Abstract
To verify c-Myc can regulate the expression of lncRNA H19 directly in non-small cell lung cancer (NSCLC) and clarify the molecular mechanism on how lncRNA H19 promote the cell cycle progression of NSCLC. The mRNA levels of lncRNA H19 in NSCLC tissues and cells, the adjacent tissues and normal cells were determined by RT-PCR. The expression change of lncRNA H19 in NSCLC cells after transfection with pcDNA3.1-c-Myc or c-Myc-siRNA was determined by RT-PCR, respectively. Targeted role of c-Myc on the promoter of H19 was studied by luciferase reporter assay. Chromosome immune coprecipitation (ChIP) was used to confirm the relationship between c-Myc and H19. MiRNAs that have base-pairing with H19 was predicted by online software. The relationship between H19 and miR-107 was determined by disturbing and overexpressing the expression of H19. The influence of the changes of H19 and miR-107 on cell cycle progression was determined by flow cytometry. The mRNA levels of lncRNA H19 in NSCLC tissues and cells were significantly higher than the adjacent tissues and normal cells, respectively. The expression of H19 increased or decreased accordingly with the overexpression and knockdown of c-Myc. The activity of the promoter of H19 was strengthened by c-Myc. While the expression of miR-107 increased or decreased with the overexpression and knockdown of H19, respectively. The number of cells in G2/M stage decreased significantly with the knockdown of H19 and miR-107 compared with the control group. Our study demonstrates that lncRNA H19, which is induced by c-Myc, is up-regulated in NSCLC. H19 influences the mitotic progression of NSCLC cell lines.
Keywords: c-Myc, LncRNA H19, MiR-107, NSCLC, cell cycle
Introduction
Non-small-cell lung cancer (NSCLC) is the leading cause of cancer death worldwide [1]. Patients with metastatic non-small-cell lung cancer have a substantial symptom burden and have a low survival rate following standard surgical treatment [2]. Previous studies showed the accumulated genomic damage can promote the progression of NSCLC [3]. However, the pathophysiological mechanism contributing to NSCLC is still largely unknown. Therefore, it is of urgent importance to understand the roles of novel molecules involved in this process.
Besides protein-coding mRNAs, eukaryotic transcriptomes include many long non-protein-coding RNAs (lncRNAs) of unknown function that are transcribed away from protein-coding loci [4]. Long non-coding RNAs (lncRNAs) are emerging as new players in the cancer paradigm demonstrating potential roles in both oncogenic and tumor suppressive pathways [5]. The human genome is replete with lncRNA, many of which are transcribed and likely to have a functional role [6]. In liver cancer (HULC), it is highly up-regulated and plays an important role in tumorigenesis [7]. Small noncoding microRNAs (miRNAs) can contribute to cancer development and progression and are differentially expressed in normal tissues and cancers. Previous studies showed miRNAs can target a number of protein-coding genes, but few report whether miRNAs/lncRNAs can also target lncRNAs/miRNAs. In recent years, a competitive RNA (ceRNA) hypothesis has been proposed and several studies have suggested the interaction between lncRNA and miRNA in cancer [8,9]. LncRNAs can act as “miRNA sponge” and sequester miRNAs to inactivate these small regulatory RNAs.
LncRNA H19 is essential for human tumor growth and is up-regulated in hypoxic stress and in some tumors [10]. Zhuang et al. reported the lncRNA H19 was upregulated and play important roles in gastric cancer tumorigenesis [11]. Up-regulated lncRNA H19 contributes to the proliferation of gastric cancer cells [12]. Luo et al. reported lncRNA H19 can increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression [13]. MicroRNA-107 (miR-107) has been demonstrated to regulate proliferation and apoptosis in many types of cancers. In gastric cancer, it can regulates tumor invasion and metastasis by targeting DICER1 [14]. In head and neck squamous cell carcinoma, microRNA-107 can function as a candidate tumor-suppressor gene by downregulating the expression of protein kinase C [15]. miRNA expression is controlled by cell-cycle-dependent transcription factors. c-myc is an inducible gene that is regulated by specific growth signals in a cell-cycle-dependent manner. It is the prototype for oncogene activation by chromosomal translocation. In contrast to the tightly regulated expression of c-myc in normal cells, c-myc is frequently deregulated in human cancers [16]. Constitutive c-myc expression suppresses cell cycle arrest, promotes entry into S phase, and results in the growth factor-independent expression of ornithine decarboxylase [17].
In this study, we aimed to study the influence of lncRNA H19 on the cell cycle progression of NSCLC and the related molecular mechanism.
Material and methods
Cell culture
Tumor tissues and adjacent tissues were obtained from 30 patients with NSCLC. NSCLC cell lines A549, L78, H460, and normal cell line 16HBE were obtained from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. The cells were routinely cultured in RPMI-1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin in a humidified cell incubator with an atmosphere of 5% CO2 at 37°C. Cells growing at an exponential rate were used for the experiments.
RNA isolation and quantitative RT-PCR
To quantitatively determine the mRNA levels of lncRNA H19 in tumor tissues, adjacent tissues, NSCLC cell lines A549, L78, H460, and normal cell line 16HBE cell, qRT-PCR was used. The cells were seeded in a 6-well plate at concentration of 1×105 cells per well and incubated in DMEM.
For RNA isolation, total RNA was extracted and isolated from tissue samples or cell lines using either the mirVana miRNA isolation kit (Ambion, Austin, TX) or the TRIzol method. Trizol of 1 mL was added and the solution was mixed homogeneously for 10 min. The mixture was then transferred into Eppendorf tubes (EP, 1.5 mL) with 200 μL chloroform. After 15 min shake, the EP tubes were centrifuged at 4°C for 15 min (12000×g). The supernate was transferred into other EP tubes and mixed with isopyknic isopropanol for 15 s. The centrifugation (4°C, 10 min, 12000×g) was carried out again and the supernate was discarded. The precipitate was washed by 75% ethonal twice and dissolved into 30 μL diethypyrocarbonate (DEPC) after dried to obtain RNA stock solution. After isolation, the concentration of RNA was assessed using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, Delaware, USA) and the RAN solution was stored at -80°C for further use.
For qRT-PCR, genes were amplified by specific oligonucleotide primer, and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as an endogenous control. The detection and quantification contained the following steps: first, reverse transcription was performed at 55°C for 30 min, initial activation for 15 min at 95°C, next 40 cycles of denaturation were conducted at 94°C for 15 s, then annealing for 30 s at 55°C, extension for 30 s at 72°C. The expression level was normalized using U6 small nuclear RNA by the 2-ΔCt method. The ΔCt values were normalized to GAPDH level.
Transfection
RNA interference (RNAi) is an evolutionarily conserved surveillance mechanism that responds to double-stranded RNA by sequence-specific silencing of homologous genes. To investigate the role of c-Myc on the expression of lncRNA H19, we used a RNAi-based strategy (c-Myc-siRNA) to specifically silence c-myc expression and c-myc -overexpression lentiviral vector (pcDNA3.1-c-Myc) (Shanghai Cancer Institute, China) to make the overexpression of c-Myc. siRNA specific for PVT1 (sense 5’-GCUUGGAGGCUGAGGAGUUTT-3’ and antisense 5’-AACUCC UCAGCCUCCAAGCTT-3’) were synthesized (Ribobio, Guangzhou, China). c-Myc-siRNA and pcDNA3.1-c-Myc transfection (50 nmol/L) was performed in cultured NSCLC cells A549. Then, qRT-PCR was used to detect relative mRNA level. Cells were then treated with cisplatin. Colorimetric water-soluble tetrazolium salt (CCK-8) assay using a Cell Counting Kit-8 (Dojindo) was used for cell viability detection and Annexin-V-FITC Apoptosis Detection Kit I (BD Pharmingen) was used for apoptosis analysis.
Luciferase reporter assays
Firefly and lung luciferase activities, as indicated by relative luminescence units (RLU) were determined using One-Glo or Dual-Glo luciferase assay kits (Promega) according to the manufacturer’s instructions. For agonist, fold of induction = firefly RLUinduced/firefly RLUuninduced. For antagonist, % of control = 100 × firefly RLU(agonist+antagonist)/firefly RLUagonist alone, all normalized to lung RLU. Both EC50 and IC50 values were generated using GraphPad Prism software. Z’ values were determined as Z’ = 1 - [(3 × SDinudced + 3 × SDuninduced)/(averageinduced – averageuninduced)” > (3 × SDinudced + 3 × SDuninduced) (averageinduced – averageuninduced)].
Chromatin immunoprecipitation (ChIP)
NSCLC cells A549 were cross-linked with 1% formaldehyde for 10 min at room temperature. The cross-linking was terminated by adding 125 mM glycine and the cells were washed twice with ice-cold PBS. The cells were solubilized in a buffer containing 10 mM Tris-HCl (pH 8.0), 1% Triton X-100, 1% sodium deoxycholate, 1 mM phenylmethanesulfonyl fluoride and protease inhibitor cocktail for 10 min at 4°C. Sonication using a Bioruptor® Sonicator (Diagenode s.a., Seraing, Belgium) was performed to shear chromatin into 500-bp fragments. The supernatant was obtained by centrifugation (16,000×g for 10 min at 4°C) and equally divided into six tubes (100 μl/tube). The appropriate antibodies (anti c-MYC antibody or rabbit IgG) was added into each tube and incubated for 3 h at 4°C. Immunoprecipitation was performed using ChIP-grade agarose beads with protein G (Cell Signaling Technology, Inc., Danvers, MA, USA), and the cells were blocked with 1% bovine albumin and 1% salmon sperm DNA. Finally, the compounds were collected and the DNA was isolated for qPCR.
Flow cytometry
To study the influence of expression changes of c-Myc and lncRNA H19 on the progression of cell cycle in NSCLC, flow cytometry was used. The NSCLC cells A549 at logarithmic phase were selected and plated in a 96-well plate at a density of 2 × 103 cells/well in supplemented RPMI 1640 and incubated for 16 h before the cells were subjected to treatment in triplicate wells. After treatment, the cells were washed twice in phosphate-buffered saline (PBS) (2.68 mM KCl, 1.47M KH2PO4, 8 mM Na2KPO4, 136.75 mM NaCl) and counted. Fifty to one hundred thousand cells were selected and centrifuged 5 min at 1000 r/min. Annexin V-FITC mixed liquor of 195 μL was added to resuspend cytotrophoblast cells and 5 μL was added to mix. Centrifugation at 1000 r/min for 5 min was performed after cultivation 10 min. Sample was obtained after discarding supernatant and 10 μL propidium iodide (PI) was added. Afterwards, the sample was stilled in dark for 30 min. Finally, the apoptosis and cell cycle were detected using flow cytometry (FCM) on the Moflo (Dako Cytomation, Glostrup, Denmark).
Statistical analysis
Data were processed using SPSS 12.0 statistical software (SPSS, Inc., Chicago, IL, USA) and recorded as the mean ± standard error of the mean. P < 0.05 was considered to indicate a statistically significant difference. In addition, one-way analysis of variance was adopted to assess the data. All of the experiments were performed in triplicate for the purposes of comparison.
Results
H19 was up-regulated in cancer tissues and cells
To explore H19 expression in NSCLC, RT-PCR was used to determine the mRNA levels of lncRNA H19 in cancer tissues of patients with NSCLC and the tissues of normal people, NSCLC cells (A549, L78 and H460) and the normal cells. As shown in Figure 1A, the mRNA level of lncRNA H19 was up-regulated in NSCLC tissues compared with the normal tissues, and the difference had statistical significance (P < 0.01). The results of NSCLC and the normal cells showed the mRNA levels of lncRNA H19 in A549, L78 and H460 cells were significantly higher than that in the normal cells 16HBE (P < 0.01) (Figure 1B). Moreover, the value in A549 cells was much higher than that in L78 and H460 cells. Therefore, A549 cells were selected for the further research. All those suggested that lncRNA H19 may be key factor for the development and progression of NSCLC.
Figure 1.
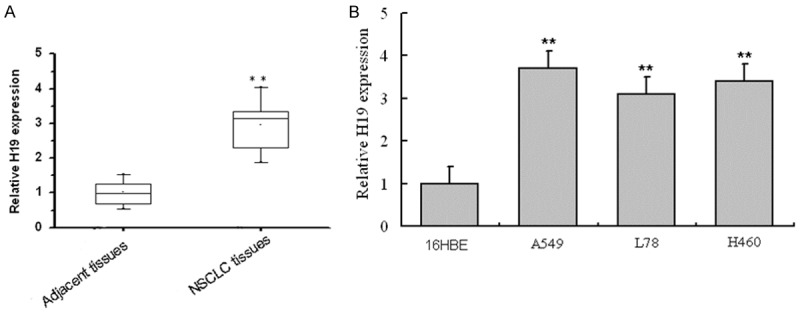
LncRNA H19 expression levels in NSCLC and the control. A: mRNA level of H19 in NSCLC tissues and adjacent tissues. **P < 0.01, compared with adjacent tissues, H19 expression level in NSCLC tissues had statistical difference; B: mRNA level of H19 in NSCLC cell line A549, L78 and H460 and the normal cell line 16HBE. **P < 0.01, compared with the normal cell line 16HBE, H19 expression level in NSCLC cell line A549, L78 and H460 had statistical difference.
C-Myc up-regulates the expression of lncRNA H19
To explore the influence of c-Myc level on the expression of lncRNA H19, pcDNA3.1-c-Myc and c-Myc siRNA was transfected into A549 cells to overexpress and knockdown c-Myc, respectively. Then, the mRNA levels of c-Myc and lncRNA H19 in the transfected A549 cells was determined by RT-PCR. The results of transfection with pcDNA3.1-c-Myc showed the mRNA expression of H19 and c-Myc in cells transfected with pcDNA3.1-c-Myc was markedly higher than that transfection with empty vector plasmid pcDNA3.1 (Figure 2A and 2B). For cells transfected with c-Myc siRNA, the mRNA level was much lower than that transfection with si-NC. Moreover, the expression of lncRNA H19 was also down-regulated after the knockdown of c-Myc (Figure 2C and 2D). Therefore, we can conclude c-Myc can up-regulate the expression of lncRNA H19.
Figure 2.
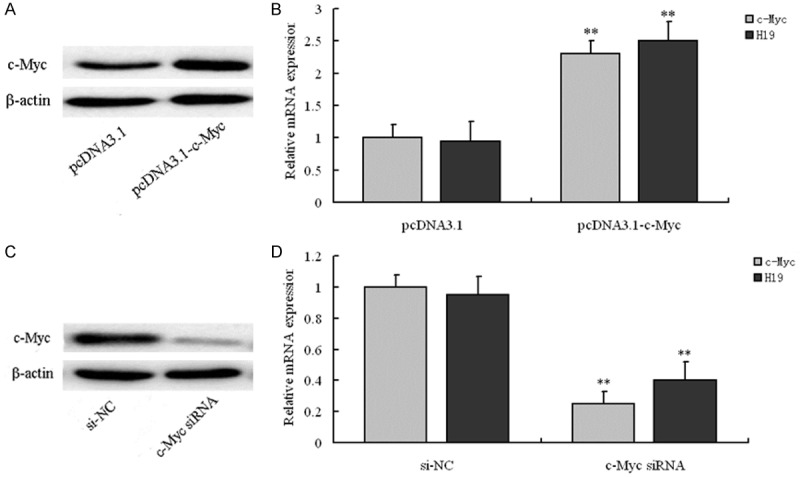
The expression changes of The c-Myc and H19 after transfection of pcDNA3.1-c-Myc and c-Myc-siRNA in NSCLC cell line A549. A: Representative images of RT-PCR results indicated pcDNA3.1-c-Myc significantly upregulated the expression of c-Myc at protein levels; B: pcDNA3.1-c-Myc significantly upregulated the expression of c-Myc and the expression of H19 was upregulated accordingly at mRNA levels; **P < 0.01, compared with pcDNA3.1 group, relative mRNA expression of c-Myc and H19 in pcDNA3.1-c-Myc had statistical difference, respectively; C: Representative images of RT-PCR results indicated c-Myc-siRNA significantly downregulated the expression of c-Myc at protein levels; D: c-Myc-siRNA significantly downregulated the expression of c-Myc and the expression of H19 was downregulated accordingly at mRNA levels; **P < 0.01, compared with si-NC group, relative mRNA expression of c-Myc and H19 in c-Myc-siRNA group had statistical difference, respectively.
C-Myc strengthen the activity of lncRNA H19 promoter
To explore the molecular about how c-Myc up-regulate the expression of lncRNA H19, luciferase reporter assay was used to explore the targeted role of c-Myc on the promoter of lncRNA H19. The schematic of H19-promoter-luciferase reporter plasmid was shown in Figure 3A. To study the influence of c-Myc on the activity of H19 promoter, 4 groups: pcDNA3.1, pcDNA3.1-c-Myc, si-NC and c-Myc siRNA were selected and relative luciferase activity in each group was determined. Compared with pcDNA3.1 groups, luciferase activity in pcDNA3.1-c-Myc group was markedly higher (P < 0.01), and the value in c-Myc siRNA group was much lower than the si-NC group (P < 0.01) (Figure 3B). The influence of c-Myc ectopic expression on the enrichment of lncRNA H19 in A549 cells was studied by employing 3 groups: Non-transfected, pcDNA3.1-c-Myc and c-Myc siRNA group. Results showed the percentage of lgG input was similar and all very low in the 3 groups. The percentage of c-Myc input was all significantly higher than lgG in each group. Moreover, the differences of c-Myc value between Non-transfected and pcDNA3.1-c-Myc group, pcDNA3.1-c-Myc and c-Myc siRNA all had statistical significance (both was P < 0.01). This indicated the ectopic expression of c-Myc would affect the enrichment of lncRNA H19 in A549 cells. So, we concluded c-Myc can up-regulate the expression of lncRNA H19 by targeting its promoter.
Figure 3.
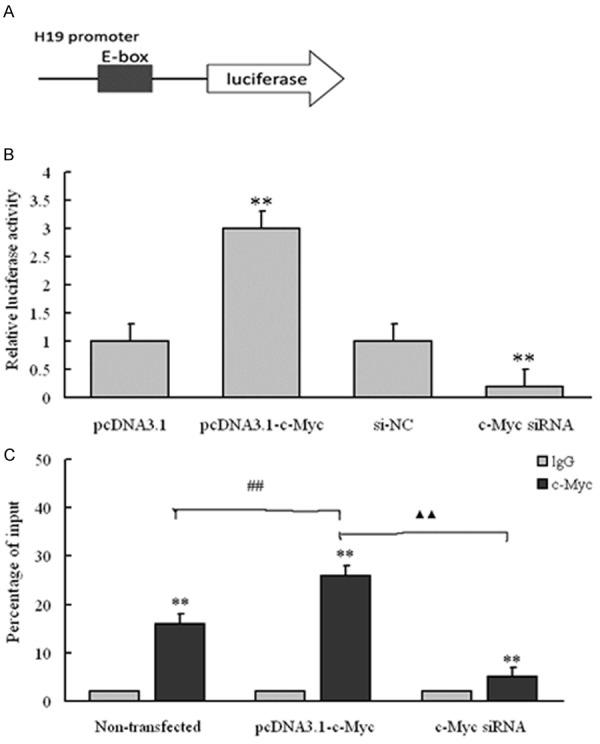
C-Myc regulates H19 promoter activity, depending on E-box element. A: Schematic of the H19-promoter-luciferase construct is depicted with locations of the E-box element and sequences of point mutation; B: Luciferase reporter assay on A549 cells cotransfected with luciferase constructs (mutant at E-box element) and pcDNA3.1-c-Myc or c-Myc-siRNA; **P < 0.01, compared with pcDNA3.1 group, relative luciferase activity in pcDNA3.1-c-Myc group had statistical difference, ##P < 0.01, compared with si-NC group, relative luciferase activity in c-Myc-siRNA group had statistical difference; C: ChIP-derived DNA was amplified by qRT-PCR using specific primers. The levels of qPCR products are expressed as a percentage of input DNA; **P < 0.01, compared with the lgG value, the percentage of c-Myc input had statistical difference in Non-transfected, pcDNA3.1-c-Myc and c-Myc-siRNA group, respectively; ##P < 0.01, compared with Non-transfected group, percentage of c-Myc input in pcDNA3.1-c-Myc had statistical difference; ▲▲P < 0.01, compared with pcDNA3.1-c-Myc group, percentage of c-Myc input in c-Myc-siRNA group had statistical difference.
LncRNA H19 influences cell cycle by regulating miR-107
With online tool Starbase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php), we obtained miR-107 was base pairing with H19. Then, the relationship between H19 and miR-107 was researched by overexpressing and interfering H19 with RT-PCR. Alignment of potential H19 base pairing with miR-107 as identified by online tool Starbase v2.0 was shown in Figure 4A. Results also showed H19 was significantly down-regulated by transfection with si-H19 (P < 0.01), while miR-107 was markedly up-regulated compared with the control (P < 0.01) (Figure 4B). When transfection with pcDNA3.1-H19, the expression of H19 was significantly increased (P < 0.01) and the value of miR-107 decreased markedly (P < 0.01) (Figure 4C). The cell-cycle distribution in A549 cells was explored in 4 groups: si-NC, si-H19, miR-107 inhibitor and si-H19+miR-107 inhibitor. Results showed the number of cells in G2/M stage in si-H19 and si-H19+miR-107 inhibitor group decreased significantly compared with the si-NC group (P < 0.01), while the value in miR-107 inhibitor group increased markedly (P < 0.01). Moreover, the values in G0/G1, S and G2/M stage between si-H19 and miR-107 inhibitor group, miR-107 inhibitor and si-H19+miR-107 inhibitor group all had statistical difference (Figure 5). Therefore, we concluded lncRNA H19 can influence the cell cycle of NSCLC cells by regulating miR-107.
Figure 4.
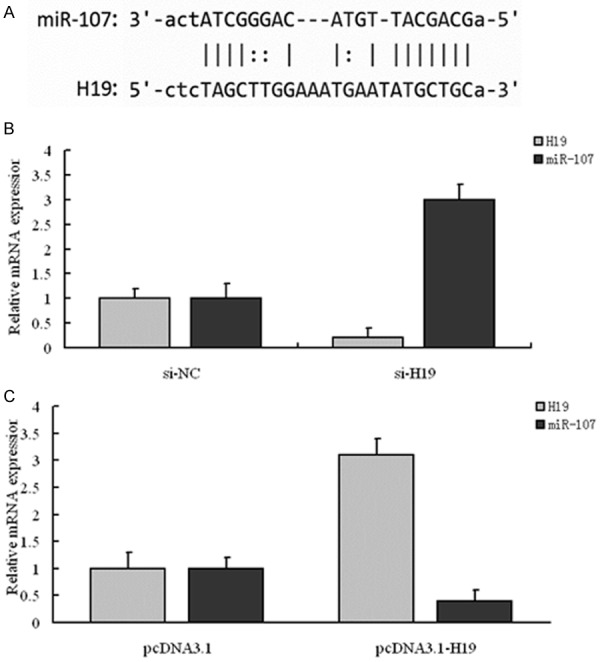
Identification of miR-107 as a target of H19. A: Alignment of potential H19 base pairing with miR-107 as identified by Starbase v2.0 (http://starbase.sysu.edu.cn/mirLncRNA.php). B: si-H19 reduced the endogenous H19 mRNA level and increased miR-107 mRNA level; **P < 0.01, compared with si-NC group, the mRNA levels of H19 and miR-107 in si-H19 group had statistical difference, respectively; C: pcDNA3.1-H19 increased the endogenous H19 mRNA level and decreased miR-107 mRNA level; **P < 0.01, compared with pcDNA3.1 group, the mRNA levels of H19 and miR-107 in pcDNA3.1-H19 group had statistical difference, respectively.
Figure 5.
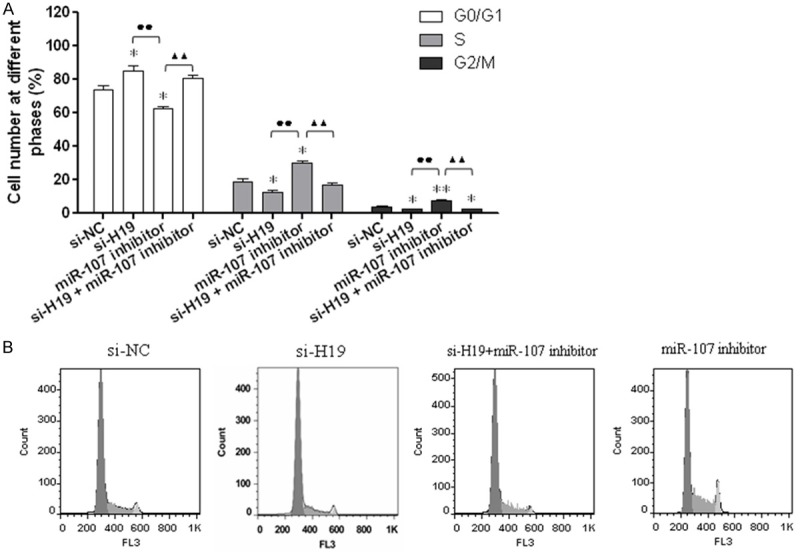
H19 influences the mitotic progression of NSCLC cell line A549 by targeting miR-107. A: Cell cycle progression of A549 after transfected with si-H19, miR-107 inhibitor, and si-H19+miR-107 inhibitor was determined by FCM analysis; *P < 0.05 and **P < 0.01, compared with si-NC group, the G0/G1, S and G2/M stage in si-H19, miR-107 inhibitor, and si-H19+miR-107 group had statistical difference, respectively. B: The distribution of A549 cell cycle; ●●P < 0.01, the difference of G0/G1, S and G2/M value between si-H19 and miR-107 group had statistical significance; ▲▲P < 0.01, the difference of G0/G1, S and G2/M value between miR-107 inhibitor and si-H19+miR-107 group had statistical significance.
Discussion
Lung cancer is the most common cause of cancer deaths worldwide, and most cases are associated with cigarette smoking [18]. The median survival of patients with untreated NSCLC is only four to five months, with a survival rate at one year of only 10 percent. Chemotherapy for NSCLC is often considered ineffective or excessively toxic, and over the past decade, a number of new agents have become available for the treatment of NSCLC, including the taxanes, gemcitabine, and vinorelbine [19]. LncRNA genes demonstrate developmental and tissue specific expression patterns, and aberrant regulation in a variety of diseases, including cancer [20]. They represent a significant untapped resource in terms of developing diagnostics and therapies. Differential or high level expression of certain cancer type-specific lncRNAs can be exploited for the development of novel biomarkers as lncRNA expression or may potentially correlate with patient response to chemotherapy. Understanding the mechanism(s) by which lncRNAs act will continue to provide novel approaches to regulating genes including the development of mimetics to compete with binding sites for miRNAs, chromatin remodelers, or DNA [5]. Yang et al. reported lncRNAs have been shown to have important regulatory roles in cancer biology, and the lncRNA H19 is up-regulated in hypoxic stress and in some tumors [10]. However, the contributions of H19 to NSCLC remain largely unknown. Therefore in this study, we aimed to study the molecular mechanism about how lncRNA H19 influence NSCLC.
First, NSCLC and adjacent tissues were separated. The mRNA levels of lncRNA H19 in those tissues, NSCLC and normal cell lines were determined by RT-PCR. Results showed the relative expression levels of H19 in NSCLC tissues and cell lines were significantly higher than those in adjacent tissues and normal cells. The imprinted H19 gene, which encodes an untranslated RNA, lies at the end of a cluster of imprinted genes [21]. It is strongly expressed in embryonic cells and in a wide-range of human cancers [22]. Hibi et al. reported that loss of imprinting (LOI) of the endogenous gene H19 was frequently found in lung cancer and choriocarcinoma, common adulthood cancers [23]. H19 transcription is up-regulated during the S-phase of growth-stimulated cells and that the H19 promoter is activated by E2F1 in breast cancer cells [24]. H19 is highly expressed from the early stages of embryogenesis to fetal life in many organs including the fetal adrenal, liver and placenta but is nearly completely downregulated postnatally [25]. Moreover, H19 noncoding RNA can be induced by c-Myc oncogene by allele-specific binding to potentiate tumorigenesis, and study showed the the promoter region of lncRNA H19 includes the binding site of c-Myc [26]. Therefore, we further explored the influence of c-Myc on the expression of lncRNA H19. PcDNA3.1-c-Myc and c-My-siRNA was transfected into NSCLC cells and RT-PCR was used to determine the expression of c-Myc and H19, respectively. Results showed the expression level of H19 increased and decreased accordingly with the overexpression and knockdown of c-Myc, which indicated c-Myc can up-regulate the expression of H19. Previous study showed the altered expression of lncRNA H19 can be duced by c-Myc in the development and progression of gastric cancer by regulating cell proliferation [27]. Matouk et al. reported the H19 non-coding RNA is essential for human tumor growth and c-Myc induced the expression of the H19 RNA [28]. Then, to explore the mechanism on how c-Myc up-regulate the expression of H19, luciferase reporter assay was used to determine the targeting role of c-Myc on the promoter of H19. Results showed c-Myc strengthen the activity of the promoter of H19 and the ectopic expression level of c-Myc influence the enrichment of H19. It indicated c-Myc can up-regulate the expression of H19 by targeting its promoter. Then, miRNA complementary base pairing with H19 was predicted by online software and miR-107 was selected. Results showed the expression of miR-107 increased with the knockdown of H19, which indicated miR-107 can be down-regulated by miR-107 and miR-107 was the target gene of H19. To explore the molecular mechanism about how H19 influence the cell cycle progression of NSCLC, NSCLC cells were transfected with si-NC, si-H19, miR-107 inhibitor and si-H19+miR-107 inhibitor, and cell cycle was determined, respectively. Results showed the number of cells in G2/M stage in si-H19 and si-H19+miR-107 inhibitor group decreased significantly compared with the si-NC group (P < 0.01), while the value in miR-107 inhibitor group increased markedly. MicroRNAs (miRNAs) are a class of ∼22-nucleotide (nt)-long noncoding RNAs expressed by all metazoan eukaryotes [29]. Dysregulation of a specific spectrum of miRNAs in human malignancies is frequently observed. Emerging evidence suggests miRNAs function as both tumor suppressors and oncogenes [30]. Rapidly accumulating evidence suggests that many miRNAs are involved in cell cycle regulation and consequentially play critical roles in carcinogenesis. The imprinted H19 noncoding RNA is a primary microRNA precursor [31]. Shi et al. reported lncRNA H19 promotes glioma cell invasion by deriving miR-675 [32]. In NSCLC cell lines, MiR-107 can induce the cell cycle arrest and suppress gene expression through either translational repression or degradation of target mRNAs [33]. In gastric cancer, it is up-regulated in gastric cancer and associated with tumour metastasis and worse prognosis [14]. MiR-107 can targets cyclin-dependent kinase 6 expression and induce cell cycle G1 arrest and inhibit invasion in gastric cancer cells [34]. All those indicated H19 can affect the mitotic progression of NSCLC cells by down-regulating the expression level of miR-107.
Our study suggests another layer of regulation that involves the lncRNAs and H19 is a c-Myc up-regulated gene that potentiates the tumorigenic phenotype of NSCLC. Moreover, H19 can promote cell cycle progression of NSCLC cells by down-regulating miR-107.
Disclosure of conflict of interest
None.
References
- 1.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Temel JS, Greer JA, Muzikansky A, Gallagher ER, Admane S, Jackson VA, Dahlin CM, Blinderman CD, Jacobsen J, Pirl WF. Early palliative care for patients with metastatic non-small-cell lung cancer. N Engl J Med. 2010;363:733–742. doi: 10.1056/NEJMoa1000678. [DOI] [PubMed] [Google Scholar]
- 3.Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, Kratzke R, Watson MA, Kelley M, Ginsburg GS. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 4.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38–55. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce C, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, Wu F, Mo YY. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juan L, Wang G, Radovich M, Schneider BP, Clare SE, Wang Y, Liu Y. Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC Med Genomics. 2013;6:S7. doi: 10.1186/1755-8794-6-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315–322. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 12.Hajjari M, Behmanesh M, Sadeghizadeh M, Zeinoddini M. Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med Oncol. 2013;30:670. doi: 10.1007/s12032-013-0670-0. [DOI] [PubMed] [Google Scholar]
- 13.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Zhang Y, Shi Y, Dong G, Liang J, Han Y, Wang X, Zhao Q, Ding J, Wu K. MicroRNA-107, an oncogene microRNA that regulates tumour invasion and metastasis by targeting DICER1 in gastric cancer. J Cell Mol Med. 2011;15:1887–1895. doi: 10.1111/j.1582-4934.2010.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datta J, Smith A, Lang JC, Islam M, Dutt D, Teknos TN, Pan Q. microRNA-107 functions as a candidate tumor-suppressor gene in head and neck squamous cell carcinoma by downregulation of protein kinase Cε. Oncogene. 2012;31:4045–4053. doi: 10.1038/onc.2011.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 17.Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkin DM, Bray F, Devesa S. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37:S4–66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 19.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 20.Silva JM, Perez DS, Pritchett JR, Halling ML, Tang H, Smith DI. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics. 2010;95:355–362. doi: 10.1016/j.ygeno.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 22.Poirier F, Chan C, Timmons P, Robertson E, Evans M, Rigby P. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development. 1991;113:1105–1114. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- 23.Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, Akiyama S, Ito K, Takagi H. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480–482. [PubMed] [Google Scholar]
- 24.Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280:29625–29636. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- 25.Lustig O, Ariel I, Ilan J, Lev-Lehman E, De-Groot N, Hochberg A. Expression of the imprinted gene H19 in the human fetus. Mol Reprod Dev. 1994;38:239–246. doi: 10.1002/mrd.1080380302. [DOI] [PubMed] [Google Scholar]
- 26.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 27.Zhang EB, Han L, Yin DD, Kong R, De W, Chen J. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Med Oncol. 2014;31:914. doi: 10.1007/s12032-014-0914-7. [DOI] [PubMed] [Google Scholar]
- 28.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 30.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. Rna. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J, Qian J, Liu N, You Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi Y, Forrest A, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng L, Xie Y, Zhang H, Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol. 2012;29:856–863. doi: 10.1007/s12032-011-9823-1. [DOI] [PubMed] [Google Scholar]


