Abstract
Poly(ADP-ribose)polymerase-1 (PARP-1) is anubiquitous, DNA repair-associated enzyme, which participates in gene expression, cell death, central nerve system (CNS) disorders and oxidative stress. According to the previous studies, PARP-1 over-activation may lead to over-consumption of ATP and even cell apoptosis. Spinal cord injury (SCI) is an inducement towards PARP-1 over-activation due to its massive damage to DNA. 3-aminobenzamide (3-AB) is a kind of PARP-1 inhibitors. The relationship among PARP-1, 3-AB, SCI and apoptosis has not been fully understood. Hence, in the present study, we focused on the effects of 3-AB on cell apoptosis after SCI. Accordingly, SCI model was constructed artificially, and 3-AB was injected intrathecally into the Sprague-Dawley (SD) rats. The results demonstrated an increase in cell apoptosis after SCI. Furthermore, PARP-1 was over-activated after SCI but inhibited by 3-AB injection. In addition, apoptosis-inducing factor (AIF) was inhibited but B-cell lymphoma-2 (Bcl-2) was up-regulated by 3-AB. Interestingly, caspase-3 was not significantly altered with or without 3-AB. In conclusion, our experiments showed that 3-AB, as a PARP-1 inhibitor, could inhibit cell apoptosis after SCI in caspase-independent way, which could provide a better therapeutic target for the treatment of SCI.
Keywords: 3-AB, PARP-1, inhibitors, SCI, apoptosis
Introduction
Poly(ADP-ribose)polymerase-1 (PARP-1) is a key protein participating in DNA repair [1], which is also seen as anuclear enzyme contributing to the chromatin integrity and cell survival [2]. PARP-1 inhibitors decrease the DNA repair by competitively binding with the DNA-repair complex [3]. Specifically, when DNA strand-breaks happen, PARP binds rapidly to the DNA strand-breaks and converts nicotinamide adenine dinucleotide (NAD) into long branched polymers of poly(ADP-ribose) (PAR), which was attached on a variety of nuclear proteins [1,4-6].
Nevertheless, excessive PARP-1 may cause a negative effect in a caspase-independent pathway-mediated cell apoptosis [7]. It is widely accepted that over-activation of PARP-1 results in the translocation of apoptosis-inducing factor (AIF) from the mitochondria to the nucleus, where AIF promotes DNA fragmentation and nuclear condensation for apoptosis [8-11]. Besides, PARP-1 over-activation is also characterized by the marked decrease of intracellular NAD and adenosine triphosphate (ATP) stores and disorders of oxidative metabolism [1,12,13], and, ultimately, leads to cell death [4,14]. In addition, the translocation of AIF destroys the mitochondrial membrane potential, which leads to less energy production and more apoptosis.
It is worth noting that PARP-1 over-activation originates from a variety of circumstances, mainly due to the massive DNA damage, including neurotoxicity, ischemia-reperfusion injury, diabetes, shock and inflammation [12,13,15]. Spinal cord injury (SCI) is another reason for massive DNA damage, which leads to both the primary injury and the subsequent secondary injury, including cell apoptosis, excessive inflammatory response, reduced spinal cord blood flow, etc [16,17]. Cell apoptosis plays a pivotal role in this secondary damage by causing degeneration of the spinal cords gradually [18,19]. Hence, supposedly, an effective inhibition of PARP-1 over-activation could reduce cell apoptosis induced by SCI, which contributes to the protection of spinal cords.
PARP inhibitors could act as competitors of NAD [20,21], accompanied by the accumulation of NAD and inhibition of PARP [22], thus preventing the DNA repair [3,23]. 3-Aminobenzamide (3-AB), one of PARP inhibitors, could attenuate the activation of PARP-1, and abolish AIF translocation [6]. To our knowledge, the role of 3-AB has not yet been studied in any animal model of SCI.
In conclusion, we constructed a SCI model in rats, and 3-AB was injected intrathecally into the spinal cords. As descriptions in this study, 3-AB was shown to inhibit PARP-1 activation and the subsequent apoptosis induced by SCI, which may provide a potential therapeutic target for SCI and neural degeneration.
Materials and methods
Animal feeding
All the experiments with animals were approved by the Institutional Animal Ethics Committee of Binzhou Medical University. The experiments were performed with Sprague-Dawley (SD) male rats (220-250 g), provided by the experimental animal center of Yantai Luye Pharma. The animals were fed in pathogen-free environment with free access to food and boiled water at a relatively constant temperature and humidity, with a 12-hour light/dark cycle. The rats were acclimatized for at least seven days since arrival and were observed for any sign of diseases before the experiment were initiated.
Animal treatment
The animals were randomly divided intodifferent groups. 3-AB was purchased from Sigma-Aldrich (St. Louis, MO, USA). There were 3 groups: sham group (n=10), SCI group (rats with SCI surgery; n=10) and 3-AB+SCI group (rats with SCI and 3-AB treatment; n=10). The 3-ABdissolved in 0.9% sodium chloride was injected intrathecally immediately after SCI with a concentration of 15 mg/kg, according to aprevious experiments [24]. The detailed procedures of SCI were as follows: rats were anesthetized with intraperitoneal injection of 4% chloral hydrate (300 mg/kg), followed by fixation in prone position. Subsequently, incision in dorsal midline, separation of muscle and exposure of spines were performed successively. The spinous process and vertebral plate were taken out along the T10 section-centered position to expose the spinal dura mater. Next, the spines were kept in horizontal position with the hit in posterior spinal veins-centered position (modified Weight dropping by Allen) to cause acute moderate injury in T 10 section. The SCI models were characterized by the retraction and flutter of double lower limbs and body, accompanied by tail-wag reflection. Hemostasis was conducted immediately after surgery, and straticulate saturation was performed. Artificial micturition was done twice a day until self-urination was recovered. The inflammation was under restrictionby injection of penicillin once a day.
Tissue collection
After 7 days, the animals were humanely sacrificed with pentobarbital sodium (500 mg/kg) and segments of spinal cord, centered with the lesion site, were harvested for formalin-fixation and paraffin-embedding in preparation for use. Specifically, a 2-cm section which centered at the lesion site of the spinal cord was removed and fixed in formalin overnight. The segment was then dehydrated through graded ethanol, embedded in paraffinand sectioned into 7 μm thick transverse sections using a cryostat microtome (Thermo Fisher Scientific, Waltham, MA, USA).
Deparaffinization and rehydration
Tissue sections were processed with the next procedures for deparaffinization and rehydration: immerged in dimethylbenzene for 15 min (repeated 3 times in 3 different containers); in 100% ethanol for 5 min (2 times in 2 different containers); in 95% ethanol for 5 min (2 times in 2 different containers); in 90% ethanol, 85% ethanol and 75% ethanol for 2 min, successively; finally rinsed in distilled water 3 times.
Hematoxylin-eosin (HE) staining
Specifically, tissue sections were stained with hematoxylin for 5 min, rinsed in running water and then immerged in hydrochloric-alcohol solution for 15 sec to differentiate. The following steps were performed, successively: microscopic examination; rinsed in running water and distilled water; dehydrated through graded ethanol; stained with 0.5% eosin solution; immerged in dimethylbenzene for 5 min 2 times for transparency and finally mounted with neutral balsam. The sections were then observed under an optical microscope (Olympus FV2000, Tokyo, Japan) and photographed.
TUNEL assay
Apoptotic cells were identified via TUNEL assay, using the in situ cell death detection kit, POD according to the manufacturer’s protocol (Roche, Basel, Switzerland). Briefly, cover-slips were subjected to fixation and permeabilization, followed by incubation with the TUNEL reaction mixture containing terminal deoxynucleoitidyl transferase (TdT) and fluorescein-labeled dUTPat 37°C for 1 hour in a dark and humid chamber. Subsequently, slips were rinsed in PBS 3 times, and incubated with 50 μl of converter-POD at 37°C for 30 min. Finally, diaminobenzidine (DAB) was used as the chromogen and added after PBS rinse again for an incubation at 25°C for 10 min. Hematoxylin was used as the counterstain. Dehydration, transparency and mounting were performed, successively. An optical microscope (Olympus) was used for the further analysis.
Immunohistochemistry (IHC)
The sections were immerged in 0.01 M citrate buffer in boiling water for 2 min, for antigen retrieval, and cooled down to room temperature by itself. Subsequently, the sections were immerged in 0.3% H2O2 for 15 min; rinsed in distilled water for 3 times; immerged in PBS buffer for 5 min for endogenous peroxidase blocking. Next, tissue sections were incubated with primary antibodies at 4°C over night in a humidified chamber. The slides were then incubated with secondary antibody at 37°C for 20 min, followed by incubation with horseradish peroxidase (HRP; Sigma-Aldrich) at 28°C for 15 min and PBS wash again. Antibody binding was visualized with freshly prepared 3,3’-DAB (Sigma-Aldrich), and counterstained with hematoxylin (Sigma-Aldrich), followed by graded alcohol dehydration and mounting as previously described. Slides were observed and recorded under 400× objectives with a microscope (Olympus).
Real-time quantitative PCR
Total RNA was extracted from the tissues using TriPure RNA Isolation Reagent (Roche). cDNA was synthesized using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative PCR was performed using FastStart Universal SYBR Green Master (Roche) on the LightCycler 480 Real-Time PCR System (Roche). All the procedures were following standard manufacturer’s instructions. β-actin was served as a reference gene. Each sample was measured in triplicate, and the relative expression was calculated using the 2-ΔΔCt method. The sequences of the involved primers were shown in Table 1.
Table 1.
List of primer sequences for real-time quantitative PCR
| Primers | Forward | Reverse |
|---|---|---|
| PARP-1 | 5’-ACGCACAATGCCTATGAC-3’ | 5’-CCAGCGGAACCTCTACAC-3’ |
| AIF | 5’-AAAGGGAAGGAGGAGGTCCCGA-3’ | 5’-CCACTGTGGGGCTTCAGTCTTCA-3’ |
| Bcl-2 | 5’-GGATTGTGGCCTTCTTTGAG-3’ | 5’-CCAAACTGAGCAGAGTCTTC-3’ |
| Caspase-3 | 5’-GGACCTGTGGACCTGAAAAA-3’ | 5’-CGGGATCTGTTTCTTTGAAT-3’ |
| β-actin | 5’-TACAACCTCCTTGCAGCTCC-3’ | 5’-GGATCTTCATGAGGT AGTCAGTC-3’ |
Western blot
Proteins were extracted using RIPA buffer containing 1 mM Phenylmethane sulfonyl fluoride (PMSF, Sigma-Aldrich). Briefly, tissues were immerged in RIPA buffer on ice with occasional shaking and mixing for 30 min. Subsequently, the proteins were collected and stored in a centrifuge tube. Electrophoresis was performed using 10% polyacrylamide gels (PAGE) containing 0.1% sodium dodecyl sulfonate (SDS). Proteins were electro-transferred to nitrocellulose (NC) membrane and blots were incubated for 2 hours at room temperature with primary antibodies, followed by incubation with corresponding biotin-labeled secondary antibodies. The antigen-antibody complexes were detected using the ECL system (Amersham Biosciences, Buckinghamshire, UK).
Statistical analysis
Data were expressed as mean ± Standard Deviation (SD). Differences between groups were calculated using one-way ANOVA. The *P<0.05 and **P<0.01 were regarded as significance.
Results
Spinal cord has some pathologic changes after sci
We first confirmed the successful construction of SCI models. As shown in Figure 1B, spinal cord demonstrated a pathologic status compared with sham group in a normal status (Figure 1A). Specifically, there was a significant decrease in the quantity of neurons in SCI group compared with sham group, because neurons tended to become smaller, even form vacuoles (black arrows). Furthermore, the glial cells (nucleus stained blue) had an obvious proliferation in SCI group (red arrows), and the red blood cells (stained dark red) became visible in SCI group (green arrows), which was possibly due to the hemorrhage after SCI surgery. In contrast, 3-AB addition showed a counteractive and opposite effect toward SCI surgery, characterized by fewer vacuoles and glial cells, and scarce red blood cells (Figure 1C).
Figure 1.
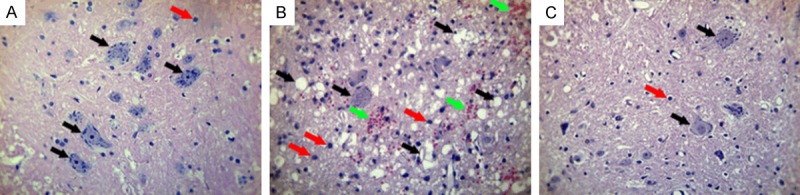
HE staining for spinal cord tissues. A. Sham group in a normal status; B. SCI group showed smaller neurons and more vacuoles (black arrows), more glial cells (red arrows), and visible red blood cells (green arrows); C. 3-AB + SCI group showed fewer vacuoles and glial cells, and scarce red blood cells.
3-AB reduces the number of apoptotic cells
Next, the apoptosis was investigated to compare the effects of SCI and 3-AB. We compared Figure 2A and 2B, there was a significant increase of apoptotic cells (nucleus stained brown) in SCI group (Figure 2B). In contrast, as shown in Figure 2C, 3-AB + SCI group demonstrated less apoptosis, indicating that 3-AB could inhibit the apoptosis induced by SCI surgery. Figure 2D also confirmed this phenomenon statistically.
Figure 2.
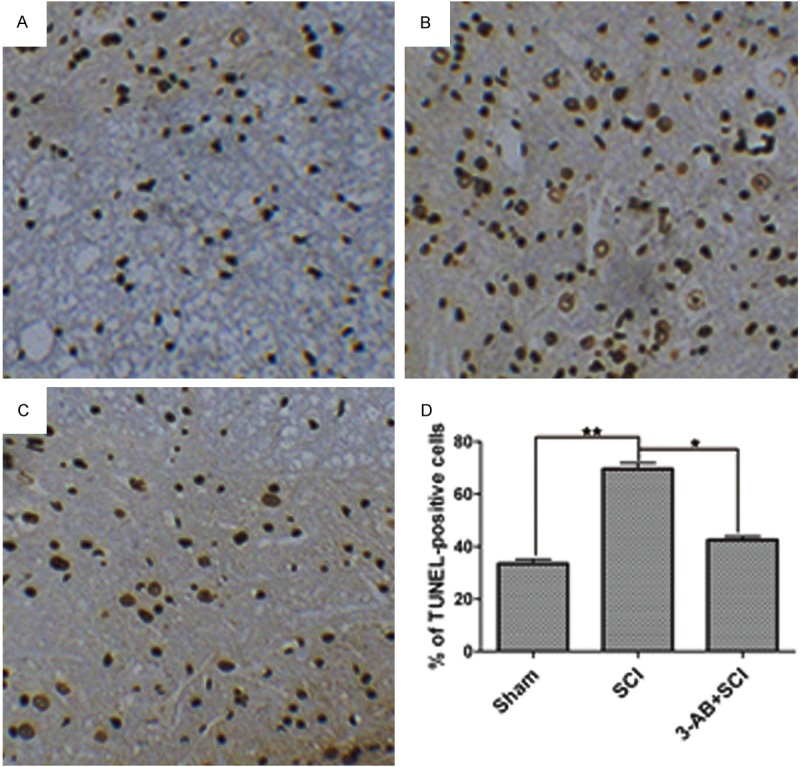
TUNEL assay for apoptosis. A. Sham group in a normal status; B. SCI group showed a significant increase of apoptotic cells; C. 3-AB decreased the apoptotic cells; D. The statistical results. Each column represents mean ± SD; n = 3 per group; *P < 0.05 and **P < 0.01.
Parp-1 expression is promoted by sci, but inhibited by 3-Ab
PARP-1 was the central enzyme participating in DNA repair. As shown in Figure 3A, PARP-1expression was significantly up-regulated after SCI surgery, confirming the over-activation of PARP-1, but down-regulated by 3-AB in mRNA level. The Western blot results also confirmed the similar alteration of PARP-1 expression (Figure 3B). Moreover, the IHC results showed that PARP-1 deposition was more visible (arrows) in SCI group, but became hardly visible after 3-AB treatment (Figure 3C).
Figure 3.
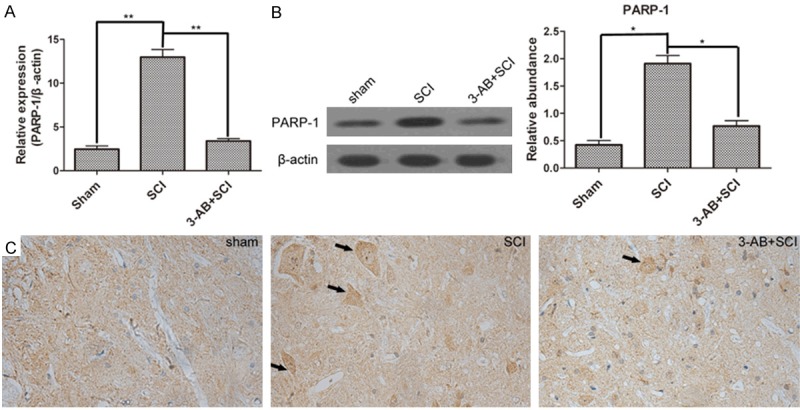
The expression of PARP-1. A. Real-time quantitative PCR results showed that PARP-1 was over-activated by SCI, but inhibited by 3-AB; B. Western blot and corresponding densitometric analysis confirmed the variation in protein level; C. IHC results showed more distribution of PARP-1 in spinal cord tissues of SCI group. Each column represents mean ± SD; n = 3 per group; *P < 0.05 and **P < 0.01; arrows: PARP-1 deposition.
AIF and Bcl-2 demonstrates opposite variations induced by SCI and 3-AB
AIF translocation is the following step after PARP-1 over-activation. Bcl-2 is a gene that inhibits cell apoptosis. First of all, SCI promoted the expression of AIF, but inhibited Bcl-2 activation. In mRNA level, 3-AB significantly decreased the expression of AIF (Figure 4A), possibly mainly due to the inhibition of PARP-1. Correspondingly, 3-AB up-regulated the expression of Bcl-2 (Figure 5A). In protein level, AIF expression was reduced, and Bcl-2 expression was promoted in the presence of 3-AB (Figures 4B and 5B). The IHC results showed widespread AIF in of SCI group (arrows), but AIF was much less after 3-AB addition (Figure 4C). On the contrary, Bcl-2 showed a richer, bigger and darker sedimentation in 3-AB + SCI group (Figure 5C). In conclusion, 3-AB could up-regulate the expression of Bcl-2 and inhibit AIF expression.
Figure 4.
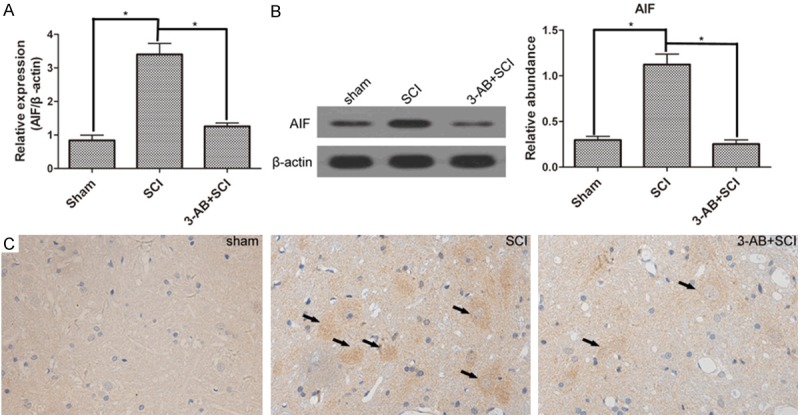
The expression of AIF. AIF expression was promoted by SCI. but inhibited by 3-AB both in mRNA level (A) and protein level (B); (C) IHC results showed more deposition of AIF in SCI group, but less in 3-AB + SCI group. Each column represents mean ± SD; n = 3 per group; *P < 0.05; arrows: AIF deposition.
Figure 5.
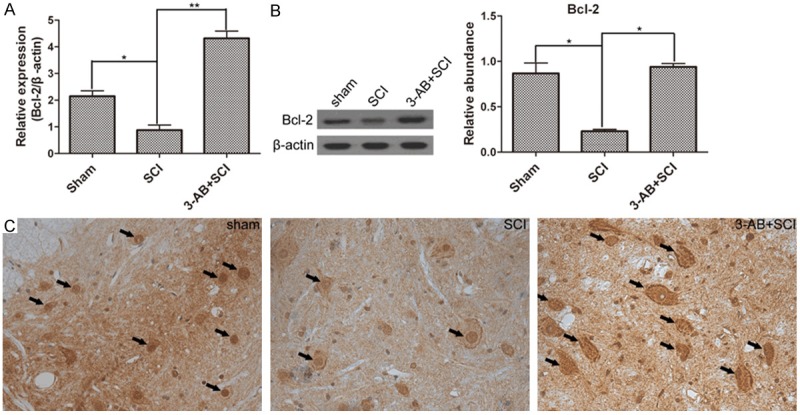
The expression of Bcl-2. Especially, the Bcl-2 expression was decreased by SCI, but significantly increased by 3-AB treatment in mRNA level (A) and protein level (B); (C) IHC results demonstrated much more deposition of Bcl-2 in 3-AB + SCI. Each column represents mean ± SD; n = 3 per group; *P < 0.05 and **P < 0.01; arrows: AIF deposition.
Caspase-3 was not significantly altered in the present study
Cleaved caspase-3 is an active state participating in cell apoptosis. In this study, cleaved caspase-3 expression was not significantly changed neither in the presence nor in the absence of 3-AB, and neither in mRNA level nor in protein level (Figure 6). In other words, caspase-3 was not activated during this process, suggesting that the apoptosis was caspase-independent.
Figure 6.
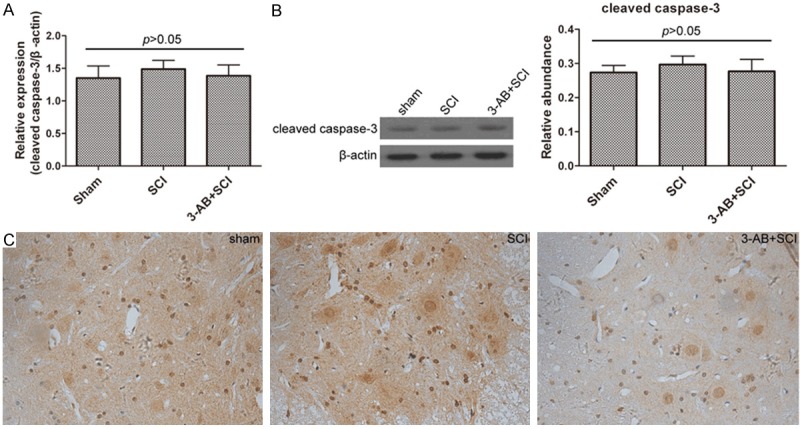
The expression of cleaved caspase-3 was not altered by SCI or 3-AB. A. Real-time quantitative PCR results; B. Western blot and corresponding densitometric analysis; C. IHC results. Each column represents mean ± SD; n = 3 per group.
Discussion
3-AB was an inhibitor of PARP-1, which has been previously reported to be involved in the pathogenesis of oxidative stress [25], cerebral ischemia [26] and Parkinson’s disease [27]. However, the role of 3-AB in SCI has not been reported. Cell apoptosis is a main symptom of SCI, which may lead to more disorders and dysfunctions of CNS [28]. In the present study, the apoptosis was induced by SCI surgery, which also involved the activation of PARP-1 and AIF, and the inactivation of Bcl-2. As reported, the over-activation of PARP-1 could result in promoting cell apoptosis [6]. 3-AB was also employed in this experiment. In conclusion, 3-AB was proved to inhibit the apoptosis induced by SCI surgery, mainly due to the inhibition of PARP-1 expression. Hence, 3-AB contributed to the neuroprotection of spinal cords, which may provide a better therapeutic target for the new drugs of neurodegenerative diseases, including SCI.
PARP-1 is a widespread enzyme in humans mainly participating in DNA damage repair [14]. Specifically, when activated by DNA damage, PARP-1 converts NAD into branched PAR on target proteins. This process may lead three important effects on the CNS, depending on the cell type and the extent of DNA damage: (1) PARP-1 activation and DNA repair; (2) transcription factors activation and inflammation; (3) massive DNA damage (such as SCI) may lead to extensive PARP-1 activation, which promotes neuronal apoptosis via NAD consumption and release of AIF from the mitochondria [29]. Moreover, excessive PARP-1 activation has been proved to be involved in the pathogenesis of CNS disorders such as ischemia-reperfusion injury, excitotoxicity injury, traumatic injury and inflammation [30,31]. The inhibitors of PARP-1, such as 3-AB, could inhibit PARP-1 activation by competitively interfering with the binding of NAD to the active site; in other words, inhibitors act as the competitors of NAD. Accordingly, based on the functions of PARP-1 inhibitors, we designed this study with a desire that 3-AB could attenuate the bad effects of apoptosis induced by SCI, a kind of massive DNA damage frequently leading to PARP-1 over-activation.
Cleaved caspase-3, consisting of two segments, p12 and p17, is an active state of caspase-3. In the classic apoptotic pathway (caspase dependent apoptotic process), PARP-1 is proteolytically cleaved into two segments, namely, p89 and p21, and inactivated by caspase-3, which is widely used as an apoptotic marker [32]. PARP-1 cleavage has weakened the capability of DNA repair, which is thought to prevent the excessive ATP consumption by PARP-1, and thereby providing the sufficient energy required for the apoptotic process.
Whereas, in the present study, the results showed that the apoptosis induced by SCI was a caspase-3-independent process. Actually, this apoptosis is PARP-1-dependent cell death (named Parthanatos) [33]. The main characteristics include fast activation of PARP-1, accumulation of PAR and variation of mitochondrial membrane permeability. Specifically, AIF tends to translocate from mitochondria to nucleus with the over-activation of PARP-1, leading to the chromatin condensation and DNA fragmentation, which ultimately causes cell apoptosis. Bcl-2 is a gene that inhibits cell apoptosis through regulating the permeability of mitochondrial membrane. In conclusion, our results demonstrated that the apoptosis induced by SCI was regulated by the caspase-3-independent (PARP-1-dependent) apoptosis pathway; furthermore, the acknowledged inhibitor of PARP-1, 3-AB, was proved to be effective in inhibiting this apoptosis process. This discovery may contribute to a better neuroprotection of spinal cords, in case of accidents or trauma, and subsequent SCI pathogenesis.
Disclosure of conflict of interest
None.
References
- 1.Herceg Z, Wang ZQ. Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat Res. 2001;477:97–110. doi: 10.1016/s0027-5107(01)00111-7. [DOI] [PubMed] [Google Scholar]
- 2.Jagtap P, Szabó C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 3.Yadav L, Khan S, Shekh K, Jena G. Influence of 3-aminobenzamide, an inhibitor of poly(ADP-ribose) polymerase, in the evaluation of the genotoxicity of doxorubicin, cyclophosphamide and zidovudine in female mice. Mutat Res Genet Toxicol Environ Mutagen. 2014;770:6–15. doi: 10.1016/j.mrgentox.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Beneke S, Diefenbach J, Bürkle A. Poly(ADP-ribosyl) ation inhibitors: promising drug candidates for a wide variety of pathophysiologic conditions. Int J Cancer. 2004;111:813–818. doi: 10.1002/ijc.20342. [DOI] [PubMed] [Google Scholar]
- 5.Chiarugi A. Poly(ADP-ribose) polymerase: killer or conspirator? The ‘suicide hypothesis’ revisited. Trends Pharmacol Sci. 2002;23:122–129. doi: 10.1016/S0165-6147(00)01902-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang SJ, Wang SH, Song ZF, Liu XW, Wang R, Chi ZF. Poly(ADP-ribose) polymerase inhibitor is neuroprotective in epileptic rat via apoptosis-inducing factor and Akt signaling. Neuroreport. 2007;18:1285–1289. doi: 10.1097/WNR.0b013e32826fb3a5. [DOI] [PubMed] [Google Scholar]
- 7.Meli E, Pangallo M, Picca R, Baronti R, Moroni F, Pellegrini-Giampietro DE. Differential role of poly(ADP-ribose) polymerase-1 in apoptotic and necrotic neuronal death induced by mild or intense NMDA exposure in vitro. Mol Cell Neurosci. 2004;25:172–180. doi: 10.1016/j.mcn.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeffler M, Daugas E, Susin Sa, Zamzami N, Métivier D, Nieminen AL, Brothers G, Penninger JM, Kroemer G. Dominant cell death induction by extramitochondrially targeted apoptosis-inducing factor. FASEB J. 2001;15:758–767. doi: 10.1096/fj.00-0388com. [DOI] [PubMed] [Google Scholar]
- 10.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 11.Li GY, Osborne NN. Oxidative-induced apoptosis to an immortalized ganglion cell line is caspase independent but involves the activation of poly(ADP-ribose) polymerase and apoptosis-inducing factor. Brain Res. 2008;1188:35–43. doi: 10.1016/j.brainres.2007.10.073. [DOI] [PubMed] [Google Scholar]
- 12.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci U S A. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szabó C, Dawson VL. Role of poly(ADP-ribose) synthetase in inflammation and ischaemia-reperfusion. Trends Pharmacol Sci. 1998;19:287–298. doi: 10.1016/s0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- 14.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Virág L, Szabó C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 16.Dumont RJ, Okonkwo DO, Verma S, Hurlbert RJ, Boulos PT, Ellegala DB, Dumont AS. Acute spinal cord injury, part I: pathophysiologic mechanisms. Clin Neuropharmacol. 2001;24:254–264. doi: 10.1097/00002826-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Liu D, Huang Y, Jia C, Li Y, Liang F, Fu Q. Administration of Antagomir-223 Inhibits Apoptosis, Promotes Angiogenesis and Functional Recovery in Rats with Spinal Cord Injury. Cell Mol Neurobiol. 2015;35:483–91. doi: 10.1007/s10571-014-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh K, Ikata T, Katoh S, Hamada Y, Nakauchi K, Sano T, Niwa M. Induction and its spread of apoptosis in rat spinal cord after mechanical trauma. Neurosci Lett. 1996;216:9–12. doi: 10.1016/0304-3940(96)12999-2. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Wu F, Kong X, Yang J, Chen H, Deng L, Cheng Y, Ye L, Zhu S, Zhang X. Nerve growth factor improves functional recovery by inhibiting endoplasmic reticulum stress-induced neuronal apoptosis in rats with spinal cord injury. J Transl Med. 2014;12:130. doi: 10.1186/1479-5876-12-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandya KG, Patel MR, Lau-Cam CA. Comparative study of the binding characteristics to and inhibitory potencies towards PARP and in vivo antidiabetogenic potencies of taurine, 3-aminobenzamide and nicotinamide. J Biomed Sci. 2010;17:S16. doi: 10.1186/1423-0127-17-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao YJ, Wang JH, Fu B, Ma MX, Li BX, Huang Q, Yang BF. Effects of 3-aminobenzamide on expressions of poly(ADP ribose) polymerase and apoptosis inducing factor in cardiomyocytes of rats with acute myocardial infarction. Chin Med J. 2009;122:1322–1327. [PubMed] [Google Scholar]
- 22.Czapski GA, Cakala M, Gajkowska B, Strosznajder JB. Poly(ADP-ribose) polymerase-1 inhibition protects the brain against systemic inflammation. Neurochem Int. 2006;49:751–755. doi: 10.1016/j.neuint.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Germain M, Affar EB, D’Amours D, Dixit VM, Salvesen GS, Poirier GG. Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis evidence for involvement of caspase-7. J Biol Chem. 1999;274:28379–28384. doi: 10.1074/jbc.274.40.28379. [DOI] [PubMed] [Google Scholar]
- 24.Czapski GA, Cakala M, Kopczuk D, Strosznajder JB. Effect of poly(ADP-ribose) polymerase inhibitors on oxidative stress evoked hydroxyl radical level and macromolecules oxidation in cell free system of rat brain cortex. Neurosci Lett. 2004;356:45–48. doi: 10.1016/j.neulet.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 25.Cole KK, Perez-Polo JR. Poly(ADP-ribose) polymerase inhibition prevents both apoptotic-like delayed neuronal death and necrosis after H2O2 injury. J Neurochem. 2002;82:19–29. doi: 10.1046/j.1471-4159.2002.00935.x. [DOI] [PubMed] [Google Scholar]
- 26.Moroni F, Meli E, Peruginelli F, Chiarugi A, Cozzi A, Picca R, Romagnoli P, Pellicciari R, Pellegrini-Giampietro D. Poly(ADP-ribose) polymerase inhibitors attenuate necrotic but not apoptotic neuronal death in experimental models of cerebral ischemia. Cell Death Differ. 2001;8:921–932. doi: 10.1038/sj.cdd.4400884. [DOI] [PubMed] [Google Scholar]
- 27.Wu XL, Wang P, Liu YH, Xue YX. Effects of Poly(ADP-ribose) Polymerase Inhibitor 3-Aminobenzamide on Blood-Brain Barrier and Dopaminergic Neurons of Rats with Lipopolysaccharide-Induced Parkinson’s Disease. J Mol Neurosci. 2014;53:1–9. doi: 10.1007/s12031-013-0175-5. [DOI] [PubMed] [Google Scholar]
- 28.Lan WB, Lin JH, Chen XW, Wu CY, Zhong GX, Zhang LQ, Lin WP, Liu WN, Li X, Lin JL. Overexpressing neuroglobin improves functional recovery by inhibiting neuronal apoptosis after spinal cord injury. Brain Res. 2014;1562:100–108. doi: 10.1016/j.brainres.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Kauppinen T, Swanson R. The role of poly (ADP-ribose) polymerase-1 in CNS disease. Neuroscience. 2007;145:1267–1272. doi: 10.1016/j.neuroscience.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 30.Besson VC, Croci N, Boulu RG, Plotkine M, Marchand-Verrecchia C. Deleterious poly(ADP-ribose) polymerase-1 pathway activation in traumatic brain injury in rat. Brain Res. 2003;989:58–66. doi: 10.1016/s0006-8993(03)03362-6. [DOI] [PubMed] [Google Scholar]
- 31.Koh SH, Chang DI, Kim HT, Kim J, Kim MH, Kim KS, Bae I, Kim H, Kim DW, Kim SH. Effect of 3-aminobenzamide, PARP inhibitor, on matrix metalloproteinase-9 level in plasma and brain of ischemic stroke model. Toxicology. 2005;214:131–139. doi: 10.1016/j.tox.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Ménissier-de Murcia J. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis Lesson from an uncleavable mutant. J Biol Chem. 1998;273:33533–33539. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- 33.Kuzhandaivel A, Nistri A, Mladinic M. Kainate-mediated excitotoxicity induces neuronal death in the rat spinal cord in vitro via a PARP-1 dependent cell death pathway (Parthanatos) Cell Mol Neurobiol. 2010;30:1001–1012. doi: 10.1007/s10571-010-9531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


