Abstract
Objective: to observe relationship between chromosome imbalance and taxol resistance in nasopharyngeal carcinoma (NPC). Methods: three taxol-resistant sub-lines were established through repeated exposure of escalating doses of paclitaxel to NPC cell lines (CNE-1, HNE-2 and 5-8F). The change of copy number of chromosomes was investigated by the genome-wide analyses of comparative genomic hybridization (CGH). Gene profiles of both parental and resistant cell lines were determined by cDNA microarray. Cell viability was assayed by colony formation assay. Results: The taxol resistant sub-lines (CNE1/Taxol, HNE2/Taxol and 5-8F/Taxol) developed displayed an average 5~8-fold higher IC50 value than their parental cells. The common losses of chromosome 18, 10q11-qter and gains of chromosome 12, 3q21-qter, 5p13-pter and 20q11-qter were observed by CGH in all of 6 NPC cell lines. A common gain region of chromosome 8q21-qter was identified in taxol resistant sub-lines. 15 genes of 762 transcripts on this chromosome region were consistently up-regulated detected by cDNA microarray in three taxol resistant sub-lines, and functionally clustered into various groups, including genes related to vascular formation vascular formation (ANGPT1), apoptosis (MYC, TOP1MT), cell adhesion and cell cycle (PPP1R16A, SDC2, CA2, ANKRD46), gene regulation (HRSP12, ZNF696, SLC39A4, POP1), metabolism (PYCRL). Inhibition of ANGPT1 expression significantly increased the sensitivity of CNE-1/taxol to paclitaxol. Conclusion: The common gain of chromosome 8q21-qter in taxol resistant sublines predicates that potential candidate genes on this region may contribute to taxol resistant phenotype. ANGPT1 may be associated with taxol resistance of NPC cells.
Keywords: Angiopoietin, chromosome imbalance, drug resistance, nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor that occurrs in the lining of nasopharynx with a multifactorial etiology. It is a rare cancer in the world, but has a high incidence rate in southeast Asians and southern provinces of China [1]. Because NPC is highly sensitive to radiotherapy and chemotherapy, a combination of radio- and chemo-therapy is the most acceptable therapeutic approach. Currently, the rate of five-year survival is approximate 60% after treatments [2]. Although some patients initially respond to chemotherapy, the majority of patients with advanced NPC fail to respond to the treatments because of the development of drug resistance [3,4]. It is therefore necessary to elucidate the mechanism of drug resistance and to develop methods to reverse drug resistance in NPC.
Paclitaxel, a prototypic taxane compound and well-known anti-neoplastic agent, specifically binds to the β-tubulin subunit of microtubulin, which promotes the polymerization of tubulin and disrupts microtubule dynamics. This blocks the cell cycle at G2/M, and results in programmed cell death [5]. Paclitaxel is one of the most active agents used in the clinical treatment of breast-, ovarian-, lung-, bladder-, prostate- and head and neck cancers, and it is currently being used for advanced NPC [6-8]. Although paclitaxel has been shown to prolong patient survival, the frequent occurrence of drug resistance either at the onset or during the course of treatment has rendered the benefit of this drug. Indeed, intrinsic and acquired drug resistances represent major obstacles in the successful treatment of many solid tumors. Molecular investigations on various human malignancies have implicated the regions of genomic aberrations and the changes in gene expressions in relation to drug insensitivity [9-11].
Comparative genomic hybridization (CGH) developed in 1992 has been widely used in cancer research to identify novel regions of genomic amplification and/or deletion [12,13]. CGH copy number profiles may facilitate identification of important new drug resistant genes located at the hotpots of the chromosomal alterations. Some studies have applied CGH to detect chromosomal imbalances specifically involved in the resistance to chemotherapeutic agents such as 5-fluorouracil, vinblastine, doxorubicin, docetaxel and cisplatin [14-18]. In this study, we identified the common region of genomic amplification in NPC paclitaxol-resistant cells by CGH, and then analyzed gene mRNA expression profile in this narrow region by cDNA microarray, and tried to uncover the paclitaxol resistant-related molecular events.
Materials and methods
Materials
Taxol was obtained from Bristol-MyersSquibb (Princeton, New Jersey). Bradford assay kits and chemiluminescent western detection kits were purchased from Bio-Rad (Hercules, California). RNA extraction kits and reverse transcription polymerase chain reaction kits were obtained from Life Technologies (Gaithersburg, Maryland). Other molecular reagents were purchased from Sigma (St Louis, Missouri). Antibodies against ANGPT1, HRSP12, CA2, and PYCRL were from Santa Cruz Biotechnology (Santa Cruz, California). Affymetrix GeneChip® human genome U133 Plus 2.0 array were from Affymetrix Inc. (Santa Clara, CA, USA).
Cell lines and paclitaxel-resistant sub-lines
Three human nasopharyngeal carcinoma cell lines, CNE-1, HNE-2, 5-8F, were used and kindly provided by the Cancer Research Institute of Central South University (Changsha, China). Cells were maintained in RPMI-1640 medium containing 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin in a humidified atmosphere containing 5% CO2 at 37°C. Cells were replated 48 hr before use. The taxol-resistant sub-lines (CNE-1/taxol, HNE-2/taxol, 5-8F/taxol) were established by exposing parental cells to gradually increasing concentrations of taxol in our previous studies, and maintained in the above medium containing 1 nmol/L paclitaxel. The half-inhibition concentration (IC50) was determined using the colony formation assay [19], and the resistance index of drug-resistant variants of cells divided by the IC50 of resistant cells. All cells were subcultured at 5-day intervals.
Comparative genomic hybridization (CGH)
Comparative genomic hybridization was performed as previously described [13]. DNA was extracted from 10×106 cells in monolayer cultures. Briefly, 1 μg of tumor DNA was labeled with Spectrum Green-dUTP using a dedicated nick-translation kit (Vysis, Downers Grove, IL), and 1 μg of normal male DNA with Spectrum Red (Vysis) was used as reference, mixed with 80 μg of Cot-1 DNA (Invitrogen) in 15 μl hybridization buffer (formamide 50%/NaH2PO4 40 mM/SDS 0.1%/dextran sulphate 10%/2X SSC) and hybridized together onto denatured metaphase spreads from normal male lymphocytes (Vysis, Inc.). Images were captured with an epifluorescence Leica DMRB microscope fitted with a Photometrix Cool Snap fx digital camera, and a minimum of 10 metaphases was analyzed with Quips software (Vysis). Based on green-to-red ratios, copy number alterations were classified as losses (ratios ≤0.75), or amplifications (≥1.25).
Gene expression profiling
Total RNA sample was isolated from exponentially growing cells using Trizol™ Reagent (Life Technologies, Carlsbad, CA) and purified on an RNeasy affinity column (Qiagen, Valencia, CA). Gene expression profiling was performed using the Genome U133 Plus 2.0 arrays with methodology as recommended in the Gene Chip Expression Analysis Technical Manual (Affymetrix Inc., Santa Clara, CA). Arrays were scanned on the Affymetrix Gene Chip scanner, and raw data was processed by MAS5 in the Affymetrix Gene Chip Operating Software (GCOS) to generate quantitative signals and qualitative Detection Calls.
Real time RT-PCR
Total RNA was performed as described above, and RNA concentration was determined by UV spectrophotometry. Reverse transcription of 1 μg RNA was performed using an RT-PCR kit (Life Technologies) in a 20 μl reaction. Two microlitres of cDNA from 10× dilution of the RT reaction was used for PCR. The primers were as follows: angiopoietin 1 (ANGPT1, 326 bp) forward primer, 5’-ACTGTGCAGATGTATATCA AGC-3’; reverse primer, 5’-GTGGAATCTGTCATACTGTGAA-3’; β-actin (540 bp) forward primer, 5’-GGACCTGACTGACTACCTC-3’; reverse primer, 5’-TCATACT CCTGCTTGCTG-3’. CA2 (249 bp) forward primer, 5’-GTACGGCAAACACAACG GAC-3’; reverse primer 5’-CTGTAAGTGCCATCCAGGGG-3’; HRSP12 (250 bp) forward primer 5’-GCATGGACCCTTCAAGTGGA-3’; reverse primer 5’-ATTCGG CTGCCTTTGGGTAA-3’; PYCRL (183 bp) forward primer, 5’-CTGTGTGGTCCA GGAAGGG-3’; Reverse 5’-TCGGAGAATGCACACACGAA-3’. Reactions were performed using a Mastercycler gradient PCR machine (Eppendorf). Amplification data was normalized to β-actin, and quantification of gene expression was performed using ΔΔCT calculation, where CT is the threshold cycle; the amount of target gene, normalized to β-actin and relative to the calibrator (control cells, CNE-1), is given as 2-ΔΔCT [20].
Western blot
The cells were lysed in a lysis buffer at 4°C for 30 minutes. Insoluble material was removed by centrifugation. The protein concentration was determined using the Pierce® BCA Protein Assay Kit (Pierce Biotechnology, Rockford, USA). Twenty μg of protein was separated by 12% SDS-PAGE and then transferred to polyvinyl difluoride membranes (Millipore, Bedford, MA) by electroblotting. After blocking with 5% non-fat dry milk, the blots were incubated with primary antibodies (against ANGPT1, HRSP12, CA2, PYCRL and GAPDH), and were developed with alkaline phosphatase-conjugated secondary antibodies using a chemiluminescent substrate. A densitometer was used for quantification of signal on the films.
Treatment with ANGPT1 and assessment of cell viability
Specific siRNA of ANGPT1 was designed and synthesized by Shenggong (Shanghai, China). The efficiency of gene inhibition was verified by transfection assay. The sequence of siRNA was 5’-CUUCUCGACUUGAGAUACAdTdT-3’, 3’-dTdT GAAGAGCUGAA CUCUAUGU-5’. The scramble siRNA was used as control. CNE-1/taxol cells were grown in RPMI media and plated at 2×105 cells/well in 6-well plates. The cells were incubated in medium without antibiotics for 16 hours and then replaced with OPTM 1 medium without serum for 1 hour. A mixture of 200 nM siRNA and Oligofectamine (Invitrogen, Gaithersburg, Maryland) was added to each well for 4 hours, after which RPMI media containing 30% serum were added. Cells were harvested 48 hours later, and total RNA and protein were extracted. Quantitative RT-PCR was used to confirm the ANGPT1 mRNA level (see above), and western blot was used to confirm the ANGPT1 protein level. Cells with ANGPT1-siRNA also were assayed for cell growth inhibition by taxol as our previously described.
Statistical analysis
Data analysis was performed with SPSS software (version 17.0, Chicago, IL). Differences in IC50 values between parental cells and taxol resistant sub-lines were evaluated by a paired nonparametric analysis using the Wilcoxon signed rank test. The overall genomic instability, as suggested from the CGH derived copy number aberrations including gains and losses, in the parental cells and taxol resistant sub-lines were compared by the two-tailed unpaired Student’s t-test. Pearson’s test was employed to assess the relationship between the expression levels of ANGPT1 and the degrees of drug resistance as represented by the IC50 value. The significance level was considered when p-value was less than 0.05.
Results
Conformation of resistance to taxol in the taxol-resistant sub-lines
Three taxol resistant sub-lines derived from the parental sensitive cell lines (CNE-1, HNE-2, 5-8F) were established by stepwise selection in paclitaxel, and were designated as CNE1/Taxol, HNE2/Taxol and 5-8F/Taxol. The effect of taxol on three NPC cell lines and three taxol-resistant sub-lines was examined by colony formation assay [21]. The IC50 value of taxol was 1.33±0.08 nM for CNE-1, 0.92±0.21 nM for HNE-2, 1.58±0.11 nM for 5-8F, 11.24±0.26 nM for CNE-1/taxol, 6.53±0.22 nM for HNE-2/taxol, and 8.85±1.95 nM for 5-8F/taxol, as determined by the growth inhibition curve (Figure 1). The three taxol resistant lines (5-8F/taxol, HNE-2/taxol, CNE-1/taxol) were 8.5, 7.1 and 5.6-fold less sensitive than their parental NPC cells (5-8F, HNE-2, CNE-1) in response to taxol, respectively.
Figure 1.
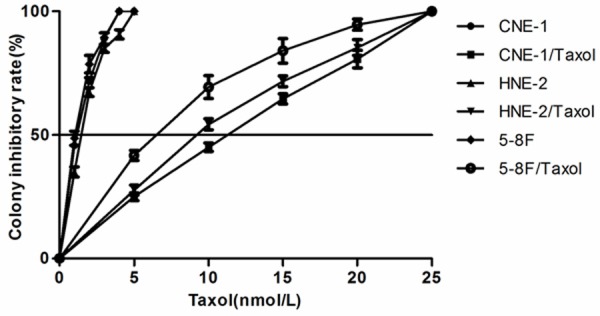
Growth inhibitation curve of both taxol-resistant sublines and their parental cell lines. The cell viability in the presence of paclitaxel was determined by the colony formation.
Metaphase CGH abnormalities of both NPC cells and taxol-resistant cells
High quality imagines of CGH were obtained for each sample as showing in Figure 2. The chromosomal losses and gains were identified by analysis of metaphase CGH in three NPC cell lines and three taxol resistant NPC cell lines. Losses and gains involving a subset of chromosomes were identified in all parental and taxol resistant cell lines (Table 1). The common losses of chromosome 18, 10q11-qter and gains of chromosome 12, 3q21-qter, 5p13-pter and 20q11-qter were observed in all of 6 NPC cell lines. Other observed alterations included losses on subsets of chromosome 2, 4, 5, 8, 19, X and gains on subsets of chromosome 1, 6, 7, 8, 9, 11, 13, 14, 16, 17, 20 in different NPC cell lines. The common gains of chromosomal region 8q21-qter in 3 taxol resistant lines, not in 3 parental cell lines, were observed (Figure 3).
Figure 2.
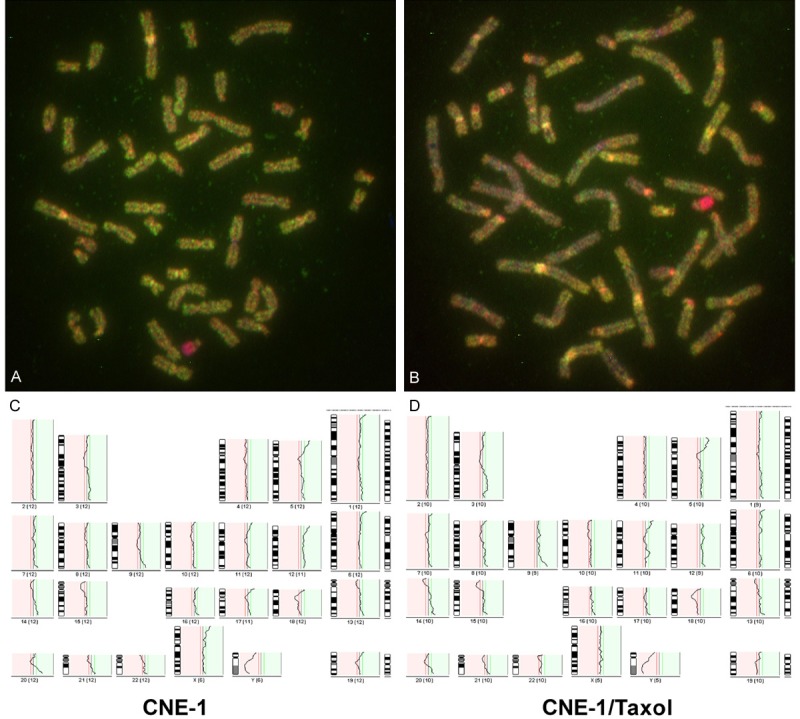
A representative images of CGH analysis. A. A photo of metaphase CGH in CNE-1 cells; B. A photo of metaphase CGH in CNE-1/taxol cells; C. The pattern of chromosome imbalance in CNE-1; D. The pattern of chromosome imbalance in CNE-1/taxol cells.
Table 1.
Genomic imbalances in NPC parental cell lines and corresponding taxol resistant sublines
| Chr. | CNE-1 | HNE-2 | 5-8F | CNE-1/T | HNE-2/T | 5-8F/T |
|---|---|---|---|---|---|---|
| Losses | ||||||
| 2 | - | - | 2p21-qter | - | - | 2p12-qter |
| 4 | 4p14-qter | 4p14-qter | 4p14-qter | 4q22-pter | 4p14-qter | |
| 8 | - | - | 8q22-pter | - | - | - |
| 10 | 10q11-qter | 10q11-qter | 10q11-pter | 10q11-qter | 10q11-qter | 10q11-pter |
| 15 | - | - | 15q21-pter | - | 15 | - |
| 18 | 18 | 18 | 18 | 18 | 18 | 18 |
| 19 | - | - | - | 19q11-qter | - | - |
| x | Xq11-qter | Xq21-qter | Xp11-qter | X | X | Xq11-qter |
| Gains | ||||||
| 1 | 1q24-ter | 1q24-ter | 1q24-qter | 1q24-qter | 1q31-qter | |
| 3 | 3q21-qter | 3q21-qter | 3q21-qter | 3q21-qter | 3q21-qter | 3q21-qter |
| 5 | 5p13-pter | 5p13-pter | 5p13-pter | 5p13-pter | 5p13-pter | 5p13-pter |
| 7 | 6q13-pter | |||||
| 7p12-pter | ||||||
| 8 | 8q21-qter | 8q21-qter | 8q21-qter | |||
| 9 | 9q33-qter | 9q33-qter | 9p21-qter | 9p21-qter | 9q21-qter | |
| 10 | ||||||
| 11 | 11p14-pter | 11p14-pter | 11q22-qter | 11p14-pter | ||
| 12 | 12 | 12 | 12 | 12 | 12 | 12 |
| 13 | 13q21-qter | |||||
| 14 | 14q13-qter | 14q13-qter | 14q13-qter | |||
| 16 | 16 | 16p12-pter | ||||
| 17 | 17q21-qter | |||||
| 20 | 20q11-qter | 20q11-qter | 20q11-qter | 20q11-qter | 20q11-qter | 20q11-qter |
Figure 3.
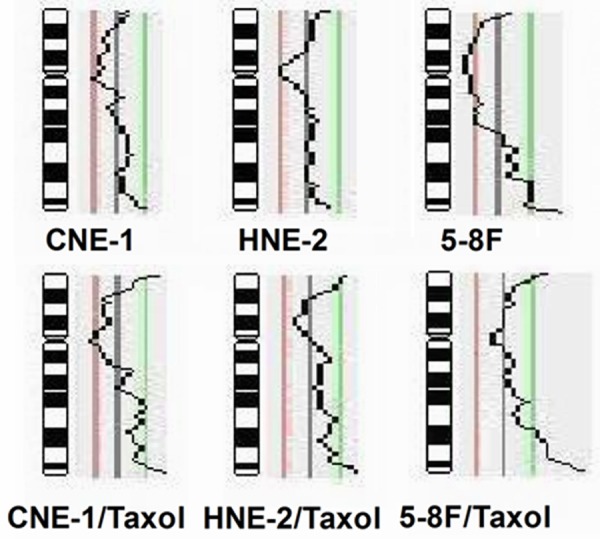
Gains and losses of chromosome 8 identified by CGH analyses in taxol resistant sublines (CNE-1/Taxol, HNE-2/Taxol, 5-8F/Taxol) and their parental cell lines (CNE-1, HNE-2, and 5-8F). It shows a common gain of 8q21-qter amplicon in three taxol resistant sublines.
Gene expression profile on chromosomal regional 8q21-qter
It has been known that a relative copy number increase of genes is commonly associated with gene over-expression [22]. For this purpose, expression profiling was carried out for three NPC cell lines and three taxol resistant cell lines using Affymetrix platform. Seven hundreds and sixty-two transcripts of HU 133 Av2 array are allocated on chromosomal region 8q21-qter. The levels of mRNA expressions of targeted 762 transcripts in all of samples were analyzed by using DNA-chip data analyzer software. The transcripts were filtered as a set, using the following criteria: the standard deviation mean was between 0.1 and 10, and present call in arrays was ≥40%. Then the relative mRNA expression levels of genes were further screened for fold changes ≥1.5, P-value ≤0.05; and signal intensity ≥40. The clustering analysis showed that 15 transcripts were consistently up-regulated, but none of transcripts consistently down-regulated, in all of 3 taxol resistant cell lines (Table 2). These differential genes were functionally clustered into vascular formation (ANGPT1), apoptosis (MYC, TOP1MT), cell adhesion and cell cycle (PPP1R16A, SDC2, CA2, ANKRD46), gene regulation (HRSP12, ZNF696, SLC39A4, POP1), metabolism (PYCRL) and three unknown genes.
Table 2.
Potential transcripts on chromosome 8q21-qter related to taxol-resistant in nasopharyngeal carcinoma
| ID1 | Gene title | Name | Fold change2 | Functions | ||
|---|---|---|---|---|---|---|
|
| ||||||
| A | B | C | ||||
| NM_002467 | v-myc myelocytomatosis viral oncogene homolog | MYC | 1.85 | 1.63 | 1.59 | Apoptosis, G1-S cell cycle,Wnt signaling |
| AW592604 | Topoisomerase I | TOP1MT | 1.81 | 1.92 | 1.54 | Inhibitor of apoptosis |
| NM_001146 | Angiopoietin 1 | ANGPT1 | 7.82 | 1.65 | 4.48 | Angiogenesis, inhibitor of apoptosis |
| AI742931 | Protein-phosphatase 1, subunit 16A | PPP1R16A | 1.95 | 1.53 | 1.63 | control of microtubule dynamics during mitosis |
| U79297 | Ankyrin repeat domain 46 | ANKRD46 | 1.73 | 1.48 | 4.71 | Protein interaction,cell cycle, cancer progression |
| AI380298 | Syndecan 2 | SDC2 | 2.35 | 1.97 | 1.71 | cell proliferation, migration |
| M36532 | Carbonic anhydrase II | CA2 | 4.57 | 1.58 | 3.78 | Carbon dioxide transport cell proliferation |
| NM_030895 | Zinc finger protein 696 | ZNF696 | 2.18 | 1.85 | 1.50 | Zinc ion binding, gene regulation |
| NM_017767 | Solute carrier family 39, member 4 | SLC39A4 | 1.81 | 4.35 | 1.86 | Zinc ion binding, gene regulation |
| D31765 | Processing of precursor 1 | POP1 | 4.18 | 1.51 | 1.67 | Ribonuclease MRP complex |
| N54448 | Heat responsive protein 12 | HRSP12 | 5.09 | 1.96 | 2.02 | Regulation translational termination |
| NM_023078 | Pyrroline-carboxylate reductase like | PYCRL | 2.85 | 2.79 | 4.40 | Proline biosynthetic process |
| AU150691 | Hypothetical protein | ? | 2.07 | 1.68 | 2.38 | Unknown |
| H78106 | Hypothetical protein | ? | 1.64 | 2.23 | 3.41 | Unknown |
| BF445343 | NudC domain 1 | NUDCD1 | 2.13 | 1.90 | 1.74 | Unknown |
an identification number in public gene database.
fold change of mRNA expression level expressed as taxol-resistant cells divided by parental cells.
A: CNE-1/Taxol divided by CNE-1; B: 5-8/F/Taxol divided by 5-8/F; C: HNE-2/Taxol divided by HNE-2.
Confirmation of microarray-detected alteration in above transcripts
Four of fifteen genes screened by above methods were selected for further conformation by real time RT-PCR, and the expression levels of these genes in arrays were a greater than two-fold increase in at least two sub-lines than their parental cell lines. The results showed that the mRNA expression levels of the selected transcripts ANGPT1, HRSP12, CA2 and PYCRL were significantly higher in CNE-1/taxol, HNE-2/taxol, and 5-8F/taxol cells than their parental cells, which strongly confirmed the accuracy of the Affymetrix microarray platform (Figure 4A). In addition, we investigated the protein abundances of the corresponding genes by Western blot, and the results demonstrated that the protein abundance of ANGPT1 was consistently increased in three taxol resistant NPC sub-lines, and the average intensity value in resistant lines was 2.2-fold greater than that in parental lines (Figure 4A). However, the protein abundances of CA2, HRSP12, and PYCRL were variable in the resistant sub-lines (Figure 4B).
Figure 4.
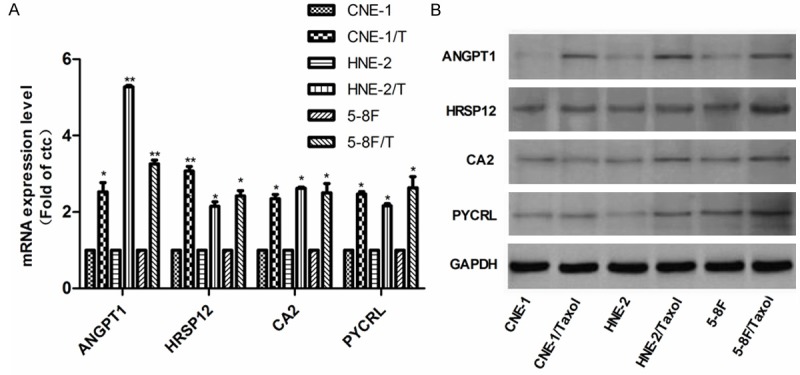
A. Validation of gene expression level from cDNA microarray. The mRNA expression level of ANGPT1, HRSP12, CA2 and PCYRL was determined by real time RT-PCR in three NPC lines and three taxol resistant cell lines. B. The protein expression levels of ANGPT1, HRSP12, CA2 and PYCRL in three NPC lines and three taxol resistant cell lines. *P<0.05, **P<0.01.
Relationship between ANGPT1 expression and drug sensitivity
A consistent increased ANGPT1 expression in taxol-resistant NPC cells suggested that ANGPT1 might play a critical role in taxol resistance of cancer. To understand the relationship between the ANGPT1 expression and taxol-resistant phenotype, a linear correlation analysis was performed between ANGPT1 mRNA expression level and the sensitivity of taxol on each of the acquired taxol-resistant NPC cell lines and its parental cell line. The ANGPT1 level was significantly and positively correlated to the IC50 value in taxol-resistant cell lines. The co-efficiency was R=0.7251 between mRNA level and IC50 value (P<05).
RNAi of ANGPT1 resulted in an increased sensitivity of taxol on taxol-resistant cells
To further understand the role of these differential genes in taxol-resistance of NPC cells, we knocked down the expression of ANGPT1 by RNA interference, and the cell growth inhibition was determined by the colony formation assay. A 2.3-fold decrease in mRNA level and 2.1-fold decrease in protein abundnce were detected in CNE-1/Taxol cells transiently transfected with ANGPT1 siRNA (Figure 5A and 5B). The sensitivity of taxol on CNE-1/Taxol with ANGPT1 siRNA was significantly higher than that on the control of CNE-1/Taxol cells. The IC50 value to taxol was changed from 11.24±0.26 nM to 7.08±0.31 nM after CNE-1/Taxol cells were transfected with ANGPT1 siRNA (Figure 5C).
Figure 5.
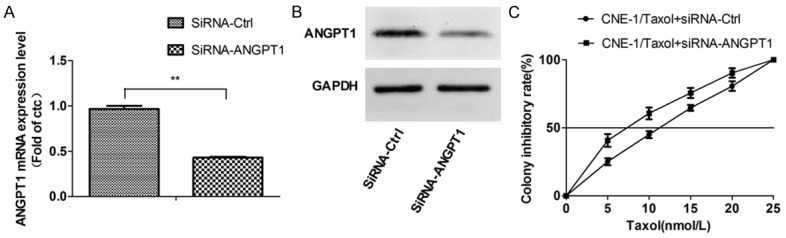
Inhibition of ANGPT1 by RNA interference resulted in an increased sensitivity of paclitaxel in CNE-1/taxol cells. A. A change of ANGPT1 mRNA level was determined by real time RT-PCR. B. Protein expression was detected by western blot; C. The growth inhibition curve was assayed by the colony formation assay. **P<0.01.
Discussion
Chromosome re-assortments, catalyzed by cancer- and cell line-specific aneuploidy, has been proposed for the phenotype of drug resistance [23]. Specific karyotypic alterations are sufficient for drug resistance via new transcriptomes of cooperative genes, independent of gene mutation [24]. Cancer cells differ from normal cells in clonal and non-clonal karyotypic alterations [25], and they acquire new karyotypic alterations and/or they lose old ones, such as common gain of 7q21 and common loss of 10q [16]. In this study, the chromosomal imbalance profiles of NPC identified by metaphase-based CGH were complex and characterized by rich gains and low-level losses that affected almost all chromosomes except chromosome 21 and 22. The common losses of chromosome 18, 10q11-qter and gains of chromosome 12, 3q21-qter, 5p13-pter, 20q11-qter were observed in all of 6 NPC cell lines. However, there was no gene related to NPC pathogenesis uncovered in these regions till today, and it deserves future study. Interestingly, a common gain of chromosome segment in three acquired taxol-resistant NPC cell lines was found on 8q21-qter as compared with their parental cell lines. It indicates that this specific karyotype might represent a common mechanism of acquired taxol resistance in NPC cells.
It has been reported that frequent gains of chromosome 8q were detected by array-based CGH in taxane-resistant breast cancer, which predicted poor prognosis of patients to taxane-based chemotherapy [26]. Genomic amplification at 8q24.22-q24.23 was also identified in carboplatin-resistant ovarian carcinoma [27]. Bergamaschi, et al. have reported that the gain on chromosome 8q is associated with higher proliferation rate of cancer cells, and this association is partly explained by increased expression of c-myc gene located on 8q24. However, only fewer of tumors with c-myc amplification had increased expression in breast cancer [27]. Here, our data demonstrates that a consensus gain is on chromosome 8q21-qter in three taxol-resistant NPC cell lines, and it is consistent with findings of above reports in ovarian and breast cancer. Following these data, we checked gene mRNA expression level of MYC from the data of cDNA microarray, and all three taxol-resistant NPC cell lines had increased expression of MYC as compared with their parental cell lines. It suggests that MYC may be a target in the chromosome 8q21-qter amplicon in NPC, which is associated with taxol-resistance.
It has been known that the copy number imbalance is in relation to gene expression level, and the relative copy number increases are commonly associated with over-expression whereas the copy number decreases are often accompanied with under-expression [21]. In this study we combined two separate analysis methods, parallel CGH analysis that detects genomic alterations with cDNA expression array to provide information at the transcriptional level, to address genomic imbalance and concurrent expression alterations associated with acquired taxol resistance in NPC cells. Gain of 8q21-qter was identified as commonly acquired aberrations by CGH in CNE-1/taxol, HNE-2/taxol and 5-8F/taxol cell lines. cDNA microarray analysis of 762 transcripts allocated on 8q21-qter demonstrated that 15 of 762 transcripts were consistently up-regulated in a setting criterion (fold change ≥1.5) in all three taxol-resistance NPC cell lines, involving in angiogenesis (ANGPT1), apoptosis (MYC, TOP1MT), cell adhesion and cell cycle (PPP1R16A , SDC2, CA2, ANKRD46), gene regulation (HRSP12, ZNF696, SLC39A4, POP1), metabolism (PYCRL) and three unknown genes. The common gained 8q21-qter interval paralleled with up-regulation of these genes might play important role in the taxol resistance of NPC.
Regional activation of chromosome 7q 21.11-13 has also been reported to be related paclitaxel resistance in ovarian cancer, and MDR1 gene on this region is a well-recognized multi-drug resistant gene [28]. However, our data does not show the change of copy number on this chromosome region, which is consistent with our previous report that the expression level MDR1 gene was not changed in NPC taxol resistant cell sub-lines [21]. Here, we validated the mRNA expression changes of 4 (ANGPT1, CA2, HRSP12, PYCRL) of 15 genes on chromosome 8q21-qter in both parental and taxol resistant NPC sub-lines by real time RT-PCR, which was consistent with cDNA microarray results. However, among four genes, the consistent change of protein abundance with mRNA expression was detected only in ANGPT1 by Western blot, and the protein abundance change of the other three genes was variable. The other eleven genes were not further conformed because their mRNA expression levels were less than 2-fold changes. Therefore, ANGPT1 might be valuable for further study.
ANGPT1 is an activating glycoprortein ligand, and acts through the endothelial receptor tyrosine kinase Tie 2 not only to regulate vessel maturation in angiogenesis, but also to inhibit apoptosis and promote cancer progression [29,30]. It has a higher expression in many malignancies, such as ovarian cancer [31], hepatocellullar cancer, oral cancer, esophageal cancer, head neck cancer, etc [32-34]. Higher expression of ANGPT1 is associated with advanced clinical stages of tumors in several reports [32-34]. Although its expression is considered to be restricted to vascular endothelial cells and hematopoietic progenitors, several studies shows that ANGPT1 was expressed in tumor cells [35]. Liu’s report shows that the RNA interference of ANGPT1 suppresses the angiogenesis and the growth of esophageal cancer [36]. Several targeting drugs of ANGPT1-Tie2 pathway were used for treatment of malignancies in clinical trials, such as AMG-386, CVX-060, CEP-11981, etc [37]. In this study, our data showed that ANGPT1 expressed in NPC cells, and a significant increased expression was detected in taxol-resistant cell lines as compared with their parental cells. A significant correlation of ANGPT1 expression level with sensitivity of taxol was also demonstrated. RNA interference of ANGPT1 significantly in-creased the sensitivity of paclitaxel on taxol-resistant NPC cells. It indicates that ANGPT1 may be associated with taxol resistance of NPC cells. It needs further studies for elucidating the role and the underlying mechanism.
Conclusion
The common gain of chromosome 8q21-qter in taxol resistant sub-lines predicates that the potential candidate genes on this region may contribute to taxol resistant phenotype. ANGPT1 may be associated with taxol resistance of NPC cells.
Acknowledgements
This work was supported by NSF of Hunan province (12JJ2053) and Hunan province science and technology plan projects (2013FJ3039).
Disclosure of conflict of interest
None.
References
- 1.Huang DP. Epidemiology of nasopharyngeal carcinoma. Ear Nose Throat J. 1990;69:222–225. [PubMed] [Google Scholar]
- 2.Ou SH, Zell JA, Ziogas A, Anton-Culver H. Epidemiology of nasopharyngeal carcinoma in the United States: improved survival of Chinese patients within the keratinizing squamous cell carcinoma histology. Ann Oncol. 2007;18:29–35. doi: 10.1093/annonc/mdl320. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Masters JR, Wong YC, Lo AK, Tsao SW. Mechanism of differential sensitivity to cisplatin in nasopharyngeal carcinoma cells. Anticancer Res. 2001;21:403–408. [PubMed] [Google Scholar]
- 4.Cheung HW, Jin DY, Ling MT, Wong YC, Wang Q, Tsao SW, Wang X. Mitotic arrest deficient 2 expression induces chemosensitization to a DNA-damaging agent, cisplatin, in nasopharyngeal carcinoma cells. Cancer Res. 2005;65:1450–1458. doi: 10.1158/0008-5472.CAN-04-0567. [DOI] [PubMed] [Google Scholar]
- 5.Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–80. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- 6.Tan EH, Khoo KS, Wee J, Fong KW, Lee KS, Lee KM, Chua ET, Tan T, Khoo-Tan HS, Yang TL, Au E, Tao M. Phase II trial of a paclitaxel and carboplatin combination in Asian patients with metastatic nasopharyngeal carcinoma. Ann Oncol. 1999;10:235–237. doi: 10.1023/a:1008390929826. [DOI] [PubMed] [Google Scholar]
- 7.Leong SS, Wee J, Rajan S, Toh CK, Lim WT, Hee SW, Tay MH, Poon D, Tan EH. Triplet combination of gemcitabine, paclitaxel, and carboplatin followed by maintenance 5-fluorouracil and folinic acid in patients with metastatic nasopharyngeal carcinoma. Cancer. 2008;113:1332–1337. doi: 10.1002/cncr.23687. [DOI] [PubMed] [Google Scholar]
- 8.Hussain M, Gadgeel S, Kucuk O, Du W, Salwen W, Ensley J. Paclitaxel, cisplatin, and 5-fluorouracil for patients with advanced or recurrent squamous cell carcinoma of the head and neck. Cancer. 1999;86:2364–2369. [PubMed] [Google Scholar]
- 9.Osterberg L, Levan K, Partheen K, Delle U, Olsson B, Sundfeldt K, Horvath G. Specific copy number alterations associated with docetaxel/carboplatin response in ovarian carcinomas. Anticancer Res. 2010;30:4451–4458. [PubMed] [Google Scholar]
- 10.Huanchun Y, Shulan Z, Jing L. Genetic imbalance related to cisplatin-based chemoresistance in epithelial ovarian cancer. Eur J Gynaecol Oncol. 2009;30:181–185. [PubMed] [Google Scholar]
- 11.Wang J, Tai LS, Tzang CH, Fong WF, Guan XY, Yang M. 1p31, 7q21 and 18q21 chromosomal aberrations and candidate genes in acquired vinblastine resistance of human cervical carcinoma KB cells. Oncol Rep. 2008;19:1155–1164. [PubMed] [Google Scholar]
- 12.Kallioniemi A. CGH microarrays and cancer. Curr Opin Biotechnol. 2008;19:36–40. doi: 10.1016/j.copbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 14.Bayani J, Squire JA. Comparative genomic hybridization. Curr Protoc Cell Biol. 2005;Chapter 22:Unit 22.6. doi: 10.1002/0471143030.cb2206s25. [DOI] [PubMed] [Google Scholar]
- 15.Wilson C, Yang J, Strefford JC, Summersgill B, Young BD, Shipley J, Oliver T, Lu YJ. Overexpression of genes on 16q associated with cisplatin resistance of testicular germ cell tumor cell lines. Genes Chromosomes Cancer. 2005;43:211–216. doi: 10.1002/gcc.20173. [DOI] [PubMed] [Google Scholar]
- 16.McDonald SL, Stevenson DA, Moir SE, Hutcheon AW, Haites NE, Heys SD, Schofield AC. Genomic changes identified by comparative genomic hybridisation in docetaxel-resistant breast cancer cell lines. Eur J Cancer. 2005;41:1086–1094. doi: 10.1016/j.ejca.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Pang E, Hu Y, Chan KY, Lai PB, Squire JA, Macgregor PF, Beheshti B, Albert M, Leung TW, Wong N. Karyotypic imbalances and differential gene expressions in the acquired doxorubicin resistance of hepatocellular carcinoma cells. Lab Invest. 2005;85:664–674. doi: 10.1038/labinvest.3700254. [DOI] [PubMed] [Google Scholar]
- 18.Rooney PH, Stevenson DA, Marsh S, Johnston PG, Haites NE, Cassidy J, McLeod HL. Comparative genomic hybridization analysis of chromosomal alterations induced by the development of resistance to thymidylate synthase inhibitors. Cancer Res. 1998;58:5042–5045. [PubMed] [Google Scholar]
- 19.Li W, Tan G, Ma Y, Li H, He G. Inhibition of alpha folate receptor resulting in a reversal of taxol resistance in nasopharyngeal carcinoma. Otolaryngol Head Neck Surg. 2012;146:250–258. doi: 10.1177/0194599811426260. [DOI] [PubMed] [Google Scholar]
- 20.Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Peng X, Li W, Tan G. Reversal of taxol resistance by cisplatin in nasopharyngeal carcinoma by upregulating thromspondin-1 expression. Anticancer Drugs. 2010;21:381–388. doi: 10.1097/CAD.0b013e3283363980. [DOI] [PubMed] [Google Scholar]
- 22.Hashemi J, Worrall C, Vasilcanu D, Fryknas M, Sulaiman L, Karimi M, Weng WH, Lui WO, Rudduck C, Axelson M, Jernberg-Wiklund H, Girnita L. Molecular characterization of acquired tolerance of tumor cells to picropodophyllin (PPP) PLoS One. 2011;6:e14757. doi: 10.1371/journal.pone.0014757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duesberg P, Stindl R, Hehlmann R. Origin of multidrug resistance in cells with and without multidrug resistance genes: chromosome reassortments catalyzed by aneuploidy. Proc Natl Acad Sci U S A. 2001;98:11283–11288. doi: 10.1073/pnas.201398998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duesberg P, Li R, Sachs R, Fabarius A, Upender MB, Hehlmann R. Cancer drug resistance: the central role of the karyotype. Drug Resist Updat. 2007;10:51–58. doi: 10.1016/j.drup.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Duesberg P, Li R, Fabarius A, Hehlmann R. The chromosomal basis of cancer. Cell Oncol. 2005;27:293–318. doi: 10.1155/2005/951598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han S, Park K, Shin E, Kim HJ, Kim JY, Kim JY, Gwak G. Genomic change of chromosome 8 predicts the response to taxane-based neoadjuvant chemotherapy in node-positive breast cancer. Oncol Rep. 2010;24:121–128. [PubMed] [Google Scholar]
- 27.Osterberg L, Levan K, Partheen K, Staaf J, Sundfeldt K, Horvath G. High-resolution genomic profiling of carboplatin resistance in early-stage epithelial ovarian carcinoma. Cytogenet Genome Res. 2009;125:8–18. doi: 10.1159/000218744. [DOI] [PubMed] [Google Scholar]
- 28.Wang YC, Juric D, Francisco B, Yu RX, Duran GE, Chen GK, Chen X, Sikic BI. Regional activation of chromosomal arm 7q with and without gene amplification in taxane-selected human ovarian cancer cell lines. Genes Chromosomes Cancer. 2006;45:365–374. doi: 10.1002/gcc.20300. [DOI] [PubMed] [Google Scholar]
- 29.Augustin HG, Koh GY, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat Rev Mol Cell Biol. 2009;10:165–177. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 30.Singh H, Tahir TA, Alawo DO, Issa E, Brindle NP. Molecular control of angiopoietin signalling. Biochem Soc Trans. 2011;39:1592–1596. doi: 10.1042/BST20110699. [DOI] [PubMed] [Google Scholar]
- 31.Brunckhorst MK, Xu Y, Lu R, Yu Q. Angiopoietins promote ovarian cancer progression by establishing a procancer microenvironment. Am J Pathol. 2014;184:2285–2296. doi: 10.1016/j.ajpath.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chien CY, Su CY, Chuang HC, Fang FM, Huang HY, Chen CM, Chen CH, Huang CC. Angiopoietin-1 and -2 expression in recurrent squamous cell carcinoma of the oral cavity. J Surg Oncol. 2008;97:273–277. doi: 10.1002/jso.20930. [DOI] [PubMed] [Google Scholar]
- 33.Zheng MH, Veelken F, Labbe D, Bloch W, Michel O. Expression of angiopoietin-1 and 2 in squamous cell carcinoma of the head and neck areas and normal mucosa. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2005;40:371–375. [PubMed] [Google Scholar]
- 34.Torimura T, Ueno T, Kin M, Harada R, Taniguchi E, Nakamura T, Sakata R, Hashimoto O, Sakamoto M, Kumashiro R, Sata M, Nakashima O. Overexpression of angiopoietin-1 and angiopoietin-2 in hepatocellular carcinoma. J Hepatol. 2004;40:799–807. doi: 10.1016/j.jhep.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 35.Mitsutake N, Namba H, Takahara K, Ishigaki K, Ishigaki J, Ayabe H, Yamashita S. Tie-2 and angiopoietin-1 expression in human thyroid tumors. Thyroid. 2002;12:95–99. doi: 10.1089/105072502753522310. [DOI] [PubMed] [Google Scholar]
- 36.Liu XH, Bai CG, Yuan Y, Gong DJ, Huang SD. Angiopoietin-1 targeted RNA interference suppresses angiogenesis and tumor growth of esophageal cancer. World J Gastroenterol. 2008;14:1575–1581. doi: 10.3748/wjg.14.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H, Bhat A, Woodnutt G, Lappe R. Targeting the ANGPT-TIE2 pathway in malignancy. Nat Rev Cancer. 2010;10:575–585. doi: 10.1038/nrc2894. [DOI] [PubMed] [Google Scholar]


