | Hemimegalencephaly | ↑ | ND | Aronica et al., 2007 | |
| Medial Temporal Cortex | Alzheimer's disease | ↑ | = | An et al., 2003 | |
| AMYGDALA | |||||
| Central | Rat | Cue-induced alcohol reinstatement | ↑ | ND | Barak et al., 2013 |
| Basolateral | Extinction after retrieval of FC | ↑ | ND | Tedesco et al., 2014 | |
| HYPOTHALAMUS | |||||
| Paraventricular | Mouse | Salt | ↑ | ↑ | Knight et al., 2012 |
| Leucine deprivation | ↓ | ND | Xia et al., 2012 | ||
| Arcuate | Fasting | ND | ↑ | Knight et al., 2012 | |
| Fasted | ND | ↑ | Villanueva et al., 2009 | ||
| Leucine deprivation | ↓ | ND | Xia et al., 2012 | ||
| Dorsomedial | ND | ↑ | Knight et al., 2012 | ||
| Preoptic area | ND | ↑ | Knight et al., 2012 | ||
| Ventrolateral | Resident-intruder | ↑ | ↑ | Knight et al., 2012 | |
| Premammillary | Cat odor | ND | ↑ | Knight et al., 2012 | |
| Suprachiasmatic | Light | ↑ | ↑ | Cao et al., 2011; Knight et al., 2012 | |
| Circadian cycle | ND | ↑ | Cao et al., 2011 | ||
| Supraoptic | Dehydration | ND | ↑ | Knight et al., 2012 | |
| Salt | ND | ↑ | Knight et al., 2012 | ||
| Arcuate | Rat | Fasted | ND | ↓ | Cota et al., 2006 |
| PERIAQUEDUCTAL GRAY | |||||
| Mouse | Resident-intruder | ↑ | ND | Knight et al., 2012 | |
| SPINAL CORD | |||||
| Rat | Inflammatory pain (carrageenan) | ↑ | ND | Norsted Gregory et al., 2010 | |
| Neurogenic inflammation (capsaicin) | ↑ | ND | Géranton et al., 2009 | ||
| DORSAL ROOT GANGLIA | |||||
| Rat | Inflammatory pain (carrageenan) | ↑ | ND | Norsted Gregory et al., 2010 | |
| Neuropathic pain (SNI) | ↑ | ND | Géranton et al., 2009 | ||
| NS | Human | Glioblastoma | ↑ | = | Harter et al., 2015 |
| SEGA | ↑ | ND | Chan et al., 2004 | ||
NS, not specified; ND, not determined; FC, fear conditioning; SEGA, subependymal giant cell astrocytoma.
Genetic mouse models displaying altered rpS6 phosphorylation
Most of the full or conditional knockout mice for the key components of the mTORC1 pathway display altered rpS6 phosphorylation (Table 4). Interestingly, the vast majority of mutant mice in which dysregulation of rpS6 phosphorylation has been demonstrated correspond to mouse models for various neurological and neurodevelopmental disorders, including Tuberous sclerosis, Down syndrome, Rett syndrome, Angelman syndrome, and Fragile X syndrome, among others. Most of these pathologies share common features such as autism, intellectual disability, and epilepsy, which might be rescued by mTORC1 inhibitors under certain circumstances. The phosphorylation of rpS6 is also altered in neurodegenerative diseases such as Huntington disease and in mouse models of psychiatric disorders such as schizophrenia (Table 4). Other genetic mouse models showing hormonal perturbations as leptin deficiency also display altered rpS6 phosphorylation in the hypothalamus (Table 4).
Table 4.
rpS6 phosphorylation in genetic mouse models.
| Model | Brain area/cell-type | S235/236 | S240/244 | References |
|---|---|---|---|---|
| KNOCKOUT | ||||
| Akt3 | Whole brain | ↓ | ↓ | Easton et al., 2005 |
| Tsc2+∕− | Hippocampus | ↑ | ND | Ehninger et al., 2008 |
| Hdc | Striatum | ↑ | = | Rapanelli et al., 2014 |
| Fmr1 | Hippocampus/pyramidal cells | ↑ | ↑ | Bhattacharya et al., 2012 |
| S6K2 | Hippocampus | ↓ | = | Antion et al., 2008a |
| Mecp2 | Cortex/principal cells | ↓ | ↓ | Ricciardi et al., 2011 |
| Cerebellum | ↓ | ↓ | Ricciardi et al., 2011 | |
| Hippocampus | ↓ | = | Ricciardi et al., 2011 | |
| miR-199a-2 | Cortex/principal cells | ↓ | ND | Tsujimura et al., 2015 |
| Hippocampus/pyramidal cells | ↓ | ND | Tsujimura et al., 2015 | |
| Cdkl5 | Cortex/principal cells | ↓ | ↓ | Amendola et al., 2014 |
| DAT | Nucleus accumbens | ↑ | ND | Sutton and Caron, 2015 |
| Lepob∕ob | Hypothalamus (arcuate) | ND | ↑ | Villanueva et al., 2009 |
| Leprdb∕db | Hypothalamus (arcuate) | ND | ↑ | Villanueva et al., 2009 |
| CONDITIONAL KNOCKOUT | ||||
| Tsc1Syn1 | Cortex/principal cells | ND | ↑ | Meikle et al., 2007 |
| Hippocampus | ND | ↑ | Meikle et al., 2007 | |
| Tsc1GFAP | Cortex | ↑ | ND | Parker et al., 2011 |
| Cortex | ↑ (NS) | ↑ (NS) | Zeng et al., 2008, 2011 | |
| Hippocampus | ↑ (NS) | ↑ (NS) | Zeng et al., 2008, 2011 | |
| Tsc1AAV−CreGFP | Hippocampus/pyramidal cells | ND | ↑ | Bateup et al., 2011 |
| Tsc1MCH | Hypothalamus | ↑ | ND | Knight et al., 2012 |
| Tsc1L7 | Cerebellum/purkinje cells | ↑ (NS) | ↑ (NS) | Tsai et al., 2012 |
| Tsc1Emx1 | Cortex/principal cells | ↑ | ND | Magri et al., 2011 |
| Tsc1CaMKII | Cortex | ↑ | ND | McMahon et al., 2012 |
| Hippocampus | ↑ | ND | McMahon et al., 2012 | |
| Tsc1Temporal# | Cortex | ND | ↑ | Feliciano et al., 2011 |
| Tsc2GFAP1 | Cortex | ↑ (NS) | ↑ (NS) | Zeng et al., 2011 |
| Hippocampus | ↑ (NS) | ↑ (NS) | Zeng et al., 2011 | |
| Tsc2hGFAP | Cortex/principal cells | ND | ↑ | Way et al., 2009 |
| Hippocampus/principal cells | ND | ↑ | Way et al., 2009 | |
| Tsc2CaMKII | Hippocampus/principal cells | ND | ↑ | Rozas et al., 2015 |
| Rheb1Nestin | Hypothalamus | ↓ | ↓ | Zou et al., 2011; Yang et al., 2014 |
| Cortex | ↓ | ↓ | Zou et al., 2011 | |
| Cerebellum | ↓ | ↓ | Zou et al., 2011 | |
| Hippocampus | ↓ | ↓ | Zou et al., 2011 | |
| RhebGFAP | Hippocampus/granule cells | ND | ↑ | Banerjee et al., 2011 |
| PtenGFAP* | Hippocampus/granule cells | ↑ | ↑ | Kwon et al., 2003; Chalhoub et al., 2006; Lugo et al., 2013 |
| Cerebellum | ↑ | ND | Kwon et al., 2003 | |
| PtenGFAP*/S6K1 ko | Hippocampus/granule cells | ↑ | ND | Chalhoub et al., 2006 |
| PtenNestin | Hippocampus/granule cells | ↑ | ND | Kwon et al., 2006; Zhou et al., 2009 |
| Cortex | ↑ | ND | Zhou et al., 2009 | |
| PtenDATCreERT2 | Ventral midbrain/DA neurons | ↑ | ND | Domanskyi et al., 2011 |
| RictorNestin | Whole brain | = | ND | Thomanetz et al., 2013 |
| Dicer1CaMKCreERT2 | Hypothalamus/arcuate nucleus | ↑ | ND | Vinnikov et al., 2014 |
| TARGETED MUTATION | ||||
| Ube3Atm1Alb∕J | Cerebellum/purkinje cells | ↑ | ↑ | Sun et al., 2015 |
| Tsc2DRG | Hippocampus | ↑ | = | Chévere-Torres et al., 2012 |
| Ts1Cje | Hippocampus/principal cells | ↑ | ND | Troca-Marín et al., 2011 |
| N171-N82Q | Striatum | ↓ | ND | Ravikumar et al., 2004; Lee et al., 2015 |
| Cortex | ↓ | ND | Ravikumar et al., 2004 | |
| Cortex | ↓ | = | Fox et al., 2010 | |
| 3xTg-AD | Whole brain | ↑ | ND | Caccamo et al., 2015 |
| VIRAL-MEDIATED GENE TRANSFER | ||||
| DISC1-shRNA | Hippocampus/granule cells | ↑ | ND | Kim et al., 2009; Zhou et al., 2013 |
| S6K1CA | Prefrontal cortex | ↑ | ND | Dwyer et al., 2015 |
| S6K1DN | Prefrontal cortex | ↑ | ND | Dwyer et al., 2015 |
The Gfap-Cre mice used in these studies lead to Pten deletion in a subset of neuronal cells, including the majority of granule cells in the dentate gyrus and cerebellum.
In utero electroporation to express Cre recombinase and remove Tsc1 in Tsc1floxed∕mutant heterozygous embryos at E16. NS, not specified; ND, not determined; DA, dopaminergic.
Physiological role of rpS6 phosphorylation
Role in overall mRNA translation
Despite the growing number of reports analyzing the phosphorylation of rpS6, its biological significance still remains controversial. One of the first hypotheses put forward suggested that rpS6 phosphorylation played a role in translation initiation. Thus, an early in vitro study reported a correlation between the phosphorylation of rpS6 and enhanced translation under certain experimental conditions (Thomas et al., 1982). Moreover, the 40S subunit with a highest proportion of phosphorylated rpS6 was preferentially found into polysomes compared to subpolysomal fractions (Duncan and McConkey, 1982). However, several studies rapidly called into question this hypothesis. Indeed, although localized at the mRNA/tRNA binding site junction between the small and large ribosomal subunits (Nygård and Nilsson, 1990), increased rpS6 phosphorylation is not sufficient to mobilize small ribosomal subunits into protein synthesis (Kruppa and Clemens, 1984; Tas and Martini, 1987). Finally, the generation of rpS6 knockin mice, in which the five phosphorylated serines were replaced by alanines, constituted a valuable tool to determine whether rpS6 phosphorylation and the protein synthesis were causally linked (Ruvinsky et al., 2005). Unexpectedly, protein synthesis is increased in mouse embryo fibroblasts (MEF) of phospho-deficient mice. Moreover, a similar (Ruvinsky et al., 2005) or even increased (Chauvin et al., 2014) proportion of ribosomes engaged in translation were found in the liver of rpS6 knockin mice. Together, these puzzling observations suggest a negative role of rpS6 phosphorylation on global protein synthesis or the presence of feedback mechanisms taking place in this mouse model.
Role in TOP mRNA translation
The phosphorylation of rpS6 through the mTORC1/S6K axis was believed for many years to exert an effect on the translation of a specific subset of mRNAs bearing a 5′ terminal oligopyrimidine tract (TOP). However, this long-lasting model has been challenged by subsequent studies showing that MEFs from the double mutant S6K1/2 as well as from the rpS6 knockin mouse lines exhibit normal TOP translation (Tang et al., 2001; Stolovich et al., 2002; Ruvinsky et al., 2005). Further work demonstrated that insulin-induced TOP translation requires the PI3K/TSC/Rheb/mTOR pathway but is independent of the S6K/rpS6 axis (Patursky-Polischuk et al., 2009).
Despite these evidences, the involvement of rpS6 phosphorylation in the control of translation in the nervous system is still controversial. Indeed, several findings in hippocampal neurons and slices correlated increased rpS6 phosphorylation with enhanced global and TOP-encoded protein synthesis following different forms of synaptic plasticity (Kelleher et al., 2004; Klann and Dever, 2004; Tsokas et al., 2005, 2007; Antion et al., 2008a,b) or in a mouse model of fragile X syndrome (Bhattacharya et al., 2012). By contrast, such correlations have not been observed in vivo in the striatum where the pharmacologically-induced rpS6 phosphorylation by d-amphetamine, haloperidol, or papaverine relies on the activation of the cAMP/PKA/DARPP-32 pathway (Biever et al., 2015). Interestingly, the direct binding of mRNA to the small ribosomal subunit decreases or increases after cAMP-dependent or cAMP-independent phosphorylation of rpS6, respectively (Burkhard and Traugh, 1983). Therefore, one cannot exclude that when an upstream signaling cascade other than PKA is engaged, for example mTORC1/S6K or ERK/RSK, rpS6 phosphorylation might have different physiological roles. Nevertheless, despite compelling studies indicating that rpS6 phosphorylation is dispensable for efficient global and TOP mRNA translation, the role of the phosphorylation in the translation of a specific subset of mRNAs remains to be fully addressed by high-throughput sequencing analyses of total and polysomal RNAs combined with proteomic approaches.
Extratranslational functions
Another possibility is that rpS6 phosphorylation, within or outside the ribosome, exerts functions unrelated to mRNA translation, for example by interacting with other cellular proteins, which might become active or inactive upon the binding with rpS6. Indeed, co-immunoprecipitation studies suggest either a direct or indirect interaction of rpS6 with several extraribosomal proteins, including heat-shock protein 90 (Kim et al., 2006), alphavirus non-structural protein (Montgomery et al., 2006), DAPK (Schumacher et al., 2006), huntingtin (Culver et al., 2012), and mTOR complex 2 (mTORC2) (Yano et al., 2014). In the latter, rpS6 phosphorylation has been proposed to have a role in cardioprotective signaling by amplifying mTORC2-mediated Akt phosphorylation (Yano et al., 2014). In the mouse liver, Chauvin and colleagues recently uncovered the involvement of rpS6 phosphorylation in the control of the ribosome biogenesis (RiBi) transcriptional program by S6Ks (Chauvin et al., 2014). This program regulates the expression of nucleolar proteins required for ribosomal RNA synthesis, cleavage, post-transcriptional modifications, ribosome assembly, and export (Lempiäinen and Shore, 2009). Whether all these translation-unrelated responses occur in the brain merits further study.
Extraribosomal functions
Surprisingly, extraribosomal functions have been attributed to several ribosomal proteins (Wool, 1996; Warner and McIntosh, 2009). For instance, the ribosomal protein rpL13a, when phosphorylated, is released from the 60S ribosomal subunit and acts as a silencer of targeted mRNAs (Mazumder et al., 2003). In this regard, few studies suggested an extraribosomal role of rpS6 phosphorylation (Kim et al., 2014; Son et al., 2015; Xiao et al., 2015). Recent work in the plant Arabidopsis Thaliana proposes a role of rpS6 in rRNA synthesis and rDNA transcription via its interaction with the histone deacetylase AtHD2B (Kim et al., 2014) and the histone chaperon AtNAP1 (Son et al., 2015), respectively, an effect that might be dependent on the phosphorylation state of rpS6. Finally, the ubiquitylation and proteasomal degradation of phosphorylated rpS6 following its subsequent interaction with PALL has been identified as a critical mechanism regulating efferocytosis in drosophila (Xiao et al., 2015). To date, such extraribosomal functions of rpS6 in the nervous system have not been described.
Concluding remarks
Since the pioneering studies performed four decades ago, many progresses have been made regarding the identification of signaling events leading to rpS6 phosphorylation. Although rpS6 phosphorylation is still considered as a readout of mTORC1/S6K activity, caution should be taken with this assumption since other intracellular cascades largely contribute to the regulation of rpS6 phosphorylation, as exemplified with the PKA/DARPP-32/PP-1 pathway in the striatum. One should also keep in mind that the different sites of phosphorylation can be regulated independently in various brain areas or different cell-types within a same brain region. Although rpS6 phosphorylation has been and will remain a valuable hallmark of neuronal activity, understanding its biological role in the brain is undoubtedly one of the major challenges of the coming years.
Author contributions
AB, EV, and EP wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Inserm, Fondation pour la Recherche Médicale (EV), and a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation (EP). AB is supported by the Fonds National de la Recherche, Luxembourg (Grant 3977033). EP is a recipient of Marie Curie Intra-European Fellowship IEF327648. The authors apologize to all colleagues whose work was not discussed in this Review.
References
- Amendola E., Zhan Y., Mattucci C., Castroflorio E., Calcagno E., Fuchs C., et al. (2014). Mapping pathological phenotypes in a mouse model of CDKL5 disorder. PLoS ONE 9:e91613. 10.1371/journal.pone.0091613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W. L., Cowburn R. F., Li L., Braak H., Alafuzoff I., Iqbal K., et al. (2003). Up-regulation of phosphorylated/activated p70 S6 kinase and its relationship to neurofibrillary pathology in Alzheimer's disease. Am. J. Pathol. 163, 591–607. 10.1016/S0002-9440(10)63687-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antion M. D., Hou L., Wong H., Hoeffer C. A., Klann E. (2008a). mGluR-dependent long-term depression is associated with increased phosphorylation of S6 and synthesis of elongation factor 1A but remains expressed in S6K-deficient mice. Mol. Cell. Biol. 28, 2996–3007. 10.1128/MCB.00201-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antion M. D., Merhav M., Hoeffer C. A., Reis G., Kozma S. C., Thomas G., et al. (2008b). Removal of S6K1 and S6K2 leads to divergent alterations in learning, memory, and synaptic plasticity. Learn. Mem. 15, 29–38. 10.1101/lm.661908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica E., Boer K., Baybis M., Yu J., Crino P. (2007). Co-expression of cyclin D1 and phosphorylated ribosomal S6 proteins in hemimegalencephaly. Acta Neuropathol. 114, 287–293. 10.1007/s00401-007-0225-6 [DOI] [PubMed] [Google Scholar]
- Bailey J., Ma D., Szumlinski K. K. (2012). Rapamycin attenuates the expression of cocaine-induced place preference and behavioral sensitization. Addict. Biol. 17, 248–258. 10.1111/j.1369-1600.2010.00311.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandi H. R., Ferrari S., Krieg J., Meyer H. E., Thomas G. (1993). Identification of 40 S ribosomal protein S6 phosphorylation sites in Swiss mouse 3T3 fibroblasts stimulated with serum. J. Biol. Chem. 268, 4530–4533. [PubMed] [Google Scholar]
- Banerjee S., Crouse N. R., Emnett R. J., Gianino S. M., Gutmann D. H. (2011). Neurofibromatosis-1 regulates mTOR-mediated astrocyte growth and glioma formation in a TSC/Rheb-independent manner. Proc. Natl. Acad. Sci. U.S.A. 108, 15996–16001. 10.1073/pnas.1019012108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak S., Liu F., Ben Hamida S., Yowell Q. V., Neasta J., Kharazia V., et al. (2013). Disruption of alcohol-related memories by mTORC1 inhibition prevents relapse. Nat. Neurosci. 16, 1111–1117. 10.1038/nn.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup H. S., Takasaki K. T., Saulnier J. L., Denefrio C. L., Sabatini B. L. (2011). Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J. Neurosci. 31, 8862–8869. 10.1523/JNEUROSCI.1617-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baybis M., Yu J., Lee A., Golden J. A., Weiner H., McKhann G. II, et al. (2004). mTOR cascade activation distinguishes tubers from focal cortical dysplasia. Ann. Neurol. 56, 478–487. 10.1002/ana.20211 [DOI] [PubMed] [Google Scholar]
- Belandia B., Brautigan D., Martín-Pérez J. (1994). Attenuation of ribosomal protein S6 phosphatase activity in chicken embryo fibroblasts transformed by Rous sarcoma virus. Mol. Cell. Biol. 14, 200–206. 10.1128/MCB.14.1.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P. B., Castano A. M., O'Leary H., Simpson K., Browning M. D., Benke T. A. (2013). Phosphorylation of FMRP and alterations of FMRP complex underlie enhanced mLTD in adult rats triggered by early life seizures. Neurobiol. Dis. 59, 1–17. 10.1016/j.nbd.2013.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertran-Gonzalez J., Chieng B. C., Laurent V., Valjent E., Balleine B. W. (2012). Striatal cholinergic interneurons display activity-related phosphorylation of ribosomal protein S6. PLoS ONE 7:e53195. 10.1371/journal.pone.0053195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A., Kaphzan H., Alvarez-Dieppa A. C., Murphy J. P., Pierre P., Klann E. (2012). Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron 76, 325–337. 10.1016/j.neuron.2012.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biever A., Puighermanal E., Nishi A., David A., Panciatici C., Longueville S., et al. (2015). PKA-dependent phosphorylation of ribosomal protein S6 does not correlate with translation efficiency in striatonigral and striatopallidal medium-sized spiny neurons. J. Neurosci. 35, 4113–4130. 10.1523/JNEUROSCI.3288-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonito-Oliva A., Pallottino S., Bertran-Gonzalez J., Girault J. A., Valjent E., Fisone G. (2013). Haloperidol promotes mTORC1-dependent phosphorylation of ribosomal protein S6 via dopamine- and cAMP-regulated phosphoprotein of 32 kDa and inhibition of protein phosphatase-1. Neuropharmacology 72, 197–203. 10.1016/j.neuropharm.2013.04.043 [DOI] [PubMed] [Google Scholar]
- Bowling H., Zhang G., Bhattacharya A., Pérez-Cuesta L. M., Deinhardt K., Hoeffer C. A., et al. (2014). Antipsychotics activate mTORC1-dependent translation to enhance neuronal morphological complexity. Sci. Signal. 7:ra4. 10.1126/scisignal.2004331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster A. L., Lugo J. N., Patil V. V., Lee W. L., Qian Y., Vanegas F., et al. (2013). Rapamycin reverses status epilepticus-induced memory deficits and dendritic damage. PLoS ONE 8:e57808. 10.1371/journal.pone.0057808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard S. J., Traugh J. A. (1983). Changes in ribosome function by cAMP-dependent and cAMP-independent phosphorylation of ribosomal protein S6. J. Biol. Chem. 258, 14003–14008. [PubMed] [Google Scholar]
- Caccamo A., Branca C., Talboom J. S., Shaw D. M., Turner D., Ma L., et al. (2015). Reducing ribosomal protein S6 kinase 1 expression improves spatial memory and synaptic plasticity in a mouse model of Alzheimer's disease. J. Neurosci. 35, 14042–14056. 10.1523/JNEUROSCI.2781-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Anderson F. E., Jung Y. J., Dziema H., Obrietan K. (2011). Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience 181, 79–88. 10.1016/j.neuroscience.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub N., Kozma S. C., Baker S. J. (2006). S6k1 is not required for Pten-deficient neuronal hypertrophy. Brain Res. 1100, 32–41. 10.1016/j.brainres.2006.05.013 [DOI] [PubMed] [Google Scholar]
- Chan J. A., Zhang H., Roberts P. S., Jozwiak S., Wieslawa G., Lewin-Kowalik J., et al. (2004). Pathogenesis of tuberous sclerosis subependymal giant cell astrocytomas: biallelic inactivation of TSC1 or TSC2 leads to mTOR activation. J. Neuropathol. Exp. Neurol. 63, 1236–1242. [DOI] [PubMed] [Google Scholar]
- Chauvin C., Koka V., Nouschi A., Mieulet V., Hoareau-Aveilla C., Dreazen A., et al. (2014). Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33, 474–483. 10.1038/onc.2012.606 [DOI] [PubMed] [Google Scholar]
- Chen L., Hu L., Dong J. Y., Ye Q., Hua N., Wong M., et al. (2012). Rapamycin has paradoxical effects on S6 phosphorylation in rats with and without seizures. Epilepsia 53, 2026–2033. 10.1111/epi.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Atkins C. M., Liu C. L., Alonso O. F., Dietrich W. D., Hu B. R. (2007). Alterations in mammalian target of rapamycin signaling pathways after traumatic brain injury. J. Cereb. Blood Flow Metab. 27, 939–949. 10.1038/sj.jcbfm.9600393 [DOI] [PubMed] [Google Scholar]
- Chévere-Torres I., Kaphzan H., Bhattacharya A., Kang A., Maki J. M., Gambello M. J., et al. (2012). Metabotropic glutamate receptor-dependent long-term depression is impaired due to elevated ERK signaling in the DeltaRG mouse model of tuberous sclerosis complex. Neurobiol. Dis. 45, 1101–1110. 10.1016/j.nbd.2011.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota D., Matter E. K., Woods S. C., Seeley R. J. (2008). The role of hypothalamic mammalian target of rapamycin complex 1 signaling in diet-induced obesity. J. Neurosci. 28, 7202–7208. 10.1523/JNEUROSCI.1389-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota D., Proulx K., Smith K. A., Kozma S. C., Thomas G., Woods S. C., et al. (2006). Hypothalamic mTOR signaling regulates food intake. Science 312, 927–930. 10.1126/science.1124147 [DOI] [PubMed] [Google Scholar]
- Culver B. P., Savas J. N., Park S. K., Choi J. H., Zheng S., Zeitlin S. O., et al. (2012). Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J. Biol. Chem. 287, 21599–21614. 10.1074/jbc.M112.359307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P. B., Pullen N., Pearson R. B., Kozma S. C., Thomas G. (1998). Phosphorylation sites in the autoinhibitory domain participate in p70(s6k) activation loop phosphorylation. J. Biol. Chem. 273, 14845–14852. 10.1074/jbc.273.24.14845 [DOI] [PubMed] [Google Scholar]
- Domanskyi A., Geissler C., Vinnikov I. A., Alter H., Schober A., Vogt M. A., et al. (2011). Pten ablation in adult dopaminergic neurons is neuroprotective in Parkinson's disease models. FASEB J. 25, 2898–2910. 10.1096/fj.11-181958 [DOI] [PubMed] [Google Scholar]
- Duncan R., McConkey E. H. (1982). Rapid alterations in initiation rate and recruitment of inactive RNA are temporally correlated with S6 phosphorylation. Eur. J. Biochem. 123, 539–544. 10.1111/j.1432-1033.1982.tb06565.x [DOI] [PubMed] [Google Scholar]
- Dwyer J. M., Maldonado-Avilés J. G., Lepack A. E., Dileone R. J., Duman R. S. (2015). Ribosomal protein S6 kinase 1 signaling in prefrontal cortex controls depressive behavior. Proc. Natl. Acad. Sci. U.S.A. 112, 6188–6193. 10.1073/pnas.1505289112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton R. M., Cho H., Roovers K., Shineman D. W., Mizrahi M., Forman M. S., et al. (2005). Role for Akt3/protein kinase Bgamma in attainment of normal brain size. Mol. Cell. Biol. 25, 1869–1878. 10.1128/MCB.25.5.1869-1878.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D., Han S., Shilyansky C., Zhou Y., Li W., Kwiatkowski D. J., et al. (2008). Reversal of learning deficits in a Tsc2+/- mouse model of tuberous sclerosis. Nat. Med. 14, 843–848. 10.1038/nm1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciano D. M., Su T., Lopez J., Platel J. C., Bordey A. (2011). Single-cell Tsc1 knockout during corticogenesis generates tuber-like lesions and reduces seizure threshold in mice. J. Clin. Invest. 121, 1596–1607. 10.1172/JCI44909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Bandi H. R., Hofsteenge J., Bussian B. M., Thomas G. (1991). Mitogen-activated 70K S6 kinase. Identification of in vitro 40 S ribosomal S6 phosphorylation sites. J. Biol. Chem. 266, 22770–22775. [PubMed] [Google Scholar]
- Fox J. H., Connor T., Chopra V., Dorsey K., Kama J. A., Bleckmann D., et al. (2010). The mTOR kinase inhibitor Everolimus decreases S6 kinase phosphorylation but fails to reduce mutant huntingtin levels in brain and is not neuroprotective in the R6/2 mouse model of Huntington's disease. Mol. Neurodegener. 5, 26. 10.1186/1750-1326-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangarossa G., Ceolin L., Paucard A., Lerner-Natoli M., Perroy J., Fagni L., et al. (2014). Repeated stimulation of dopamine D1-like receptor and hyperactivation of mTOR signaling lead to generalized seizures, altered dentate gyrus plasticity, and memory deficits. Hippocampus 24, 1466–1481. 10.1002/hipo.22327 [DOI] [PubMed] [Google Scholar]
- Gangarossa G., Di Benedetto M., O'sullivan G. J., Dunleavy M., Alcacer C., Bonito-Oliva A., et al. (2011). Convulsant doses of a dopamine D1 receptor agonist result in Erk-dependent increases in Zif268 and Arc/Arg3.1 expression in mouse dentate gyrus. PLoS ONE 6:e19415. 10.1371/journal.pone.0019415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangarossa G., Perroy J., Valjent E. (2013). Combinatorial topography and cell-type specific regulation of the ERK pathway by dopaminergic agonists in the mouse striatum. Brain Struct. Funct. 218, 405–419. 10.1007/s00429-012-0405-6 [DOI] [PubMed] [Google Scholar]
- Gangarossa G., Valjent E. (2012). Regulation of the ERK pathway in the dentate gyrus by in vivo dopamine D1 receptor stimulation requires glutamatergic transmission. Neuropharmacology 63, 1107–1117. 10.1016/j.neuropharm.2012.06.062 [DOI] [PubMed] [Google Scholar]
- Géranton S. M., Jiménez-Díaz L., Torsney C., Tochiki K. K., Stuart S. A., Leith J. L., et al. (2009). A rapamycin-sensitive signaling pathway is essential for the full expression of persistent pain states. J. Neurosci. 29, 15017–15027. 10.1523/JNEUROSCI.3451-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert D., Topolnik L., Azzi M., Huang L., Badeaux F., Desgroseillers L., et al. (2008). Forskolin induction of late-LTP and up-regulation of 5' TOP mRNAs translation via mTOR, ERK, and PI3K in hippocampal pyramidal cells. J. Neurochem. 106, 1160–1174. 10.1111/j.1471-4159.2008.05470.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Ishida-Takahashi R., Villanueva E. C., Fingar D. C., Münzberg H., Myers M. G., Jr. (2007). The long form of the leptin receptor regulates STAT5 and ribosomal protein S6 via alternate mechanisms. J. Biol. Chem. 282, 31019–31027. 10.1074/jbc.M702838200 [DOI] [PubMed] [Google Scholar]
- Greengard P. (2001). The neurobiology of dopamine signaling. Biosci. Rep. 21, 247–269. 10.1023/A:1013205230142 [DOI] [PubMed] [Google Scholar]
- Gressner A. M., Wool I. G. (1974). The phosphorylation of liver ribosomal proteins in vivo. Evidence that only a single small subunit protein (S6) is phosphorylated. J. Biol. Chem. 249, 6917–6925. [PubMed] [Google Scholar]
- Gu Q. H., Yu D., Hu Z., Liu X., Yang Y., Luo Y., et al. (2015). miR-26a and miR-384-5p are required for LTP maintenance and spine enlargement. Nat. Commun. 6, 6789. 10.1038/ncomms7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guegan T., Cutando L., Gangarossa G., Santini E., Fisone G., Martinez A., et al. (2013). Operant behavior to obtain palatable food modifies ERK activity in the brain reward circuit. Eur. Neuropsychopharmacol. 23, 240–252. 10.1016/j.euroneuro.2012.04.009 [DOI] [PubMed] [Google Scholar]
- Harter P. N., Jennewein L., Baumgarten P., Ilina E., Burger M. C., Thiepold A. L., et al. (2015). Immunohistochemical assessment of phosphorylated mTORC1-pathway proteins in human brain tumors. PLoS ONE 10:e0127123. 10.1371/journal.pone.0127123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich K. A., Toledo S. P. (1989). Insulin receptors mediate growth effects in cultured fetal neurons. II. Activation of a protein kinase that phosphorylates ribosomal protein S6. Endocrinology 125, 1458–1463. 10.1210/endo-125-3-1458 [DOI] [PubMed] [Google Scholar]
- Hemmings H. C., Jr., Nairn A. C., Aswad D. W., Greengard P. (1984). DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Purification and characterization of the phosphoprotein from bovine caudate nucleus. J. Neurosci. 4, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House C., Wettenhall R. E., Kemp B. E. (1987). The influence of basic residues on the substrate specificity of protein kinase C. J. Biol. Chem. 262, 772–777. [PubMed] [Google Scholar]
- Huang X., Zhang H., Yang J., Wu J., McMahon J., Lin Y., et al. (2010). Pharmacological inhibition of the mammalian target of rapamycin pathway suppresses acquired epilepsy. Neurobiol. Dis. 40, 193–199. 10.1016/j.nbd.2010.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. A., Shanware N. P., Chang H., Tibbetts R. S. (2011). Regulation of ribosomal protein S6 phosphorylation by casein kinase 1 and protein phosphatase 1. J. Biol. Chem. 286, 8688–8696. 10.1074/jbc.M110.141754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka Y., Kakiya N., Nawa H., Takei N. (2008). Leucine induces phosphorylation and activation of p70S6K in cortical neurons via the system L amino acid transporter. J. Neurochem. 106, 934–942. 10.1111/j.1471-4159.2008.05438.x [DOI] [PubMed] [Google Scholar]
- Iyer A. M., van Scheppingen J., Milenkovic I., Anink J. J., Adle-Biassette H., Kovacs G. G., et al. (2014). mTOR Hyperactivation in down syndrome hippocampus appears early during development. J. Neuropathol. Exp. Neurol. 73, 671–683. 10.1097/NEN.0000000000000083 [DOI] [PubMed] [Google Scholar]
- Jansen F. E., Notenboom R. G., Nellist M., Goedbloed M. A., Halley D. J., de Graan P. N., et al. (2004). Differential localization of hamartin and tuberin and increased S6 phosphorylation in a tuber. Neurology 63, 1293–1295. 10.1212/01.WNL.0000140702.08902.D7 [DOI] [PubMed] [Google Scholar]
- Jansen L. A., Mirzaa G. M., Ishak G. E., O'Roak B. J., Hiatt J. B., Roden W. H., et al. (2015). PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain 138, 1613–1628. 10.1093/brain/awv045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher R. J., III, Govindarajan A., Jung H. Y., Kang H., Tonegawa S. (2004). Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell 116, 467–479. 10.1016/S0092-8674(04)00115-1 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Duan X., Liu C. Y., Jang M. H., Guo J. U., Pow-Anpongkul N., et al. (2009). DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 63, 761–773. 10.1016/j.neuron.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Park H. G., Kim H. S., Ahn Y. M., Kim Y. S. (2010). Effects of neonatal MK-801 treatment on p70S6K-S6/eIF4B signal pathways and protein translation in the frontal cortex of the developing rat brain. Int. J. Neuropsychopharmacol. 13, 1233–1246. 10.1017/S1461145709991192 [DOI] [PubMed] [Google Scholar]
- Kim S. H., Yu H. S., Park H. G., Ha K., Kim Y. S., Shin S. Y., et al. (2013). Intracerebroventricular administration of ouabain, a Na/K-ATPase inhibitor, activates mTOR signal pathways and protein translation in the rat frontal cortex. Prog. Neuropsychopharmacol. Biol. Psychiatry 45, 73–82. 10.1016/j.pnpbp.2013.04.018 [DOI] [PubMed] [Google Scholar]
- Kim T. S., Jang C. Y., Kim H. D., Lee J. Y., Ahn B. Y., Kim J. (2006). Interaction of Hsp90 with ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Mol. Biol. Cell 17, 824–833. 10.1091/mbc.E05-08-0713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. K., Kim S., Shin Y. J., Hur Y. S., Kim W. Y., Lee M. S., et al. (2014). Ribosomal protein S6, a target of rapamycin, is involved in the regulation of rRNA genes by possible epigenetic changes in Arabidopsis. J. Biol. Chem. 289, 3901–3912. 10.1074/jbc.M113.515015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann E., Dever T. E. (2004). Biochemical mechanisms for translational regulation in synaptic plasticity. Nat. Rev. Neurosci. 5, 931–942. 10.1038/nrn1557 [DOI] [PubMed] [Google Scholar]
- Knight Z. A., Tan K., Birsoy K., Schmidt S., Garrison J. L., Wysocki R. W., et al. (2012). Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151, 1126–1137. 10.1016/j.cell.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh P. O. (2013). Ferulic acid attenuates focal cerebral ischemia-induced decreases in p70S6 kinase and S6 phosphorylation. Neurosci. Lett. 555, 7–11. 10.1016/j.neulet.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Kressler D., Hurt E., Bassler J. (2010). Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683. 10.1016/j.bbamcr.2009.10.009 [DOI] [PubMed] [Google Scholar]
- Krieg J., Hofsteenge J., Thomas G. (1988). Identification of the 40 S ribosomal protein S6 phosphorylation sites induced by cycloheximide. J. Biol. Chem. 263, 11473–11477. [PubMed] [Google Scholar]
- Kroczynska B., Kaur S., Katsoulidis E., Majchrzak-Kita B., Sassano A., Kozma S. C., et al. (2009). Interferon-dependent engagement of eukaryotic initiation factor 4B via S6 kinase (S6K)- and ribosomal protein S6K-mediated signals. Mol. Cell. Biol. 29, 2865–2875. 10.1128/MCB.01537-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa J., Clemens M. J. (1984). Differential kinetics of changes in the state of phosphorylation of ribosomal protein S6 and in the rate of protein synthesis in MPC 11 cells during tonicity shifts. EMBO J. 3, 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C. H., Luikart B. W., Powell C. M., Zhou J., Matheny S. A., Zhang W., et al. (2006). Pten regulates neuronal arborization and social interaction in mice. Neuron 50, 377–388. 10.1016/j.neuron.2006.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C. H., Zhu X., Zhang J., Baker S. J. (2003). mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc. Natl. Acad. Sci. U.S.A. 100, 12923–12928. 10.1073/pnas.2132711100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K. O., Liang Z., Fei E., Huang H., Ip N. Y. (2015). Cyclin-dependent kinase 5 (Cdk5)-dependent phosphorylation of p70 ribosomal S6 kinase 1 (S6K) is required for dendritic spine morphogenesis. J. Biol. Chem. 290, 14637–14646. 10.1074/jbc.M114.627117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K. O., Wong A. S., Cheung M. C., Xu P., Liang Z., Lok K. C., et al. (2012). TrkB phosphorylation by Cdk5 is required for activity-dependent structural plasticity and spatial memory. Nat. Neurosci. 15, 1506–1515. 10.1038/nn.3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastick S. M., Nielsen P. J., McConkey E. H. (1977). Phosphorylation of ribosomal protein S6 in suspension cultured HeLa cells. Mol. Gen. Genet. 152, 223–230. 10.1007/BF00693074 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Tecedor L., Chen Y. H., Monteys A. M., Sowada M. J., Thompson L. M., et al. (2015). Reinstating aberrant mTORC1 activity in Huntington's disease mice improves disease phenotypes. Neuron 85, 303–315. 10.1016/j.neuron.2014.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen H., Shore D. (2009). Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 21, 855–863. 10.1016/j.ceb.2009.09.002 [DOI] [PubMed] [Google Scholar]
- Lenz G., Avruch J. (2005). Glutamatergic regulation of the p70S6 kinase in primary mouse neurons. J. Biol. Chem. 280, 38121–38124. 10.1074/jbc.C500363200 [DOI] [PubMed] [Google Scholar]
- Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. (2005). Rheb binds and regulates the mTOR kinase. Curr. Biol. 15, 702–713. 10.1016/j.cub.2005.02.053 [DOI] [PubMed] [Google Scholar]
- Lugo J. N., Smith G. D., Morrison J. B., White J. (2013). Deletion of PTEN produces deficits in conditioned fear and increases fragile X mental retardation protein. Learn. Mem. 20, 670–673. 10.1101/lm.032839.113 [DOI] [PubMed] [Google Scholar]
- Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P. (2005). Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121, 179–193. 10.1016/j.cell.2005.02.031 [DOI] [PubMed] [Google Scholar]
- Macias M., Blazejczyk M., Kazmierska P., Caban B., Skalecka A., Tarkowski B., et al. (2013). Spatiotemporal characterization of mTOR kinase activity following kainic acid induced status epilepticus and analysis of rat brain response to chronic rapamycin treatment. PLoS ONE 8:e64455. 10.1371/journal.pone.0064455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri L., Cambiaghi M., Cominelli M., Alfaro-Cervello C., Cursi M., Pala M., et al. (2011). Sustained activation of mTOR pathway in embryonic neural stem cells leads to development of tuberous sclerosis complex-associated lesions. Cell Stem Cell 9, 447–462. 10.1016/j.stem.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Mahoney S. J., Dempsey J. M., Blenis J. (2009). Cell signaling in protein synthesis ribosome biogenesis and translation initiation and elongation. Prog. Mol. Biol. Transl. Sci. 90, 53–107. 10.1016/S1877-1173(09)90002-3 [DOI] [PubMed] [Google Scholar]
- Martin K. A., Schalm S. S., Romanelli A., Keon K. L., Blenis J. (2001). Ribosomal S6 kinase 2 inhibition by a potent C-terminal repressor domain is relieved by mitogen-activated protein-extracellular signal-regulated kinase kinase-regulated phosphorylation. J. Biol. Chem. 276, 7892–7898. 10.1074/jbc.M009972200 [DOI] [PubMed] [Google Scholar]
- Martin-Pérez J., Thomas G. (1983). Ordered phosphorylation of 40S ribosomal protein S6 after serum stimulation of quiescent 3T3 cells. Proc. Natl. Acad. Sci. U.S.A. 80, 926–930. 10.1073/pnas.80.4.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazei-Robison M. S., Koo J. W., Friedman A. K., Lansink C. S., Robison A. J., Vinish M., et al. (2011). Role for mTOR signaling and neuronal activity in morphine-induced adaptations in ventral tegmental area dopamine neurons. Neuron 72, 977–990. 10.1016/j.neuron.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B., Sampath P., Seshadri V., Maitra R. K., Dicorleto P. E., Fox P. L. (2003). Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115, 187–198. 10.1016/S0092-8674(03)00773-6 [DOI] [PubMed] [Google Scholar]
- McMahon J., Huang X., Yang J., Komatsu M., Yue Z., Qian J., et al. (2012). Impaired autophagy in neurons after disinhibition of mammalian target of rapamycin and its contribution to epileptogenesis. J. Neurosci. 32, 15704–15714. 10.1523/JNEUROSCI.2392-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre J., Chaumont-Dubel S., Mannoury la Cour C., Loiseau F., Watson D. J., Dekeyne A., et al. (2012). 5-HT(6) receptor recruitment of mTOR as a mechanism for perturbed cognition in schizophrenia. EMBO Mol. Med. 4, 1043–1056. 10.1002/emmm.201201410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle L., Talos D. M., Onda H., Pollizzi K., Rotenberg A., Sahin M., et al. (2007). A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J. Neurosci. 27, 5546–5558. 10.1523/JNEUROSCI.5540-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O. (2008). Physiological roles of ribosomal protein S6: one of its kind. Int. Rev. Cell Mol. Biol. 268, 1–37. 10.1016/S1937-6448(08)00801-0 [DOI] [PubMed] [Google Scholar]
- Meyuhas O. (2015). Ribosomal protein S6 phosphorylation: four decades of research. Int. Rev. Cell Mol. Biol. 320, 41–73. 10.1016/bs.ircmb.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Montgomery S. A., Berglund P., Beard C. W., Johnston R. E. (2006). Ribosomal protein S6 associates with alphavirus nonstructural protein 2 and mediates expression from alphavirus messages. J. Virol. 80, 7729–7739. 10.1128/JVI.00425-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. E., Xie J., Gomez E., Herbert T. P. (2009). Identification of cAMP-dependent kinase as a third in vivo ribosomal protein S6 kinase in pancreatic beta-cells. J. Mol. Biol. 389, 480–494. 10.1016/j.jmb.2009.04.020 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay N. K., Price D. J., Kyriakis J. M., Pelech S., Sanghera J., Avruch J. (1992). An array of insulin-activated, proline-directed serine/threonine protein kinases phosphorylate the p70 S6 kinase. J. Biol. Chem. 267, 3325–3335. [PubMed] [Google Scholar]
- Norsted Gregory E., Codeluppi S., Gregory J. A., Steinauer J., Svensson C. I. (2010). Mammalian target of rapamycin in spinal cord neurons mediates hypersensitivity induced by peripheral inflammation. Neuroscience 169, 1392–1402. 10.1016/j.neuroscience.2010.05.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygård O., Nilsson L. (1990). Translational dynamics. Interactions between the translational factors, tRNA and ribosomes during eukaryotic protein synthesis. Eur. J. Biochem. 191, 1–17. 10.1111/j.1432-1033.1990.tb19087.x [DOI] [PubMed] [Google Scholar]
- Parker W. E., Orlova K. A., Heuer G. G., Baybis M., Aronica E., Frost M., et al. (2011). Enhanced epidermal growth factor, hepatocyte growth factor, and vascular endothelial growth factor expression in tuberous sclerosis complex. Am. J. Pathol. 178, 296–305. 10.1016/j.ajpath.2010.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patursky-Polischuk I., Stolovich-Rain M., Hausner-Hanochi M., Kasir J., Cybulski N., Avruch J., et al. (2009). The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol. Cell. Biol. 29, 640–649. 10.1128/MCB.00980-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M., Um S. H., Mieulet V., Sticker M., Goss V. L., Mestan J., et al. (2004). S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24, 3112–3124. 10.1128/MCB.24.8.3112-3124.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx K., Cota D., Woods S. C., Seeley R. J. (2008). Fatty acid synthase inhibitors modulate energy balance via mammalian target of rapamycin complex 1 signaling in the central nervous system. Diabetes 57, 3231–3238. 10.2337/db07-1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puighermanal E., Marsicano G., Busquets-Garcia A., Lutz B., Maldonado R., Ozaita A. (2009). Cannabinoid modulation of hippocampal long-term memory is mediated by mTOR signaling. Nat. Neurosci. 12, 1152–1158. 10.1038/nn.2369 [DOI] [PubMed] [Google Scholar]
- Rapanelli M., Frick L. R., Pogorelov V., Ota K. T., Abbasi E., Ohtsu H., et al. (2014). Dysregulated intracellular signaling in the striatum in a pathophysiologically grounded model of Tourette syndrome. Eur. Neuropsychopharmacol. 24, 1896–1906. 10.1016/j.euroneuro.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., et al. (2004). Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595. 10.1038/ng1362 [DOI] [PubMed] [Google Scholar]
- Ricciardi S., Boggio E. M., Grosso S., Lonetti G., Forlani G., Stefanelli G., et al. (2011). Reduced AKT/mTOR signaling and protein synthesis dysregulation in a Rett syndrome animal model. Hum. Mol. Genet. 20, 1182–1196. 10.1093/hmg/ddq563 [DOI] [PubMed] [Google Scholar]
- Roberts S., Morelos B. S. (1979). Phosphorylation of multiple proteins of both ribosomal subunits in rat cerebral cortex in vivo. Effect of adenosine 3':5'-cyclic monophosphate. Biochem. J. 184, 233–244. 10.1042/bj1840233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux P. P., Blenis J. (2004). ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68, 320–344. 10.1128/MMBR.68.2.320-344.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux P. P., Shahbazian D., Vu H., Holz M. K., Cohen M. S., Taunton J., et al. (2007). RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 282, 14056–14064. 10.1074/jbc.M700906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas N. S., Redell J. B., Mckenna J., III, Moore A. N., Gambello M. J., Dash P. K. (2015). Prolonging the survival of Tsc2 conditional knockout mice by glutamine supplementation. Biochem. Biophys. Res. Commun. 457, 635–639. 10.1016/j.bbrc.2015.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I., Sharon N., Lerer T., Cohen H., Stolovich-Rain M., Nir T., et al. (2005). Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 19, 2199–2211. 10.1101/gad.351605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles M. J., Hervé D., Rivet J. M., Longueville S., Millan M. J., Girault J. A., et al. (2013). Transient and rapid activation of Akt/GSK-3beta and mTORC1 signaling by D3 dopamine receptor stimulation in dorsal striatum and nucleus accumbens. J. Neurochem. 125, 532–544. 10.1111/jnc.12206 [DOI] [PubMed] [Google Scholar]
- Santini E., Feyder M., Gangarossa G., Bateup H. S., Greengard P., Fisone G. (2012). Dopamine- and cAMP-regulated phosphoprotein of 32-kDa (DARPP-32)-dependent activation of extracellular signal-regulated kinase (ERK) and mammalian target of rapamycin complex 1 (mTORC1) signaling in experimental parkinsonism. J. Biol. Chem. 287, 27806–27812. 10.1074/jbc.M112.388413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E., Heiman M., Greengard P., Valjent E., Fisone G. (2009). Inhibition of mTOR signaling in Parkinson's disease prevents L-DOPA-induced dyskinesia. Sci. Signal. 2:ra36. 10.1126/scisignal.2000308 [DOI] [PubMed] [Google Scholar]
- Santini E., Sgambato-Faure V., Li Q., Savasta M., Dovero S., Fisone G., et al. (2010). Distinct changes in cAMP and extracellular signal-regulated protein kinase signalling in L-DOPA-induced dyskinesia. PLoS ONE 5:e12322. 10.1371/journal.pone.0012322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraf A., Luo J., Morris D. R., Storm D. R. (2014). Phosphorylation of eukaryotic translation initiation factor 4E and eukaryotic translation initiation factor 4E-binding protein (4EBP) and their upstream signaling components undergo diurnal oscillation in the mouse hippocampus: implications for memory persistence. J. Biol. Chem. 289, 20129–20138. 10.1074/jbc.M114.552638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A. M., Velentza A. V., Watterson D. M., Dresios J. (2006). Death-associated protein kinase phosphorylates mammalian ribosomal protein S6 and reduces protein synthesis. Biochemistry 45, 13614–13621. 10.1021/bi060413y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son O., Kim S., Shin Y. J., Kim W. Y., Koh H. J., Cheon C. I. (2015). Identification of nucleosome assembly protein 1 (NAP1) as an interacting partner of plant ribosomal protein S6 (RPS6) and a positive regulator of rDNA transcription. Biochem. Biophys. Res. Commun. 465, 200–205. 10.1016/j.bbrc.2015.07.150 [DOI] [PubMed] [Google Scholar]
- Stolovich M., Tang H., Hornstein E., Levy G., Cohen R., Bae S. S., et al. (2002). Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 22, 8101–8113. 10.1128/MCB.22.23.8101-8113.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Liu Y., Moreno S., Baudry M., Bi X. (2015). Imbalanced mechanistic target of rapamycin C1 and C2 activity in the cerebellum of Angelman syndrome mice impairs motor function. J. Neurosci. 35, 4706–4718. 10.1523/JNEUROSCI.4276-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. X., Ji X., Mao X., Xie L., Jia J., Galvan V., et al. (2014). Differential activation of mTOR complex 1 signaling in human brain with mild to severe Alzheimer's disease. J. Alzheimers Dis. 38, 437–444. 10.3233/JAD-131124 [DOI] [PubMed] [Google Scholar]
- Sutton L. P., Caron M. G. (2015). Essential role of D1R in the regulation of mTOR complex1 signaling induced by cocaine. Neuropharmacology 99, 610–619. 10.1016/j.neuropharm.2015.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N., Inamura N., Kawamura M., Namba H., Hara K., Yonezawa K., et al. (2004). Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J. Neurosci. 24, 9760–9769. 10.1523/JNEUROSCI.1427-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Hornstein E., Stolovich M., Levy G., Livingstone M., Templeton D., et al. (2001). Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 21, 8671–8683. 10.1128/MCB.21.24.8671-8683.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas P. W., Martini O. H. (1987). Are highly phosphorylated 40-S subunits preferentially utilized during protein synthesis in a cell-free system from HeLa cells? Eur. J. Biochem. 163, 561–567. 10.1111/j.1432-1033.1987.tb10904.x [DOI] [PubMed] [Google Scholar]
- Tedesco V., Ravagnani C., Bertoglio D., Chiamulera C. (2013). Acute ketamine-induced neuroplasticity: ribosomal protein S6 phosphorylation expression in drug addiction-related rat brain areas. Neuroreport 24, 388–393. 10.1097/WNR.0b013e32836131ad [DOI] [PubMed] [Google Scholar]
- Tedesco V., Roquet R. F., DeMis J., Chiamulera C., Monfils M. H. (2014). Extinction, applied after retrieval of auditory fear memory, selectively increases zinc-finger protein 268 and phosphorylated ribosomal protein S6 expression in prefrontal cortex and lateral amygdala. Neurobiol. Learn. Mem. 115, 78–85. 10.1016/j.nlm.2014.08.015 [DOI] [PubMed] [Google Scholar]
- Thomanetz V., Angliker N., Cloëtta D., Lustenberger R. M., Schweighauser M., Oliveri F., et al. (2013). Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. J. Cell Biol. 201, 293–308. 10.1083/jcb.201205030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G., Martin-Pérez J., Siegmann M., Otto A. M. (1982). The effect of serum, EGF, PGF2 alpha and insulin on S6 phosphorylation and the initiation of protein and DNA synthesis. Cell 30, 235–242. 10.1016/0092-8674(82)90029-0 [DOI] [PubMed] [Google Scholar]
- Troca-Marín J. A., Alves-Sampaio A., Montesinos M. L. (2011). An increase in basal BDNF provokes hyperactivation of the Akt-mammalian target of rapamycin pathway and deregulation of local dendritic translation in a mouse model of Down's syndrome. J. Neurosci. 31, 9445–9455. 10.1523/JNEUROSCI.0011-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P. T., Hull C., Chu Y., Greene-Colozzi E., Sadowski A. R., Leech J. M., et al. (2012). Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature 488, 647–651. 10.1038/nature11310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P., Grace E. A., Chan P., Ma T., Sealfon S. C., Iyengar R., et al. (2005). Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J. Neurosci. 25, 5833–5843. 10.1523/JNEUROSCI.0599-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P., Ma T., Iyengar R., Landau E. M., Blitzer R. D. (2007). Mitogen-activated protein kinase upregulates the dendritic translation machinery in long-term potentiation by controlling the mammalian target of rapamycin pathway. J. Neurosci. 27, 5885–5894. 10.1523/JNEUROSCI.4548-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura K., Irie K., Nakashima H., Egashira Y., Fukao Y., Fujiwara M., et al. (2015). miR-199a links MeCP2 with mTOR signaling and its dysregulation leads to rett syndrome phenotypes. Cell Rep. 12, 1887–1901. 10.1016/j.celrep.2015.08.028 [DOI] [PubMed] [Google Scholar]
- Valjent E., Bertran-Gonzalez J., Bowling H., Lopez S., Santini E., Matamales M., et al. (2011). Haloperidol regulates the state of phosphorylation of ribosomal protein S6 via activation of PKA and phosphorylation of DARPP-32. Neuropsychopharmacology 36, 2561–2570. 10.1038/npp.2011.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva E. C., Münzberg H., Cota D., Leshan R. L., Kopp K., Ishida-Takahashi R., et al. (2009). Complex regulation of mammalian target of rapamycin complex 1 in the basomedial hypothalamus by leptin and nutritional status. Endocrinology 150, 4541–4551. 10.1210/en.2009-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinnikov I. A., Hajdukiewicz K., Reymann J., Beneke J., Czajkowski R., Roth L. C., et al. (2014). Hypothalamic miR-103 protects from hyperphagic obesity in mice. J. Neurosci. 34, 10659–10674. 10.1523/JNEUROSCI.4251-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Luo Y. X., He Y. Y., Li F. Q., Shi H. S., Xue L. F., et al. (2010). Nucleus accumbens core mammalian target of rapamycin signaling pathway is critical for cue-induced reinstatement of cocaine seeking in rats. J. Neurosci. 30, 12632–12641. 10.1523/JNEUROSCI.1264-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R., McIntosh K. B. (2009). How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11. 10.1016/j.molcel.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way S. W., Mckenna J., III, Mietzsch U., Reith R. M., Wu H. C., Gambello M. J. (2009). Loss of Tsc2 in radial glia models the brain pathology of tuberous sclerosis complex in the mouse. Hum. Mol. Genet. 18, 1252–1265. 10.1093/hmg/ddp025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall R. E., Erikson E., Maller J. L. (1992). Ordered multisite phosphorylation of Xenopus ribosomal protein S6 by S6 kinase II. J. Biol. Chem. 267, 9021–9027. [PubMed] [Google Scholar]
- Wool I. G. (1996). Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 21, 164–165. 10.1016/S0968-0004(96)20011-8 [DOI] [PubMed] [Google Scholar]
- Wu J., McCallum S. E., Glick S. D., Huang Y. (2011). Inhibition of the mammalian target of rapamycin pathway by rapamycin blocks cocaine-induced locomotor sensitization. Neuroscience 172, 104–109. 10.1016/j.neuroscience.2010.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Cheng Y., Zhang Q., Xiao F., Liu B., Chen S., et al. (2012). S6K1 in the central nervous system regulates energy expenditure via MC4R/CRH pathways in response to deprivation of an essential amino acid. Diabetes 61, 2461–2471. 10.2337/db11-1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Wang H., Silva E. A., Thompson J., Guillou A., Yates J. R., Jr., et al. (2015). The Pallbearer E3 ligase promotes actin remodeling via RAC in efferocytosis by degrading the ribosomal protein S6. Dev. Cell 32, 19–30. 10.1016/j.devcel.2014.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Xie R., Zhang H., Gu L., Xie W., Cheng M., et al. (2014). PRAS40 plays a pivotal role in protecting against stroke by linking the Akt and mTOR pathways. Neurobiol. Dis. 66, 43–52. 10.1016/j.nbd.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Barna M. (2012). Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369. 10.1038/nrm3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Jiang W., Luo L., Bu J., Pang D., Wei J., et al. (2014). Genetic deletion of Rheb1 in the brain reduces food intake and causes hypoglycemia with altered peripheral metabolism. Int. J. Mol. Sci. 15, 1499–1510. 10.3390/ijms15011499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T., Ferlito M., Aponte A. M., Kuno A., Miura T., Murphy E., et al. (2014). Pivotal role of mTORC2 and involvement of ribosomal protein S6 in cardioprotective signaling. Circ. Res. 114, 1268–1280. 10.1161/CIRCRESAHA.114.303562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. C., Seo M. S., Kim S. H., Jeon W. J., Ahn Y. M., Kang U. G., et al. (2008). The effect of MK-801 on mTOR/p70S6K and translation-related proteins in rat frontal cortex. Neurosci. Lett. 434, 23–28. 10.1016/j.neulet.2008.01.020 [DOI] [PubMed] [Google Scholar]
- Yu J. J., Zhang Y., Wang Y., Wen Z. Y., Liu X. H., Qin J., et al. (2013). Inhibition of calcineurin in the prefrontal cortex induced depressive-like behavior through mTOR signaling pathway. Psychopharmacology (Berl). 225, 361–372. 10.1007/s00213-012-2823-9 [DOI] [PubMed] [Google Scholar]
- Zeng L. H., Rensing N. R., Wong M. (2009). The mammalian target of rapamycin signaling pathway mediates epileptogenesis in a model of temporal lobe epilepsy. J. Neurosci. 29, 6964–6972. 10.1523/JNEUROSCI.0066-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L. H., Rensing N. R., Zhang B., Gutmann D. H., Gambello M. J., Wong M. (2011). Tsc2 gene inactivation causes a more severe epilepsy phenotype than Tsc1 inactivation in a mouse model of tuberous sclerosis complex. Hum. Mol. Genet. 20, 445–454. 10.1093/hmg/ddq491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L. H., Xu L., Gutmann D. H., Wong M. (2008). Rapamycin prevents epilepsy in a mouse model of tuberous sclerosis complex. Ann. Neurol. 63, 444–453. 10.1002/ana.21331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Wong M. (2012). Pentylenetetrazole-induced seizures cause acute, but not chronic, mTOR pathway activation in rat. Epilepsia 53, 506–511. 10.1111/j.1528-1167.2011.03384.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Blundell J., Ogawa S., Kwon C. H., Zhang W., Sinton C., et al. (2009). Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J. Neurosci. 29, 1773–1783. 10.1523/JNEUROSCI.5685-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Li W., Huang S., Song J., Kim J. Y., Tian X., et al. (2013). mTOR Inhibition ameliorates cognitive and affective deficits caused by Disc1 knockdown in adult-born dentate granule neurons. Neuron 77, 647–654. 10.1016/j.neuron.2012.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Zhou L., Du X. X., Ji Y., Xu J., Tian J., et al. (2011). Rheb1 is required for mTORC1 and myelination in postnatal brain development. Dev. Cell 20, 97–108. 10.1016/j.devcel.2010.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]