Abstract
Aim: Three common polymorphisms in CD209 gene (-336A/G, -871A/G and -139G/A) have been reportedly associated with pulmonary tuberculosis risk. However, the findings from different studies were inconsistent. Therefore, we conducted a comprehensive meta-analysis to determine the association between CD209 gene polymorphisms and pulmonary tuberculosis susceptibility. Methods: The PubMed, SCI and Elsevier were searched up to April 18, 2015 for studies on the association of CD209 gene polymorphisms and pulmonary tuberculosis. Pooled odds ratio (ORs) and 95% confidence intervals (95% CIs) were calculated in a fixed-effects or random-effects model. Results: Twelve case-control studies with 3114 cases and 3088 controls were included. For -871A/G mutation, significant decreased pulmonary tuberculosis risk was observed in allele model (G vs. A: P = 0.009; OR = 0.70, 95% CI = 0.54-0.92), heterozygous model (AG vs. AA: P = 0.009; OR = 0.59, 95% CI = 0.40 to 0.88) and dominant model (AG+GG vs. AA: p =0.01; OR = 0.61, 95% CI = 0.42 to 0.89). For -336A/G polymorphism, no associations were found in all genetic models. In the subgroup analysis by ethnicity, statistical association was observed for Asians in GG vs. AA (P = 0.04; OR = 2.31, 95% CI = 1.05-5.09). No significant association was identified between -139G/A variation and pulmonary tuberculosis risk. Conclusions: This meta-analysis provides evidences that CD209 gene -871A/G is associated with decreased susceptibility to pulmonary tuberculosis in overall population; -336A/G polymorphism is associated with increased susceptibility of pulmonary tuberculosis in Asians. However, the -139G/A polymorphism is not associated with susceptibility to pulmonary tuberculosis.
Keywords: Meta-analysis, CD209, polymorphism, pulmonary tuberculosis
Introduction
Tuberculosis is caused by the bacillus Mycobacterium tuberculosis and is one of the most prevalent infectious diseases [1]. There were 8.6 million incident cases of tuberculosis which caused 1.3 million deaths according to the report from World Health Organization in 2012 [2]. However, only 5% to 10% of the individuals infected with Mycobacterium tuberculosis develop symptomatic tuberculosis [3]. Although host immune response and environmental factors are supposed to contribute to tuberculosis, increasing evidence has confirmed that various genetic factors affect the susceptibility to tuberculosis [4].
Human CD209 gene contains 7 exons and 6 introns and is localized on chromosome 19p13.2-3 [5]. Dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN), a C-type lectin encoded by the CD209 gene, is one of major receptors expressing on dendritic cells (DCs) and alveolar macrophages. DC-SIGN plays an important first-line role in host defense against pathogens [6]. In mature DCs, DC-SIGN also promotes the activation and proliferation of resting T cells and increases primary immune responses [7]. In addition, DC-SIGN may influence the pathogenesis of tuberculosis by activating the Raf-1-acetylation-dependent signaling pathway that is involved in the regulation of adaptive immune [8,9]. Moreover, it has also been proposed that the DC-SIGN may play a role in immune evasion to tuberculosis progression by suppress Toll-like receptor signaling leading to cytokine secretion [10].
The association between CD209 gene polymorphisms and tuberculosis risk has been extensively investigated. The most common single-nucleotide polymorphisms (SNPs) in the CD209 gene are the -336A/G (rs4804803), -871A/G (rs735239) and -139G/A (rs2287886), which are considered as potential susceptibility factors for tuberculosis [11-13]. However, the conclusions of the published studies remain conflicting. Genome-wide association studies (GWASs) were used as an important tool to improve our knowledge of many common variants and the association with different diseases. Pulmonary tuberculosis susceptibility genes have also been studied. However, these studies did not provide an association between pulmonary tuberculosis and the CD209 gene. Therefore, we chose the three widely studied polymorphisms (-336A/G, -871A/G and -139G/A) and performed a meta-analysis to provide a more precise and comprehensive estimation of the association between CD209 gene polymorphisms and pulmonary tuberculosis risk.
Materials and methods
Literature search strategy
We conducted a comprehensive search on the PubMed, SCI and Elsevier (up to April 18, 2015). The following terms were used: (“tuberculosis” or “TB”) and (“CD209” or “DC-SIGN”) and (“polymorphism” or “mutation” or “variation”). To access additional publications, we also searched the references of the retrieved literatures. There was no restriction for population, sample size, time period, or types of reports. The language of the articles was restricted to English.
Inclusion and exclusion criteria
Eligible studies in this meta-analysis had to meet the following criteria: (a) a human case-control study on the association between CD209 polymorphisms and the risks of pulmonary tuberculosis; (b) original papers containing available data in cases and controls for estimating an odds ratio (OR) and 95% confidence interval (CI); (c) studies written in English. The exclusion criteria were: (a) abstracts, letters, reviews, editorial articles or case reports; (b) studies on the association between other gene polymorphisms and pulmonary tuberculosis risks; (c) studies with HIV/TB patients; (c) studies without specific tuberculosis type.
Data extraction
Two investigators (Yi and Zhang) extracted the data independently from all of the eligible publications according to the inclusion and exclusion criteria, and reached a consensus on all items. Primary extraction data were reviewed by Zhao, in case of disagreement, a third author would assess these articles. From each study, the collected data included: first author’s name, year of publication, study location, sample size, source of control, the genotyping method, genotype distribution in cases and controls.
Statistical analysis
Pooled ORs and their 95% CI were calculated to examine the relation between CD209 polymorphisms and pulmonary tuberculosis risks in allele model, the co-dominant model, the dominant model and recessive model based on the extracted data. Heterogeneity among included studies was measured by chi-square-based Q test and P and I2 test [14]. The heterogeneity was not considered significant if I2 < 50%, and Mantel-Haenszel fix effect model was used, otherwise DerSimonian-Laird random effect model was adopted [15]. To explore the source of heterogeneity, the subgroup analysis by ethnicity was performed. Hardy-Weinberg equilibrium (HWE) was also tested in controls by asymptotic Pearson’s chi-square test [16]. Sensitivity analyses were performed by omitting certain studies each time to identify individual study’s effect on pooled results and test the reliability of the results. Publication bias was assessed by Begg’s adjusted rank correlation test and Egger’s regression asymmetry test, and the statistical significance was identified as P < 0.05 [17]. Data were analyzed using the program Review Manager 5.2 and STATA 11.0 Software (StataCorp LP, College Station, TX, USA).
Results
Studies selection process and characteristics
The flow chart was shown in Figure 1. All retrieved articles were examined carefully by reading the titles and abstracts, 17 potential studies were identified for further investigation. Based upon the inclusion and exclusion criteria, one study was excluded for data overlapping and one was excluded for not written in English. The studies not indicating the tuberculosis type were also excluded. Only case-control studies having frequency of all three genotypes were included in this meta-analysis. In Vannberg and colleagues’ study [18], the results of people in different contries were presented seperately, each group in this study was considered as a case-control study. Thus, 9 articles with 12 case-control studies [11-13,18-23] were included for data extraction (Figure 1). Among these studies, only one study enrolled subjects with pulmonary tuberculosis and extra-pulmonary tuberculosis [11] while others were focused on the association between pulmonary tuberculosis risks and CD209 polymorphisms. A total of 3114 subjects with pulmonary tuberculosis were included in this meta-analysis. 2494 subjects in 8 studies were diagnosed by culture, 532 subjects were diagnosed by smear (Table 1). The genotype distributions in controls were in agreement with HWE except for one study conducted by Naderi [20].
Figure 1.

Flow diagram of the study selection.
Table 1.
Characteristics of the case-control studies included in the meta-analysis
| First Author | Year | Population | Genotyping method | Cases/Controls | SNP studied | Male Patients (%) | Male Controls (%) | Age of cases | Age of controls | Diagnosis method | Control source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Barreiro et al. | 2006 | African (South African) | fluorescence polarization or TaqMan | 351/360 | -336A/G, -871A/G | 51.8 | 22 | 36.7±10.9 | 34.6±12.5 | Smear, culture | Healthy individuals |
| da Silva et al. | 2014 | Mixed (Brazilian) | Taq Man SNP genotyping assays | 95/164 | -336A/G, -871A/G, -139G/A | 63.2 | 34.8 | 24.8±17.6 | 21.6±2.4 | Baciloscopy or culture | Healthy unrelated individuals |
| Kobayashi et al. | 2011 | Asian (Indonesian) | Sequencing | 532/561 | -336A/G, -871A/G, -139G/A | 53.7 | DNR | 41.6±15.4 | 39.3±12.7 | Smear, radiologic, clinical symptoms | Healthy individuals |
| Naderi et al. | 2014 | Asian (Iran) | T-ARMS-PCR | 156/154 | -336A/G | 37.2 | 43.5 | 48.1±15.6 | 49.8±20.7 | Smear, culture, radiologic, clinical symptoms | Healthy individuals |
| Ogarkov et al. | 2012 | Caucasian (Russian) | Taq Man LNA technology | 101/177 | -336A/G | 76.3 | 71.8 | 42.3±12.1 | 41.9±9.2 | DNR | Healthy individuals |
| Olesen et al. | 2007 | African (Guinea-Bissau) | Taq Man SNP genotyping assays | 315/340 | -336A/G, -139G/A | 60.4 | 49.9 | 37.3 | 38.1 | Smear, culture, radiologic, clinical symptoms | Healthy unrelated donors |
| Selvaraj et al. | 2009 | Caucasian (Indian) | PCR-RFLP | 107/157 | -336A/G,-139G/A | 49.5 | 51 | 34.0±8.2 | 30.6±8.3 | Smear, culture, radiologic, clinical symptoms | Healthy individuals |
| Vannberg et al. (a) | 2008 | African (Gambian) | MALDI-TOF | 676/327 | -336A/G | DNR | DNR | DNR | DNR | Smear, culture and history | Healthy unrelated donors |
| Vannberg et al. (b) | 2008 | African (Guinean) | MALDI-TOF | 151/180 | -336A/G | DNR | DNR | DNR | DNR | Smear, culture and history | Healthy unrelated donors |
| Vannberg et al. (c) | 2008 | African (Guinea-Bissau) | MALDI-TOF | 162/141 | -336A/G | DNR | DNR | DNR | DNR | Smear, culture and history | Healthy unrelated donors |
| Vannberg et al. (d) | 2008 | African (Malawian) | MALDI-TOF | 244/295 | -336A/G | DNR | DNR | DNR | DNR | Smear, culture and history | Healthy unrelated donors |
| Zheng et al. | 2011 | Asian (China) | Sequencing | 237/244 | -336A/G, -871A/G | 65.4 | DNR | 44.6±17.7 | DNR | Culture, radiologic | Healthy individuals |
Abbreviations and definitions: T-ARMS-PCR, Tetra-amplification Refractory Mutation System -Polymerase Chain Reaction; PCR-RFLP, Restriction Fragment Length Polymorphism analysis of PCR amplified fragments; MALDI-TOF, Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry; DNR: data not reported.
Quantitative data synthesis
Four case-control studies (1208 cases and 1320 controls) evaluating the susceptibility of CD209 -871A/G polymorphism and the risk of pulmonary tuberculosis. A protective effect on pulmonary tuberculosis development was observed in allele model (G vs. A: P = 0.009; OR = 0.70, 95% CI = 0.54-0.92) (Figure 2), heterozygous model (AG vs. AA: P = 0.009; OR = 0.59, 95% CI = 0.40 to 0.88) (Figure 3) and dominant model (AG+GG vs. AA: P = 0.01; OR = 0.61, 95% CI = 0.42 to 0.89) (Figure 4).
Figure 2.
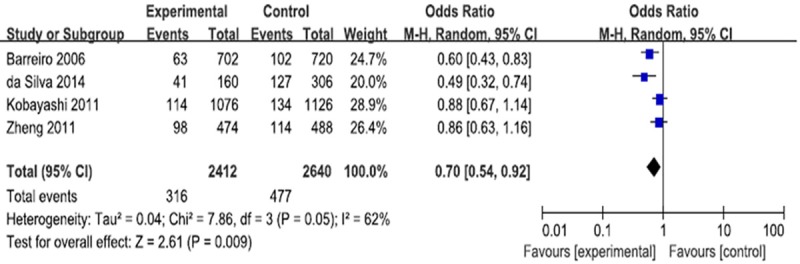
Forest plot and ORs with 95% CI of CD209-871A/G polymorphism and pulmonary tuberculosis risk (G vs. A). The squares and horizontal lines correspond to the OR and 95% CI for each study. The area of the squares reflects the weight. The diamond represents the summary OR and 95% CI.
Figure 3.
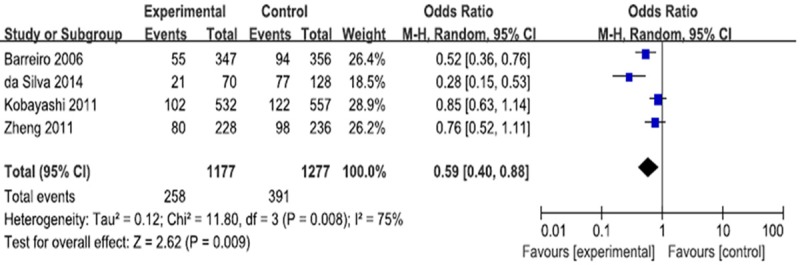
Forest plot and ORs with 95% CI of CD209-871A/G polymorphism and pulmonary tuberculosis risk (AG vs. AA).
Figure 4.

Forest plot and ORs with 95% CI of CD209-871A/G polymorphism and pulmonary tuberculosis risk (AG+GG vs. AA).
There were 12 case-control studies (3114 cases and 3088 controls) were pooled together for evaluation of the association between CD209 -336A/G polymorphism and risk of pulmonary tuberculosis. The combined results showed that no associations were found in all genetic models (G allele vs. A: p =0.32; OR=1.18, 95% CI = 0.85-1.65; GG vs. AA: P = 0.64; OR = 1.04, 95% CI = 0.87 to 1.25; AG vs. AA: P = 0.98; OR = 1.00, 95% CI = 0.89 to 1.13; AG+GG vs. AA: P = 0.92; OR = 1.01, 95% CI = 0.90 to 1.13; GG vs. AG+AA: P = 0.96; OR = 1.00, 95% CI = 0.86 to 1.17). In the subgroup analysis by ethnicity, statistical association was observed for Asians in GG vs. AA (P = 0.04; OR = 2.31, 95% CI = 1.05-5.09) (Figure 5). However, there was no association in other comparison models in African and Caucasian populations (Table 2).
Figure 5.
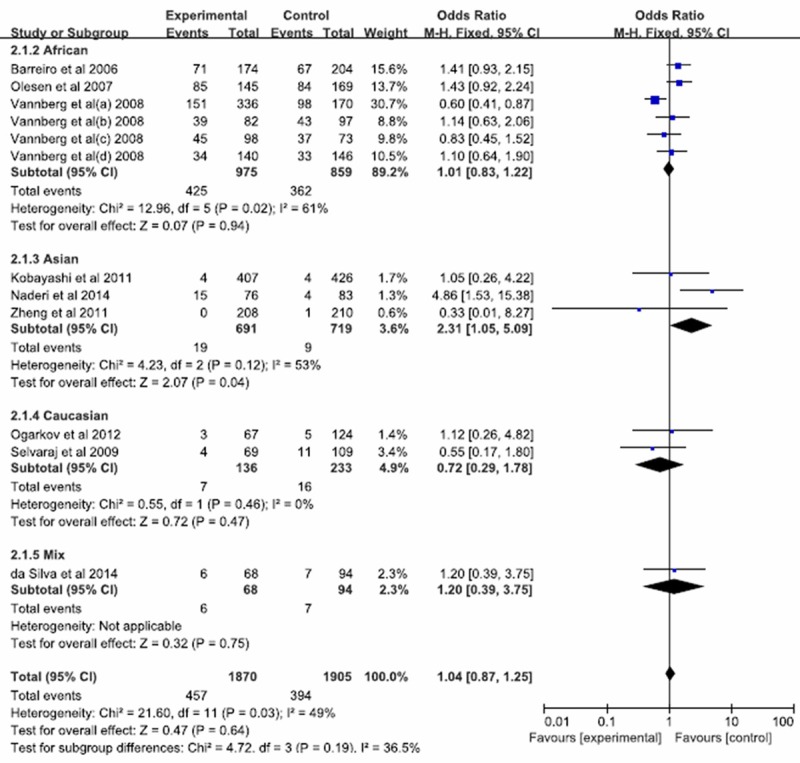
Forest plot and ORs with 95% CI of CD209 -336A/G polymorphism and pulmonary tuberculosis risk (GG vs. AA).
Table 2.
Determination of the genetic effects of CD209 polymorphisms on pulmonary TB and subgroup analysis
| Allele model | Dominant model | Reccessive model | Homozygous model | Heterozygous model | |
|---|---|---|---|---|---|
|
|
|||||
| OR (95% CI), I2 (%) | OR (95% CI), I2 (%) | OR (95% CI), I2 (%) | OR (95% CI), I2 (%) | OR (95% CI), I2 (%) | |
| -336A/G | G allele vs. A allele | GG+AG vs. AA | GG vs. AA+AG | GG vs. AA | AG vs. AA |
| African | 1.66 (0.77-3.62), 90 | 1.01 (0.87-1.18), 69 | 0.97 (0.83-1.15), 42 | 1.01 (0.83-1.22), 61 | 1.02 (0.87-1.19), 66 |
| Asian | 1.22 (0.67-2.21), 87 | 1.07 (0.86-1.32), 58 | 2.13 (0.97-4.67), 42 | 2.31 (1.05-5.09), 53 | 1.04 (0.84-1.29), 29 |
| Caucasian | 1.05 (0.77-1.44), 0 | 1.13 (0.79-1.62), 0 | 0.67 (0.27-1.66), 0 | 0.72 (0.29-1.78), 0 | 1.19 (0.82-1.73), 0 |
| Mixed | 0.52 (0.31-0.86) | 0.43 (0.24-0.78) | 1.64 (0.53-5.04) | 1.20 (0.39-3.75) | 0.34 (0.17-0.66) |
| Overall | 1.18 (0.85-1.65), 84 | 1.01 (0.90-1.13), 62 | 1.00 (0.86-1.17), 35 | 1.04 (0.87-1.25), 49 | 1.00 (0.89-1.13), 62 |
| -871A/G | G allele vs. A allele | GG+AG vs. AA | GG vs. AA+AG | GG vs. AA | AG vs. AA |
| Overall | 0.70 (0.54-0.92), 62 | 0.61 (0.42-0.89), 74 | 0.93 (0.56-1.52), 0 | 0.71 (0.43-1.17), 0 | 0.59 (0.40-0.88), 75 |
| -139G/A | A allele vs. G allele | AA+GA vs. GG | AA vs. GG+GA | AA vs. GG | GA vs. GG |
| Overall | 1.18 (0.88-1.60), 50 | 1.21 (0.99-1.49), 49 | 1.10 (0.90-1.35), 21 | 1.18 (0.88-1.60), 50 | 1.22 (0.98-1.52), 21 |
There were 4 case-control studies (1036 cases and 1210 controls) exploring CD209 -139G/A variation and pulmonary tuberculosis susceptibility. We did not find any significant association between CD209 -139G/A variation and the risk of pulmonary tuberculosis in all genetic models (A allele vs. G: P = 0.31; OR=1.12, 95% CI = 0.90-1.39; AA vs. GG: P = 0.27; OR=1.18, 95% CI = 0.88 to 1.60; GA vs. GG: P = 0.07; OR = 1.22, 95% CI = 0.98 to 1.52; AA+AG vs. GG: P = 0.07; OR = 1.21, 95% CI = 0.99 to 1.49; AA vs. AG+GG: P = 0.33; OR = 1.10, 95% CI = 0.90 to 1.35) (Table 2).
Publication bias
Begg’s funnel plot and Egger’s test were conducted to assess the risk of publication bias in the studies. The shape of the funnel plots did not reveal obvious asymmetry under the allele model (-336A/G, P = 0.63; -871A/G, P = 0.308; -139G/A, P = 1.00). Besides, the result of Egger’s test also showed no publication bias. These results indicated there is no risk of publication bias in this meta-analysis.
Test of heterogeneity
Heterogeneity among the included studies is shown in Table 2. For CD209 -871A/G mutation, The I2 showed a stable variation under all comparisons, thus the random effect model was applied. For CD209 -336A/G polymorphism, significant heterogeneity was observed in G vs. A (I2 = 84% P < 0.00001), AG vs. AA (I2 = 62% P = 0.002), AG+GG vs. AA (I2 = 62% P = 0.002). In subgroup analyses, P value for heterogeneity was still significant in African and Asian groups. The heterogeneity sources may be due to the sample size, control source and environmental factors.
Sensitivity analysis
Sensitivity analysis was performed by sequentially omitting one single study at each time for all studies. The pooled ORs showed that none of the studies alone had a significant impact on the results of all CD209 -336A/G, -871A/G and -139G/A variations, suggesting stability of the results of this meta-analysis.
Discussion
Previous studies have shown that the pathogenesis of tuberculosis is affected by environmental factors, host-pathogen interactions and genetic factors [24]. Many polymorphic genes have been reported as candidate genes in tuberculosis susceptibility [4]. Among them, SLC11A1 (formerly NRAMP1), monocyte chemoattractant protein-1 and vitamin D receptor gene polymorphisms were considered as risk factor for tuberculosis susceptibility, while the tumor necrosis factor-α (TNF-α), P2X7 and SP110 (Speckled 110) gene were not related to the susceptibility to tuberculosis [25,26]. CD209 polymorphisms including -336A/G, -871A/G and -139G/A may influence the pathogenesis of tuberculosis by increasing or decreasing the level of DC-SIGN [27].
In a previous meta-analysis of 11 publications including 14 case-control studies, it has been reported that -871A/G mutation do not affect susceptibility to tuberculosis, while the -336A/G polymorphism might be a genetic risk factor that increases tuberculosis susceptibility for Asians in two genetic models (GG vs. AA and GG vs. AG+AA) [28]. Our meta-analysis included 12 case-control studies with 3114 cases and 3088 controls indicated that CD209 -871A/G mutation is associated with resistance to pulmonary tuberculosis in overall population, while -336A/G polymorphism is not associated with the risk of pulmonary tuberculosis in overall population. In subgroup analysis by ethnicity, -336A/G polymorphism is associated with the risk of pulmonary tuberculosis in homozygous model (GG vs. AA) for Asians. However, the -139G/A variation is not associated with susceptibility to pulmonary tuberculosis. Several reasons may explain the different results between previous meta-analysis and ours. First, our meta-analysis focused on pulmonary tuberculosis. Second, our work allowed for the inclusion of two new studies. Thus, our conclusions may be more scientific for the association between CD209 polymorphisms and pulmonary tuberculosis.
Our meta-analysis indicated that Asian subjects with variant homozygous (GG) in CD209 -336A/G polymorphism may have 2.31 fold increased risk of developing pulmonary tuberculosis. This polymorphism was found to regulate DC-SIGN gene promoter activity by affecting a Sp1-like biding site [27]. For -871A/G mutation, the presence of G allele, both in heterozygous model and dominant model, was associated with a protective effect against pulmonary tuberculosis. The -139G/A variation was significantly associated with extra-pulmonary tuberculosis in the study conducted by da Silva [11]. However, we did not find any association between CD209 -139G/A variation and pulmonary tuberculosis in this meta-analysis.
This meta-analysis has some limitations. First, only studies published in English were included, it might be possible that some relevant studies published in other language were missed. Second, heterogeneity was observed in some comparisons. The genetic backgrounds, different environmental exposures may contribute to the heterogeneity. Third, one study included in the meta-analysis did not conform with HWE [20]. Despite of these limitations, our study also has some advantages. We updated the recent data for CD209 polymorphisms and focused on pulmonary tuberculosis. Besides, the methodological issues were all well investigated to provide reliable results.
In conclusion, this meta-analysis indicates that the CD209-871A/G polymorphism is associated with decreased susceptibility to pulmonary tuberculosis. For -336A/G mutation, the G allele may contribute to the occurrence of pulmonary tuberculosis in Asians. However, -139G/A variation is not associated with pulmonary tuberculosis risk. Future studies with large sample size and stringent design are required.
Acknowledgements
This study was supported by National Natural Science Foundation of China Grant 81470227, and National Key Technology R&D Program of the 12th Five-year Development Plan (2012BAI05B01).
Disclosure of conflict of interest
None.
References
- 1.Zaman K. Tuberculosis: a global health problem. J Health Popul Nutr. 2010;28:111–113. doi: 10.3329/jhpn.v28i2.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zumla A, George A, Sharma V, Herbert N Baroness Masham of Ilton. WHO’s 2013 global report on tuberculosis: successes, threats, and opportunities. Lancet. 2013;382:1765–1767. doi: 10.1016/S0140-6736(13)62078-4. [DOI] [PubMed] [Google Scholar]
- 3.Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol. 2011;2011:405310. doi: 10.1155/2011/405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 2010;90:71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Soilleux EJ, Barten R, Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 6.Tailleux L, Schwartz O, Herrmann JL, Pivert E, Jackson M, Amara A, Legres L, Dreher D, Nicod LP, Gluckman JC, Lagrange PH, Gicquel B, Neyrolles O. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med. 2003;197:121–127. doi: 10.1084/jem.20021468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geijtenbeek TB, Torensma R, van Vliet SJ, van Duijnhoven GC, Adema GJ, van Kooyk Y, Figdor CG. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 8.den Dunnen J, Gringhuis SI, Geijtenbeek TB. Innate signaling by the C-type lectin DC-SIGN dictates immune responses. Cancer Immunol Immunother. 2009;58:1149–1157. doi: 10.1007/s00262-008-0615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gringhuis SI, den Dunnen J, Litjens M, van Het Hof B, van Kooyk Y, Geijtenbeek TB. C-type lectin DC-SIGN modulates Toll-like receptor signaling via Raf-1 kinase-dependent acetylation of transcription factor NF-kappaB. Immunity. 2007;26:605–616. doi: 10.1016/j.immuni.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Geijtenbeek TB, Van Vliet SJ, Koppel EA, Sanchez-Hernandez M, Vandenbroucke-Grauls CM, Appelmelk B, Van Kooyk Y. Mycobacteria target DC-SIGN to suppress dendritic cell function. The J Exp Med. 2003;197:7–17. doi: 10.1084/jem.20021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Silva RC, Segat L, da Cruz HL, Schindler HC, Montenegro LM, Crovella S, Guimarães RL. Association of CD209 and CD209L polymorphisms with tuberculosis infection in a Northeastern Brazilian population. Mol Biol Rep. 2014;41:5449–5457. doi: 10.1007/s11033-014-3416-y. [DOI] [PubMed] [Google Scholar]
- 12.Zheng R, Zhou Y, Qin L, Jin R, Wang J, Lu J, Wang W, Tang S, Hu Z. Relationship between polymorphism of DC-SIGN (CD209) gene and the susceptibility to pulmonary tuberculosis in an eastern Chinese population. Hum Immunol. 2011;72:183–186. doi: 10.1016/j.humimm.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi K, Yuliwulandari R, Yanai H, Lien LT, Hang NT, Hijikata M, Keicho N, Tokunaga K. Association of CD209 polymorphisms with tuberculosis in an Indonesian population. Hum Immunol. 2011;72:741–745. doi: 10.1016/j.humimm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Wu R, Li B. A multiplicative-epistatic model for analyzing interspecific differences in outcrossing species. Biometrics. 1999;55:355–365. doi: 10.1111/j.0006-341x.1999.00355.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang YG, Li XB, Zhang J, Huang J, He C, Tian C, Deng Y, Wan H, Shrestha D, Yang YY, Fan H. The I/D polymorphism of angiotensin-converting enzyme gene and asthma risk: a meta-analysis. Allergy. 2011;66:197–205. doi: 10.1111/j.1398-9995.2010.02438.x. [DOI] [PubMed] [Google Scholar]
- 16.Olson JM, Foley M. Testing for homogeneity of Hardy-Weinberg disequilibrium using data sampled from several populations. Biometrics. 1996;52:971–979. [PubMed] [Google Scholar]
- 17.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vannberg FO, Chapman SJ, Khor CC, Tosh K, Floyd S, Jackson-Sillah D, Crampin A, Sichali L, Bah B, Gustafson P, Aaby P, McAdam KP, Bah-Sow O, Lienhardt C, Sirugo G, Fine P, Hill AV. CD209 genetic polymorphism and tuberculosis disease. PLoS One. 2008;3:e1388. doi: 10.1371/journal.pone.0001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barreiro LB, Neyrolles O, Babb CL, van Helden PD, Gicquel B, Hoal EG, Quintana-Murci L. Length variation of DC-SIGN and L-SIGN neck-region has no impact on tuberculosis susceptibility. Hum Immunol. 2007;68:106–112. doi: 10.1016/j.humimm.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naderi M, Hashemi M, Taheri M, Pesarakli H, Eskandari-Nasab E, Bahari G. CD209 promoter -336 A/G (rs4804803) polymorphism is associated with susceptibility to pulmonary tuberculosis in Zahedan, southeast Iran. J Microbiol Immunol Infect. 2014;47:171–175. doi: 10.1016/j.jmii.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Ogarkov O, Mokrousov I, Sinkov V, Zhdanova S, Antipina S, Savilov E. ‘Lethal’ combination of Mycobacterium tuberculosis Beijing genotype and human CD209 -336G allele in Russian male population. Infect Genet Evol. 2012;12:732–736. doi: 10.1016/j.meegid.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Olesen R, Wejse C, Velez DR, Bisseye C, Sodemann M, Aaby P, Rabna P, Worwui A, Chapman H, Diatta M, Adegbola RA, Hill PC, Østergaard L, Williams SM, Sirugo G. DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes Immunity. 2007;8:456–467. doi: 10.1038/sj.gene.6364410. [DOI] [PubMed] [Google Scholar]
- 23.Alagarasu K, Selvaraj P, Swaminathan S, Raghavan S, Narendran G, Narayanan PR. CCR2, MCP-1, SDF-1a & DC-SIGN gene polymorphisms in HIV-1 infected patients with & without tuberculosis. Indian J Med Res. 2009;130:444–450. [PubMed] [Google Scholar]
- 24.Bellamy R. Susceptibility to mycobacterial infections: the importance of host genetics. Genes Immunity. 2003;4:4–11. doi: 10.1038/sj.gene.6363915. [DOI] [PubMed] [Google Scholar]
- 25.Yi L, Cheng D, Shi H, Huo X, Zhang K, Zhen G. A meta-analysis of P2X7 gene-762T/C polymorphism and pulmonary tuberculosis susceptibility. PLoS One. 2014;9:e96359. doi: 10.1371/journal.pone.0096359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei X, Zhu H, Zha L, Wang Y. SP110 gene polymorphisms and tuberculosis susceptibility: a systematic review and meta-analysis based on 10 624 subjects. Infect Genet Evol. 2012;12:1473–1480. doi: 10.1016/j.meegid.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Sakuntabhai A, Turbpaiboon C, Casademont I, Chuansumrit A, Lowhnoo T, Kajaste-Rudnitski A, Kalayanarooj SM, Tangnararatchakit K, Tangthawornchaikul N, Vasanawathana S, Chaiyaratana W, Yenchitsomanus PT, Suriyaphol P, Avirutnan P, Chokephaibulkit K, Matsuda F, Yoksan S, Jacob Y, Lathrop GM, Malasit P, Desprès P, Julier C. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang K, Deng S, Lu W, Wang F, Jia S, Li F, Yu L, Chen M. Association between CD209 -336A/G and -871A/G polymorphisms and susceptibility of tuberculosis: a meta-analysis. PLoS One. 2012;7:e41519. doi: 10.1371/journal.pone.0041519. [DOI] [PMC free article] [PubMed] [Google Scholar]


