Abstract
Background: Recent studies demonstrate that plant homeodomain finger protein 20 (PHF20), which was initially described as an immunogenic antigen in glioblastoma, is a putative transcriptional factor, and exhibits tumor suppressor activity. However, little is known about its expression and clinical significance in lung cancer. Methods: We investigated the expression of PHF20 in 142 cases of NSCLC tissue and 30 cases of normal lung tissue by immunohistochemical staining and downregulated PHF20 expression in SPC cell. Results: PHF20 expression was significantly higher in normal lung tissues than that in NSCLC tissues. The expression of PHF20 in NSCLC was significantly correlated with histological grade, p-TNM stage and lymph node metastasis. Moreover, the loss of PHF20 expression was associated with short overall survival. We also found that the expression of PHF20 was associated with Bax expression. Additionally, PHF20 markedly inhibited cell proliferation and invasion. Conclusions: PHF20 may play an important role in NSCLC, and may serve as a potential therapeutic target of NSCLC.
Keywords: Non-small-cell lung cancer, plant homeodomain finger protein, glioma-expressed antigen 2, Bax
Introduction
Lung cancer is a common malignancy and is the leading cause of cancer deaths worldwide. Non-small-cell lung cancer (NSCLC) represents 80%-85% of lung cancer cases, and the long time survival is still unsatisfactory. The accurate molecular mechanism of lung cancer has also not fully elucidated. The initiation and progression of tumor is related to the balance of cell proliferation and apoptosis, especially the inhibition of apoptosis. Therefore, some apoptosis associated proteins may serve as useful therapeutic targets of NSCLC.
Plant homeodomain finger protein 20 (PHF20), also known as glioma-expressed antigen 2 (GLEA2), was initially described as an immunogenic antigen in glioblastoma [1,2]. Subsequent studies showed that PHF20 was likely a putative transcriptional factor, and was related to tumorgenesis [3]. Autoantibodies against PHF20 was also detected in hepatocellular carcinoma [4] and meduloblastoma [5], indicating its potential association with tumor. However, it remains controversial about the function of PHF20 in tumor proliferation and invasion. Park et al. reported that PHF20 might exhibit putative tumor suppressor activity by binding to Akt and inducing p53 expression at the transcription level [6]. Pallasch et al. found that PHF20 was expressed in 48.4% of glioblastomas, and the patients with positive PHF20 expression showed prolonged survival [2]. In contrast, Zhang et al. found PHF20 could promote NF-κB transcriptional activity by interacting with p65 in a methylation-dependent manner, and expression levels of PHF20 were significantly associated with pathological tumour grade of gliomas [7]. Taniwaki et al. reported that PHF20 was abundantly expressed both in advanced small-cell lung cancers and advanced adenocarcinomas [8].
Thus, we think that PHF20 may play dual roles as oncogene and tumor suppressor gene in human oncogenesis, depending on the specific tissue involved or the context. Recent studies showed PHF20 was a multidomain protein, of which the second Tudor domain could recognize and combine P53. Association with PHF20 promotes stabilization and activation of P53 by diminishing Mdm2-mediated P53 ubiquitylation and degradation [9,10]. Bax is the main apoptosis promoter of Bcl-2 family [11]. In p53-dependent cell apoptosis, p53 have obvious promoting effect on the expression of Bax [12,13]. In vitro, knockdown of PHF20 by RNA interference could reduce the level of P53 and Bax. Moreover, in the p53-null cells, the knockdown of PHF20 obviously reduces the Bax level as in the wild-type p53 cells [9,10].
To examine the clinical significance and biological functions of PHF20 in non-small-cell lung cancer (NSCLC), we investigated the expression of PHF20 in 142 cases of NSCLC tissue and 30 cases of normal lung tissue by immunohistochemical staining. We also investigated the role of PHF20 in regulating the proliferation and invasion of lung cancer cells.
Materials and methods
Tissue samples
142 lung cancer samples and 30 normal tissues were obtained from patients who had surgery in the First Affiliated Hospital of China Medical University during 2009 to 2011. All of the enrolled patients underwent curative surgical resection without having chemotherapy or radiation therapy. Formalin-fixed paraffin-embedded sections of tissues obtained from surgical samples were stained routinely with hematoxylin and eosin (H&E).
Cell culture
Human NSCLC cell lines H1299, H460, H157, A549, SPC, LK-2 and one human bronchial epithelium cell line (HBE) were cultured in RPMI-1640 (Gibco, Invitrogen, NY, USA) supplemented with 10% fetal bovine serum at 37°C in 5% CO2. For transfections, cells were seeded in a six-well plate 24 h before the experiment.
Immunohistochemical staining
All the resected specimens were fixed with 10% neutral-buffered formalin and embedded in paraffin blocks. Tissue blocks were cut into 4-μm slides, deparaffinized in xylene, rehydrated with graded alcohols, and immunostained with anti-PHF20 (Rabbit monoclonal antibody, Cell Signaling Technology, USA) and anti-Bax (Rabbit monoclonal antibody, Cell Signaling Technology, USA).
Sections were stained with a streptavidin-peroxidase system (KIT-9710, Ultrasensitive TM S-P, MaiXin, China). The chromogen used was diaminobenzidine tetrahydrochloride substrate (DAB kit, MaiXin, China), slightly counterstained with hematoxylin, dehydrated and mounted. For the negative controls, the primary antibody was replaced with PBS. The sections were observed under five random high power fields, and 100 cells were counted in each field. The staining was scored as follows: positive cells ≤10% (-); cells staining positive: 11-25% (+), 26-50 %(++), 51-75% (+++) and ≥76% (++++).
Small-interfering RNA experiment
PHF20 siRNA (sc-76117) was purchased from Santa Cruz Biotechnology (Santa Cruz, USA). For the siRNA transfection experiments, cells were seeded in six-well plate according to manufacturer’s instructions. Twenty-hours later, the siRNA was transfected into the cells using Lipofectamine RNA iMAX reagent (Invitrogen, Carlsbad, CA, USA). Transfected cells were incubated for another 48 h and subjected to various analyses. Following transfection, the protein level was assessed 72 h later by western blotting.
Colony formation assay
Forty-eight hours after siRNA transfection, cells were planted into 6-cm cell culture dishes (1000 cells per dish) and incubated for 14 days. Cells were then stained with Giemsa and the number of colonies with more than 50 cells was counted.
Cell invasion assay
A 24-well Transwell chamber was used with a pore size of 8 μm (Corning, NY, USA) and the inserts were coated with Matrigel (BD Bioscience) in serum-free medium. Forty-eight hours after transfection, cells were trypsinized and transferred to the upper Matrigel hamber in serum-free medium containing 3×105 cells and incubated for 16 h. Then the non-invading cells on the upper membrane surface were removed with a cotton tip, and the cells which passed through the filter were fixed with 4% paraformaldehyde and stained with hematoxylin. The number of invaded cells was counted in 10 randomly selected high power fields under microscope. Data presented are representative of three individual wells.
Western blotting
Total protein from cells were extracted in lysis buffer (Pierce, Rockford, IL, USA) and quantified by the Bradford method. The same amount of protein was separated by 10% SDS-PAGE and then electophoretically transferred to a PVDF membrane (Millipore, Bedford, MA, USA). The membrane was blocked with 5% dry milk and incubated overnight at 4°C with antibodies against PHF20 (1:500, Cell Signaling Technology, USA) and GAPDH (1:5,000; Santa Cruz Biotechnology, USA). After washing, the membrane was incubated with a secondary antibody for 2 h. Protein bands were visualized with the enhanced chemiluminescence (Pierce) and detected using BioImaging Systems (UVP, Upland, CA, USA). The relative protein levels were calculated based on GAPDH protein as a loading control.
Statistical analysis
Pearson’s chi-square test was used to analyze the relationship between PHF20 expression and the clinicopathological factors. McNemar’s test was used to compare PHF20 expression in normal lung and lung cancer tissues. Spearman’s correlation coefficient (r) was used to find out the correlation between the expressions of PHF20 and Bax. The t test was used to analyze the results of western blot. SPSS statistical software package version 13.0 (SPSS Inc., Chicago, IL, USA) was used for all analyses. P values of <0.05 were considered statistically significant.
Results
PHF20 expression in normal lung and NSCLC tissues
The expression of PHF20 was assessed by immunohistochemistry in 142 NSCLC samples and 30 adjacent non-cancer samples. PHF20 expression was found in 76.7% (23/30) of normal lung tissues, while PHF20 expression was found in 43.0% (61/142) of NSCLC tissues. In normal lung tissues, PHF20 was located in the cytoplasm and nuclei of the normal bronchial epithelium (Figure 1A) and the alveolar epithelial cells (Figure 1B), and the staining was strong and constant. In contrast, PHF20 immunostaining in cancer cells was frequently reduced compared with the adjacent normal tissue (Figure 1C, 1F). As summarized in Table 1, NSCLC showed a significantly lower PHF20 expression in NSCLC tissues than normal tissues (P<0.05).
Figure 1.
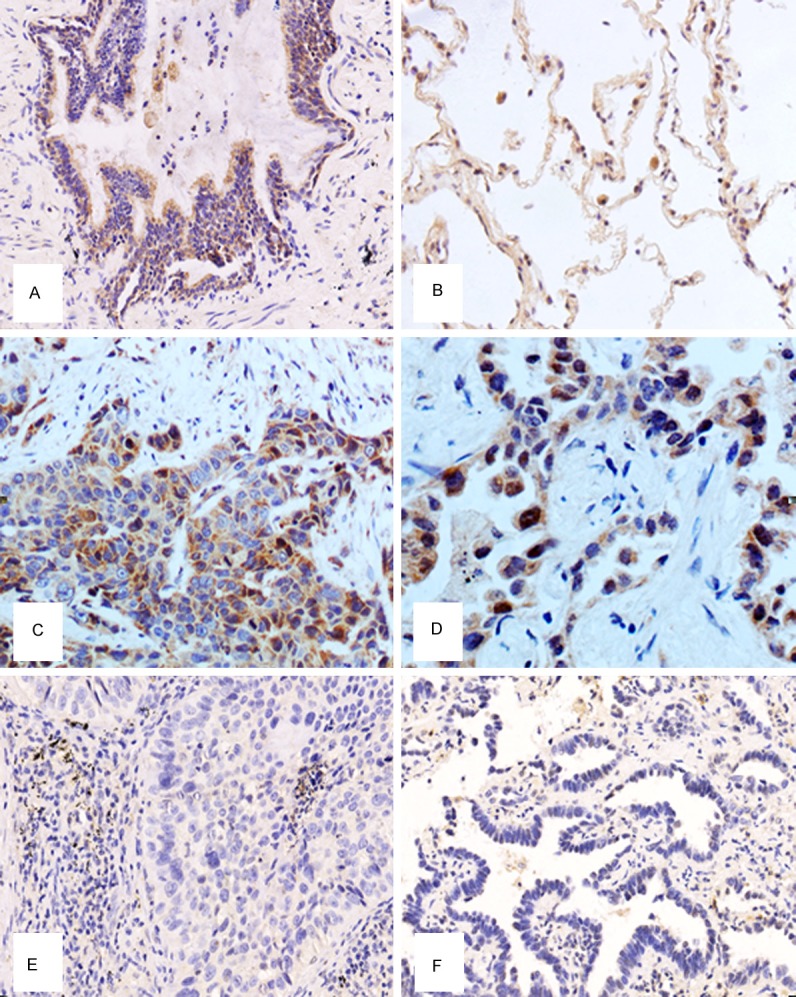
Expression of Plant homeodomain finger protein 20 (PHF20) in normal lung tissue and non-small-cell lung cancer (NSCLC). A. High PHF20 expression was observed in the normal bronchial epithelia. B. High PHF20 expression could also be seen in the normal alveolar epithelial cells. C. The positive staining of PHF20 could be seen in lung squamous cell carcinoma. D. The positive staining of PHF20 could be seen in lung adenocarcinoma. E. The negative PHF20 expression could be observed in lung squamous cell carcinoma. F. The negative PHF20 expression could be observed in lung squamous cell carcinoma.
Table 1.
PHF20 Expression pattern in normal lung tissues and NSCLC tissues
| N | PHF20 | P-value | ||
|---|---|---|---|---|
|
| ||||
| Negative | Positive | |||
| Normal | 30 | 7 (23.3%) | 23 (76.7%) | |
| Tumor | 142 | 81 (57.0%) | 61 (43.0%) | 0.0001 |
NSCLC, non-small-cell lung cancer; *P<0.05, indicate statistical significance.
Western blot study demonstrated that expression of PHF20 protein in lung cancer cell lines (SPC, A549, H1299, H157, H460, LK2) was significantly lower than that in human bronchial epithelium cell line (HBE, P<0.05, Figure 2).
Figure 2.
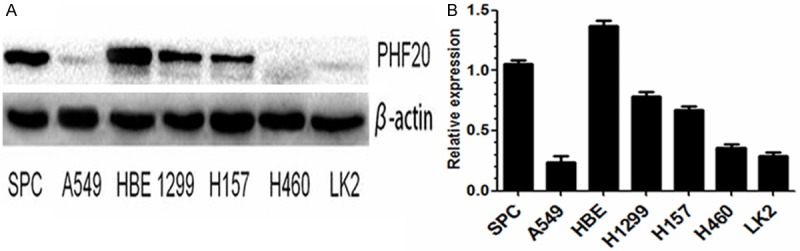
A and B. The protein level of PHF20 in 6 lung cancer cell lines (A549, SPC, H1299, H460, H157 and LK-2) and one human bronchial epithelium cell line (HBE).
Relationship between the expression of PHF20 and the clinicopathological factors in NSCLC
We analyzed the relationship between PHF20 expression and the clinicopathologic factors. As described in Table 2, PHF20 expression was significantly correlated with differentiation (P=0.015), pTNM stage (P=0.031) and nodal status (P=0.004). No significant difference was observed in age, gender, and tumor size.
Table 2.
Association of PHF20 expression in NSCLC with clinical and pathologic factors
| N | PHF20 | P-value | ||
|---|---|---|---|---|
|
| ||||
| Negative | Positive | |||
| Total | 142 | 81 (57.04%) | 61 (42.96%) | |
| Age (years) | ||||
| ≤55 | 56 | 36 (64.29%) | 20 (35.71%) | |
| >55 | 86 | 45 (52.33%) | 41 (47.67%) | 0.170 |
| Gender | ||||
| Male | 76 | 43 (56.58%) | 33 (43.42%) | |
| Female | 66 | 36 (54.55%) | 30 (45.45%) | 0.866 |
| Histological type | ||||
| Squamous cell carcinoma | 70 | 36 (51.42%) | 34 (48.58%) | |
| Adenocarcinoma | 72 | 45 (62.50%) | 27 (37.50%) | 0.631 |
| Grade | ||||
| Well | 42 | 17 (40.48%) | 25 (59.52%) | |
| Moderate -Poor | 100 | 64 (64.00%) | 36 (36.00%) | 0.015 |
| TNM stage | ||||
| I and II | 95 | 48 (50.53%) | 47 (49.47%) | |
| III and IV | 47 | 33 (70.21%) | 14 (29.79%) | 0.031 |
| Tumor size | ||||
| <3 cm | 51 | 27 (52.94%) | 24 (47.06%) | |
| ≥3 cm | 81 | 54 (66.67%) | 27 (33.33%) | 0.484 |
| Lymph node metastasis | ||||
| Yes | 75 | 34 (45.33%) | 41 (54.67%) | |
| No | 67 | 47 (70.15%) | 20 (29.85%) | 0.004 |
NSCLC, non-small-cell lung cancer; *p<0.05, indicate statistical significance.
Furthermore, we also analyzed the relationship of PHF20 expression to the overall survival rate. The result showed that the overall survival was significantly higher in patients with PHF20 positive expression than in patients with PHF20 negative expression (P=0.035, Figure 3). That is, the patients with PHF20 expression have a better prognosis. Multivariate survival analysis revealed that the nodal status of the tumor was the significant and independent prognosticator (P=0.024; Cox regression model, Table 3).
Figure 3.
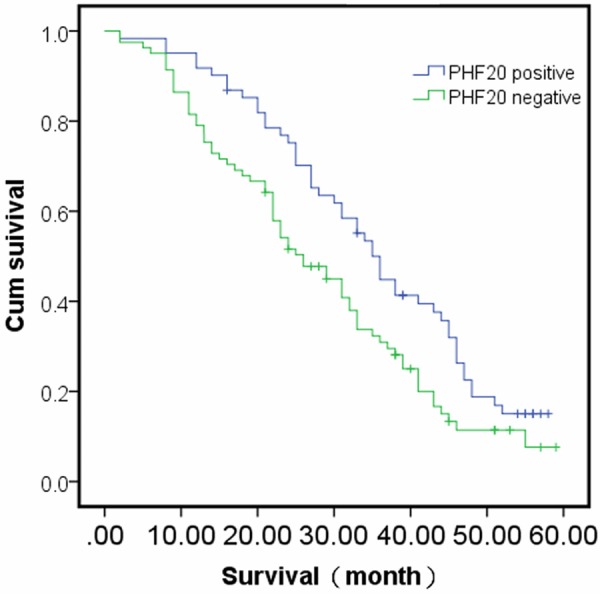
Survival curves of patients with positive and negative PHF20 expression. The patients with PHF20 expression have a better prognosis (P<0.05).
Table 3.
Multivariate analysis for predictive factors in patients with NSCLC (Cox regression model)
| Factor | P-values | RR | 95% CI |
|---|---|---|---|
| Gender | 0.316 | 0.624 | 0.248-1.569 |
| Age | 0.345 | 0.678 | 0.302-1.520 |
| Histology | 0.617 | 1.241 | 0.532-2.896 |
| Tumor status | 0.299 | 0.567 | 0.194-1.653 |
| Nodal status | 0.024 | 2.825 | 1.144-6.973 |
| PHF20 | 0.202 | 0.595 | 0.268-1.321 |
CI, confidence interval; NSCLC, non-small-cell lung cancer; RR, risk ratio.
PHF20 inhibited the proliferation of lung cancer cells
We down-expressed PHF20 in SPC cell, the protein levels of PHF20 were significantly down regulated compared with control empty vector. By colony formation assay, silence of PHF20 in SPC cells by siRNA knockdown led to a significant increase (control versus PHF20 siRNA, 95±6 versus 165±7, P=0.004) in colony numbers as well as sizes (Figure 4).
Figure 4.
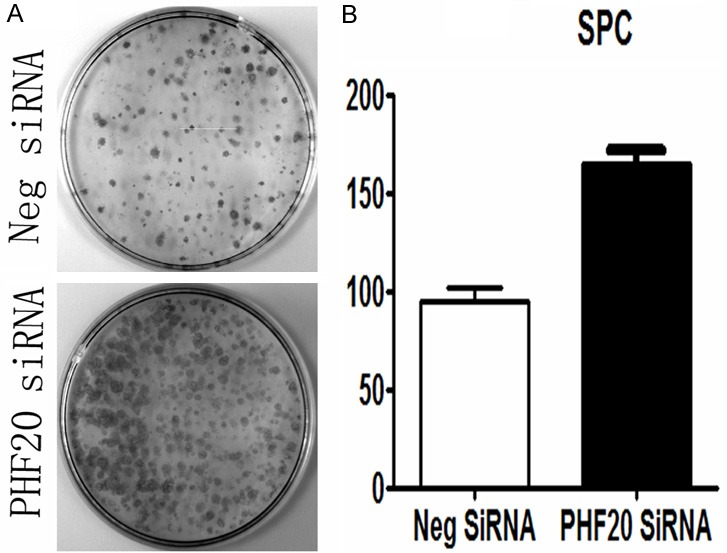
Effects of PHF20 on the proliferation of NSCLC cell lines. A. Colony formation assay indicated that PHF20 down-regulation markedly promoted proliferation in SPC cells. B. Columns, mean for three experiments; bars, SD (P<0.05).
PHF20 inhibited the invasion of lung cancer cells
To address the impact of PHF20 on NSCLC cell invasion, we performed matrigel invasion assay. As shown in Figure 5, PHF20-depleted SPC cells showed significantly stronger invasion ability (control versus PHF20 siRNA, 64±6 versus 105±6, P=0.0038) compared with control siRNA transfected cells.
Figure 5.
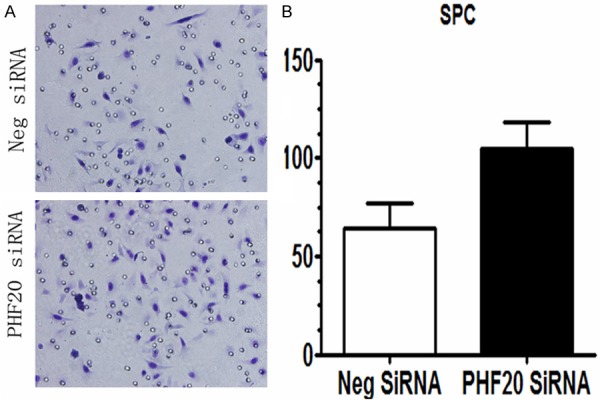
Effects of PHF20 on the invasion of NSCLC cell lines. A. Transwell assay indicated that knockdown of PHF20 in SPC cells greatly promoted tumor cell invasion. B. Graphs show the number of cells invaded through the transwell after 16 h of incubation. The number of invaded cells was counted in 10 randomly selected high power fields under microscope. Columns, mean for three experiments; bars, SD (P<0.05).
Relationship between the expressions of PHF20 and Bax in NSCLC
The relationship between the expressions of PHF20 and Bax in NSCLC is shown in Table 4. A positive correlation was found between PHF20 and Bax (P=0.001, Figure 6).
Table 4.
Relationship between the expressions of PHF20 and Bax in NSCLC
| Bax | PHF20 | N | Correlation | P-value | |
|---|---|---|---|---|---|
|
|
|
||||
| Positive | Negative | Coefficient(rs) | |||
| Positive | 61 | 22 | 83 | 0.56 | 0.001 |
| Negative | 20 | 39 | 59 | ||
NSCLC, non-small-cell lung cancer; *P<0.05, indicate statistical significance.
Figure 6.
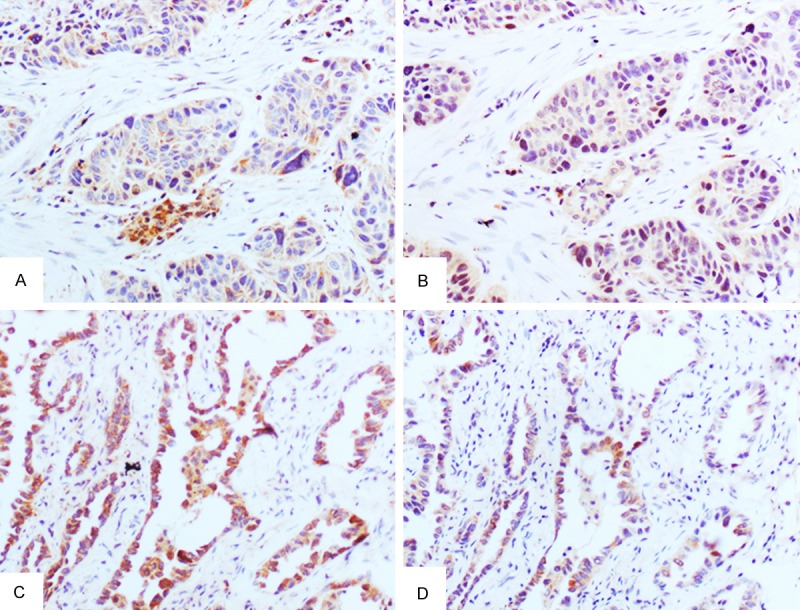
Expression of PHF20 and Bax in serial sections of NSCLC. PHF20 expression in squamous cell carcinoma (A) and adenocarcinoma (C) was positively related to Bax expression in squamous cell carcinoma (B) and adenocarcinoma (D).
Discussion
It still remains unclear that PHF20 plays a role as oncogene or tumor suppressor gene in tumorgenesis. Some studies reported that PHF20 could inhibit the tumorigenicity by inducing apoptosis. PHF20 could promote stabilization and activation of p53 by diminishing Mdm2-mediated P53 ubiquitylation and degradation [9,10]. PHF20 gene was located in chromosome 20q11, in which high-level loss of heterozygosity was often detected [14], indicating PHF20 might be a potential tumor suppressor gene. Bankovic et al. examined genomic instability in 30 patients with non-small-cell lung cancer (NSCLC) by comparing DNA fingerprints of paired tumor and normal tissues using arbitrarily primed polymerase chain reaction. The results indicated that PHF20 gene alteration was present in 8 out of 30 (26.7%) NSCLC samples. Moreover, PHF20 DNA alteration was more observed in tumors of grade 1, stage 1 and without lymph node invasion, and was also associated with poor prognosis [3]. In contrast, other studies indicated that PHF20 might function as an oncogene. Zhang et al. reported PHF20 could promote NF-κB transcriptional activity, and expression levels of PHF20 were significantly associated with pathological tumour grade of gliomas [7].
So, PHF20 may play different roles in human oncogenesis depending on the specific tissue or the context. In addition, little is known about the expression, clinical significance, potential function and mechanism of PHF20 in NSCLC. In the current study, we found PHF20 was positively expressed in 76.7% of the normal lung tissues (n=30) but only expressed in 43.0% of the lung cancer samples (n=142). The expression level of PHF20 in normal bronchial epithelium and alveolar epithelial cells was significantly higher than the expression level in cancer cells. Our result demonstrated that down-regulation of PHF20 might involve tumorgenesis of NSCLS. Our study also showed PHF20 expression was significantly correlated with differentiation (P=0.015), pTNM stage (P=0.031) and nodal status (P=0.004). That is PHF20 was more often expressed in well differentiated tumors with I and II TNM stage and without lymph node metastasis. Importantly, although multivariate survival analysis revealed that PHF20 expression was not independent prognosticator, we found that the loss of PHF20 expression was associated with short overall survival in NSCLC patients. In contract, Taniwaki et al. reported that PHF20 was abundantly expressed both in advanced small-cell lung cancers and advanced adenocarcinomas [8].
We think that the different statistic analysis and the limited sum of specimens might lead to the different result. Next, we further investigate the role of PHF20 in regulating the proliferation and invasion of lung cancer cells. Colony formation assay showed that the depletion of PHF20 in SPC cells led to a significant increase in colony numbers. Moreover, matrigel invasion assay showed that depletion of PHF20 also significantly induced invasion compared with control empty vector in lung cancer cells. These results demonstrated that loss of PHF20 expression can promote the colony formation and invasion in lung cancer. Taken together, these results indicated PHF20 might be a candidate tumor suppressor and loss of PHF20 expression was useful for predicting the poor prognosis of NSCLC patients.
Recently, PHF20 reportedly could inhibit cell proliferation through inducing cell apoptosis mediated by p53 [9,10]. Bax is the main apoptosis promoter of Bcl-2 family, and could be affected by p53 in cell apoptosis [11]. Cui et al. reported that knockdown of PHF20 could reduce the level of p53 and Bax in HCT 116 cells [9]. Moreover, in the p53-null cells, the knockdown of PHF20 could obviously reduce the Bax level as in the wild-type p53 cells. Therefore, it could be observed that PHF20 inhibits cell proliferation, DNA synthesis, and cell survival. So far reduced PHF20 expression was found in many cancers including NSCLC [16-20]. In our study, expression of PHF20 was significantly related to the Bax expression. These results indicated that loss of PHF20 might induce down-regulation of Bax, and then induce cell proliferation of NSCLC.
Collectively, the present study identified PHF20 as a candidate tumor suppressor which was down-expressed in NSCLC. The expression of PHF20 in NSCLC was significantly correlated with p-TNM stage, lymph node metastasis, differentiation and overall survival. Moreover, the expression of PHF20 in NSCLC was positively associated with Bax expression. These results strongly indicate that PHF20 may play an important role in NSCLC and PHF20 might serve as a candidate target protein for future therapeutics of NSCLC. In addition, PHF20 may function in NSCLC through associating with Bax.
Acknowledgements
This work was supported by Program for Liaoning Innovative Research Team in University (No: LC2015029).
Disclosure of conflict of interest
None.
References
- 1.Fischer U, Struss AK, Hemmer D, Pallasch CP, Steudel WI, Meese E. Glioma-expressed antigen 2 (GLEA2): a novel protein that can elicit immune responses in glioblastoma patients and some controls. Clin Exp Immunol. 2001;126:206–13. doi: 10.1046/j.1365-2249.2001.01635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pallasch CP, Struss AK, Munnia A, König J, Steudel WI, Fischer U, Meese E. Autoantibodies against GLEA2 and PHF3 in glioblastoma: tumor-associated autoantibodies correlated with prolonged survival. Int J Cancer. 2005;117:456–9. doi: 10.1002/ijc.20929. [DOI] [PubMed] [Google Scholar]
- 3.Bankovic J, Stojsic J, Jovanovic D, Andjelkovic T, Milinkovic V, Ruzdijic S, Tanic N. Identification of genes associated with non-small-cell lung cancer promotion and progression. Lung Cancer. 2010;67:151–9. doi: 10.1016/j.lungcan.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Han KJ, Pang XW, Vaughan HA, Qu W, Dong XY, Peng JR, Zhao HT, Rui JA, Leng XS, Cebon J, Burgess AW, Chen WF. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. J Immunol. 2002;169:1102–9. doi: 10.4049/jimmunol.169.2.1102. [DOI] [PubMed] [Google Scholar]
- 5.Behrends U, Schneider I, Rössler S, Frauenknecht H, Golbeck A, Lechner B, Eigenstetter G, Zobywalski C, Müller-Weihrich S, Graubner U, Schmid I, Sackerer D, Späth M, Goetz C, Prantl F, Asmuss HP, Bise K, Mautner J. Novel tumor antigens identified by autologous antibody screening of childhood medulloblastoma cDNA libraries. Int J Cancer. 2003;106:244–51. doi: 10.1002/ijc.11208. [DOI] [PubMed] [Google Scholar]
- 6.Park S, Kim D, Dan HC, Chen H, Testa JR, Cheng JQ. Identification of Akt interaction protein PHF20/TZP that transcriptionally regulates p53. J Biol Chem. 2012;287:11151–63. doi: 10.1074/jbc.M111.333922. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Zhang T, Park KA, Li Y, Byun HS, Jeon J, Lee Y, Hong JH, Kim JM, Huang SM, Choi SW, Kim SH, Sohn KC, Ro H, Lee JH, Lu T, Stark GR, Shen HM, Liu ZG, Park J, Hur GM. PHF20 regulates NF-κB signalling by disrupting recruitment of PP2A to p65. Nat Commun. 2013;4:2062. doi: 10.1038/ncomms3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniwaki M, Daigo Y, Ishikawa N, Takano A, Tsunoda T, Yasui W, Inai K, Kohno N, Nakamura Y. Gene expression profiles of small-cell lung cancers: molecular signatures of lung cancer. Int J Oncol. 2006;29:567–75. [PubMed] [Google Scholar]
- 9.Cui G, Park S, Badeaux AI, Kim D, Lee J, Thompson JR, Yan F, Kaneko S, Yuan Z, Botuyan MV, Bedford MT, Cheng JQ, Mer G. PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53. Nat Struct Mol Biol. 2012;19:916–24. doi: 10.1038/nsmb.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Park J, Piao L, Kong G, Kim Y, Park KA, Zhang T, Hong J, Hur GM, Seok JH, Choi SW, Yoo BC, Hemmings BA, Brazil DP, Kim SH, Park J. PKB-mediated PHF20 phosphorylation on Ser291 is required for p53 function in DNA damage. Cell Signal. 2013;25:74–84. doi: 10.1016/j.cellsig.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a concerved homolog, Bax that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 12.Mori S, Ito G, Usami N, Yoshioka H, Ueda Y, Kodama Y, Takahashi M, Fong KM, Shimokata K, Sekido Y. p53 apoptotic pathway molecules are frequently and simultaneously altered in non small cell lung carcinoma. Cancer. 2004;100:1673–1682. doi: 10.1002/cncr.20164. [DOI] [PubMed] [Google Scholar]
- 13.Park DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 14.Vauhkonen H, Vauhkonen M, Sajantila A, Sipponen P, Knuutila S. DNA copy number aberrations in intestinal-type gastric cancer revealed by array-based comparative genomic hybridization. Cancer Genet. Cytogenet. 2006;167:150–154. doi: 10.1016/j.cancergencyto.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Bankovic J, Stojsic J, Jovanovic D, Andjelkovic T, Milinkovic V, Ruzdijic S, Tanic N. Identification of genes associated with non-small-cell lung cancer promotion and progression. Lung Cancer. 2010;67:151–159. doi: 10.1016/j.lungcan.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Krajewski S, Blomquist C, Franssila K, Krajewska M, Wasenius VM, Niskanen E, Nordling S, Reed JC. Reduced expression of proapoptosis gene Bax is associated with poor response rates to combination chemotherapy and shorter survival in women with metastatic breast adenocarcinoma. Cancer Res. 1995;55:4471–8. [PubMed] [Google Scholar]
- 17.Krajewska M, Moss S, Krajewski S, Song K, Holt PR, Reed JC. Elevated expression of Bcl-2 and reduced Bax in primary colorectal adenocarcinoma. Cancer Res. 1996;56:1834. [PubMed] [Google Scholar]
- 18.Chresta CM, Masters JR, Hickman JA. Hypersensitivity of human testicular tumors to etoposide-induced apoptosis is associated with functional p53 and a high Bax: Bcl-2 ratio. Cancer Res. 1996;56:1834. [PubMed] [Google Scholar]
- 19.Krajewska M, Krajewski S, Epstein JI, Shabaik A, Sauvageot J, Song K, Kitada S, Reed JC. Immunohistochemical analysis of Bcl-2, Bax, Bcl-x, and mcl-1 expression in prostate cancer. Am J Pathol. 1996;148:1567. [PMC free article] [PubMed] [Google Scholar]
- 20.Milas I, Kamaki R, Hachiga T, Bubb RS, Ro JY, Langford L, Sawaya R, Putnam JB, Allen P, Cox JD, McDonnell TJ, Brock W, Hong WK, Roth JA, Milas L. Epidermal growth factor receptor, cyclo-oxygenase-2, and Bax expression in the primary non-small-cell lung cancer and brain metastasis. Clin Cancer Res. 2003;9:1070–1076. [PubMed] [Google Scholar]


