Abstract
MicroRNAs (miRNAs) play a crucial role in cancer development and progression of hepatocellular carcinoma (HCC). In this study, we aimed to analyze the role of microRNA-194 (miR-194) in HCC. We found that miR-194 expression was significantly reduced in HCC and its expression was an independent poor prognostic factor for HCC patient overall and disease-free survival rate. A significant correlation was observed between miR-194 reduction and unfavourable variables including tumor size (P = 0.0315), histologic grade (P = 0.0038), TNM stage (P = 0.0083), intrahepatic metastasis (P = 0.0184). Overexpression of miR-194 in HCC cell lines HepG2 and Hep3B inhibited cell proliferation by blocking G1-S transition and inducing apoptosis. Mitogen-activated protein kinase 4 (MAP4K4), a potential target gene of miR-194, was inversely correlated with miR-194 expression in HCC tissues and cell lines. Further studies demonstrated that miR-194 regulated the progression of HCC through directly inhibiting the expression of MAP4K4 and the restoration of MAP4K4 expression reversed the inhibitory effects of miR-194 on HCC cell proliferation. Together, our findings indicate that miR-194 may serve as a valuable prognostic marker and promising interventional therapeutic target for HCC.
Keywords: microRNA-194, hepatocellular carcinoma, MAP4K4, proliferation, interventional therapy
Introduction
Human hepatocellular carcinoma (HCC) is one of the most common cancer worldwide, especially in China, East Asia and South Africa [1]. It is estimated that there are over 700,000 new HCC cases are diagnosed every year, equaling the number of HCC-related deaths. The mortality of HCC has become higher than that in any other cancer, and the 5-year survival rate is below 5% [2]. Although recent advances in surgical techniques (resection, transplantation, or local ablation) and interventional therapy (chemoembolization) have improved survival, only about 50% of HCC patients are eligible for potential curative treatments or chemoembolization, and the incidence of recurrence remains high [3]. Thus, finding new biological marker for prognosis and the development of novel systemic therapies for advanced disease are of paramount importance.
miRNAs are a class of evolutionarily, single-stranded, noncoding RNAs that act as post-transcriptional regulators of gene expression through interacting with the 3’-untranslated region (3’UTR) of target messenger RNAs (mRNAs) by translational repression and/or cleavage [4]. miRNAs are implicated in various biological processes including cell proliferation, differentiation, metabolism, and stem cell renewal [5]. Furthermore, ample evidences have confirmed that dysregulation of miRNAs contributes to the development and progression of human malignancies, including HCC [6]. Moreover, miRNA-targeted treatment approaches reveal enormous potential in controlling the aggressive pathology of HCC [7,8]. Therefore, miRNAs have been proposed as promising prognostic markers and therapeutic targets for HCC patients.
The aim of this study was to determinate the role of miR-194 in HCC. We found that miR-194 was down-regulated in the majority of HCC tissues, and low miR-194 expression linked with poor prognostic significance in HCC patients. In vitro and in vivo studies demonstrated that miR-194 repressed cell viability and proliferation, and induced G1 arrest and apoptosis in HCC cells. Moreover, we identified that miR-194 exerted its biological function via inhibiting MAP4K4 expression. Our study elucidates the underlying mechanism by which miR-194 inhibits HCC, and proposes miR-194 as a potential role in the diagnosis and therapy of HCC.
Materials and methods
Patients and tissue samples
Fifty-six paired human HCC samples and matched adjacent normal tissues were obtained from patients who underwent curative resection in our hospital between July, 2013 and July, 2014. The samples were snap frozen in liquid nitrogen and stored at -80°C for later RNA extraction or formalin-fixed and paraffin embedded for imunohistochemistry. None of the patients were pretreated with interventional or other treatment prior to surgery. Histopathology was evaluated by two certified pathologists in our hospital. All specimens were obtained with informed consent from patients, and approved by the Ethics Committee of the First Hospital Affiliated to the Xinxiang Medical College.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using an RNeasy Mini and miRNeasy Mini Kits (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. qRT-PCR for miR-194 detection was performed using 10 ng of total RNA and TaqMan MicroRNA Assay kit (ABI, Foster City, CA, USA) on a LightCycler 480 System II (Roche, Mannhein, Germany). The relative quantification of miR-194 was calculated using the. The qRT-PCR data were normalized using 2-ΔΔCt method relative to U6 small nuclear RNA.
Cell culture and generation of stable cell lines
Human HCC cell lines HepG2 and Hep3B were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and grown in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and 100 units/ml penicillin/streptomycin at 37°C in a humidified incubator containing 5% CO2. To generate the stable miR-194-overexpressing HCC cell line, the primary miR-194 gene was amplified from human genomic DNA using Accuprime Taq polymerase (Invitrogen, Carlsbad, CA, USA) and cloned into pMSCV-PIG vectors. HepG2 and Hep3B cells were infected with retroviruses generated in phoenix cells as described previously [9]. After 72 h, the cells were selected with 2 μg/ml puromycin. Primers used to amplify primary miR-194 gene: forward, 5’-TCGAGGATCCTAAAACAGGCTGAAAATCTTT-3’, reverse, 5’-TCGAGCTAGCTAGAACATGAATAAATCGAGA-3’.
Cell viability assay
HepG2 and Hep3B were plated into 96-well plates (1×103 cells/well) and 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT, Sigma-Aldrich, St. Louis, MO, USA) assay was employed to assess cell viability at indicated time points (1, 2, 3, 4, 5 and 6 days after seeding into plates), and incubation was performed at 37°C for 2 h. The absorbance wavelength was measured at 570 nm, and 620 nm as the reference wavelength.
3H thymidine incorporation assay
For 3H thymidine incorporation assay, cells were plated onto 12-well plates at a density of 1×104 cells/well. Cells were incubated with 3H thymidine (1 μCi/ml) for 6 h before harvesting the cells with 10 mM EDTA. The amount of radioactivity was detected by liquid scintillation counting system (Beckman Coulter, Fullerton, CA, USA).
Cell cycle analysis
The cells were trypsinized and fixed in 70% cold ethanol at 4°C overnight. The fixed cells were washed with PBS, and re-suspended in PBS solution with 20 μg/ml RNase A (Sigma, St. Louis, MO, USA) and 50 μg/ml of propidium iodide (PI, Sigma) for 20 min on ice to label DNA, cell Sample acquisition and analyses were performed with a Becton Dickinson FACS/Calibur cytometer (Becton Dickinson Biosciences, Inc., NJ, USA) using the FACSuite software.
Apoptosis assay
For cell apoptosis assay, Annexin-V fluorescein isothiocyanate (FITC) Apoptosis Kit (Calbiochem, San Diego, CA, USA) and PI was used to assess the apoptotic effect of miR-194. Briefly, cells were harvested by trypsinization, washed with cold PBS, and re-suspended in 500 µl cold 1× binding buffer containing 1.25 μl annexin V-FITC, followed by 15 min incubation at room temperature in the dark. Then cells were re-suspended in 500 ml cold 1× binding buffer with 10 μl of PI, incubated for 4 hours and detected using Becton Dickinson FACS/Calibur cytometer (Becton Dickinson Biosciences).
Xenograft model
Balb/c nude mice (5~6 week(s) old, male, ~20 g) were received with ultrasound guided subcutaneous injections of 1×107 cells in the flank with 1×107 cells suspended in 100 μl of PBS. Tumor volumes were measured every week. Mice were sacrificed at 4 weeks after injection. All procedures were performed in accordance with the Guidelines of the Chinese Association for Laboratory Animal Science.
Immunohistochemistry assay
Paraffin-embedded tissue sections (4 μm thick) were deparaffinized in xylene, and rehydrated in graded alcohol. Antigen retrieval was performed in sodium citrate buffer for 2 min were placed in an autoclave cooker. Subsequently, endogenous peroxidase activity was blocked by incubation with 0.3% hydrogen peroxide for 10 min. The primary MAP4K4 monoclonal antibodies (Santa Cruz Biotech, Santa Cruz, CA) were diluted to 1:50 and incubated with the sections at 4°C in a humid chamber overnight. The biotinylated secondary antibody antibody (ZSGB-Bio, Beijing, China) was used to detect the primary antibody. Then, antigen-antibody reaction was visualized with diaminobenzidine serving as the chromogen.
Western blot assays
Western blot assay was performed as described previously [10]. Briefly, cells were lysed on ice in RIPA buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1% TritonX-100, 5 mM ethylenediaminetetraacetic acid). BCA Kit (Pierce, IL, USA) was used to detect protein concentration. Protein (30 μg) was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred onto nitrocellulose (NC) membranes. Membranes were incubated at 4°C overnight with primary antibodies: MAP4K4 (Santa Cruz Biotech, Santa Cruz, CA). GAPDH (Cell Signaling Technology, Boston, USA) was used as a control. The bands were visualized with the Fusion FX7 system (VilberLourmat, France).
Luciferase assays
The 3’UTR sequence of MAP4K4 predicted to interact with miR-194 or the mutated sequence containing the potential binding region was synthesized by PCR and inserted into the HindIII and EcoRI restriction sites downstream of the luciferase open reading frame in the pGL3-promoter Luciferase vector (Promega, Madison, WI, USA). The correct clones were confirmed by sequencing. HCC cells were seeded into a 24-well plate (1×104 per well), and were co-transfected with a luciferase reporter construct (200 ng) and miRNA mimics (50 nM) using LipofectamineTM 2000 reagent (Invitrogen, Carlsbad, CA, USA). After 48 h, luciferase assays was measured using dual luciferase system kit (Promega, Madison, WI, USA). Firefly luciferase activity was normalized against Renilla luciferase gene activity.
Ad-MAP4K4 adenovirus construction and infection
Both the Ad-MAP4K4 (lacking 3’UTR) adenovirus and the control adenovirus were constructed with the Ad-easy system as previous described [11]. HepG2 and Hep3B cells were seeded into 6-well plate at a density of 5×105 cells per well and incubated overnight until 50-70% confluent. The Ad-MAP4K4 and control adenovirus were respectively infected into cells at an MOI of 50. Further analyses performed 48 h after infection.
Statistical analysis
All data were presented as mean ± standard deviation (SD). Values analyses were carried out using SPSS 15.0 software (SPSS, Inc., Chicago, IL, USA). The relationship between miR-194 expression and clinicopathologic features of HCC was analyzed using the Fisher’s exact tests. Univariate survival analysis was performed using Kaplan-Meier method and the Log-rank test. The statistical significance between groups was analysed using Student’s t-test or one-way ANOVA. Differences were considered statistically significant at P<0.05.
Results
Clinical significance of miR-194 expression in HCC
We first determined the miR-194 expression in 56 pairs of HCC tissue and matched adjacent non-tumor tissues from HCC patients who received hepatotectomy by qRT-PCR. The miR-194 expression levels were classified either low or high according to the median of the cohort. As shown in Table 1, the results showed that miR-194 was downregulated in 40 HCC tissues compared with non-tumor tissues. Low expression of miR-194 was found to significantly correlate with unfavourable variables, including tumour size (P = 0.0124), histologic grade (P = 0.0119), TNM stage (P = 0.0286), Intrahepatic metastasis (P = 0.0184). Furthermore, the patients in low miR-194 expression group associated with worse overall survival and disease-free survival (P = 0.0252 and P = 0.0479, respectively) (Figure 1A and 1B).
Table 1.
Clinicopathologic factors and miR-194 expression in HCC
| Group | NO. | miR-194 expression | P value* | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| Nontumor tissue | 56 | 42 | 14 | 0.0001 |
| HCC tissue | 56 | 16 | 40 | |
| Sex | ||||
| Male | 32 | 15 | 17 | 0.4303 |
| Female | 24 | 14 | 10 | |
| Age | ||||
| ≤50 | 26 | 12 | 14 | 0.4210 |
| >50 | 30 | 18 | 12 | |
| Tumor size (cm) | ||||
| ≤2 | 28 | 18 | 10 | 0.0315 |
| >2 | 28 | 9 | 19 | |
| Histologic grade | ||||
| Well and moderately differentiation | 27 | 20 | 7 | 0.0038 |
| Poorly differentiation | 29 | 10 | 19 | |
| TNM stage | ||||
| INM | 25 | 18 | 7 | 0.0083 |
| III.0 | 31 | 11 | 20 | |
| Intrahepatic metastasis | ||||
| Negative | 26 | 18 | 8 | 0.0184 |
| Positive | 30 | 11 | 19 | |
Based on the American Joint Committee on Cancer/International Union Against Cancer staging manual (2015).
Differences between the variables were assessed with Fisher’s exact tests.
Figure 1.
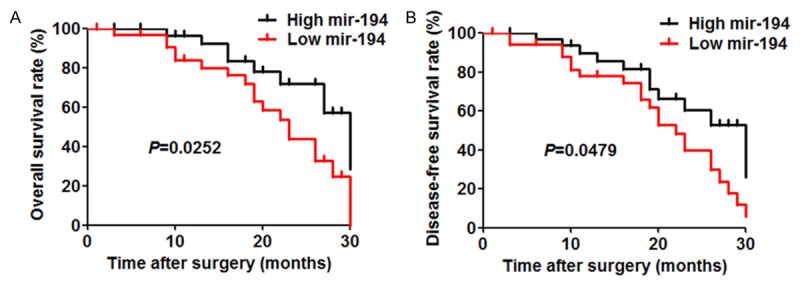
Prognostic value of miR-194 in HCC patients. A and B. Overall survival rate and disease-free survival rate of 56 HCC patients with different miR-194 expression.
miR-194 suppresses HCC cell proliferation
To further evaluate miR-194 function in HCC cells, we studied the proliferation of HCC cells (HepG2 and Hep3B) after miR-194 overexpression. The expression of miR-194 was assessed by RT-PCR after transfection (Figure 2A). The effect of miR-194 on the proliferation of cells was measured using MTT assays. MTT assays revealed that miR-194 overexpression significantly inhibited the viability of HepG2 and Hep3B cells, and this effect increased with time (Figure 2B). 3H thymidine incorporation assays also showed that miR-194 overexpression repressed HCC cell DNA synthesis (Figure 2C). Quantification of apoptosis by Annexin V/PI double labelling indicated that a remarkably higher apoptotic index was detected in miR-194-overexpressing cells relative to control cells (Figure 2D). Cell cycle analysis indicated that miR-194 overexpression caused a considerable inhibition of cell cycle progression, leading to an accumulation of cells in the G0/G1 phase compared with the control group (Figure 2E).
Figure 2.
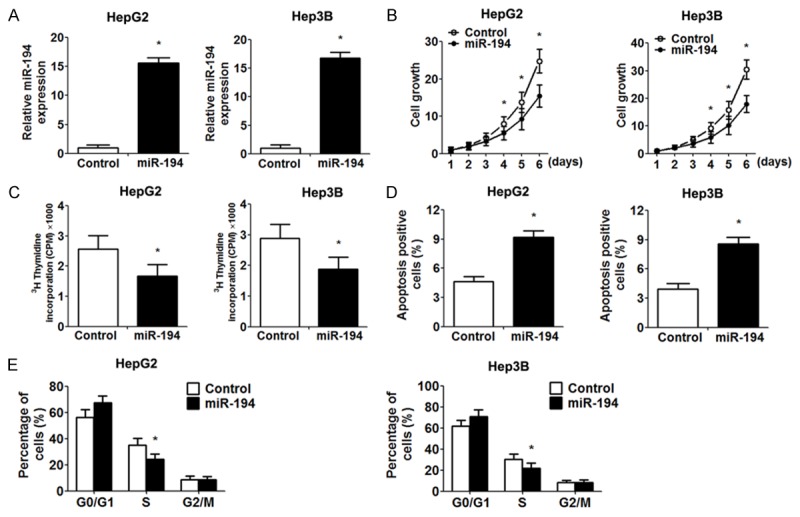
The impact of miR-194 overexpression on proliferation. A. Validation of miR-194 overexpression in HepG2 and Hep3B cells by RT-PCR analysis. B. MTT assays were performed to examine cells proliferation at the indicated time points. C. The value of 3H thymidine incorporation was measured. D. The percentage of apoptotic cells (Annexin V-FITC positive, PI negative). E. Frequencies of cells at different stages of the cell cycle. Data are expressed as the mean ± SD. *, P<0.05 vs. control group, n = 5.
miR-194 inhibits tumor growth in vivo
To get more insight into the relevance of miR-194 in vivo, miR-194-overexpressing HCC cells and control cells were injected subcutaneously into the left flank of nude mice. Consistent with the in vitro study, miR-194 significantly suppressed the xenograft tumor growth, including tumor volume and weight, compared with controls (Figure 3A-C). Expression of MAP4K4 in the xenografts was determined by immunohistochemistry. The miR-194-overexpressing-derived tumor xenografts displayed a considerably lower proportion of MAP4K4-positive cells (Figure 3D).
Figure 3.
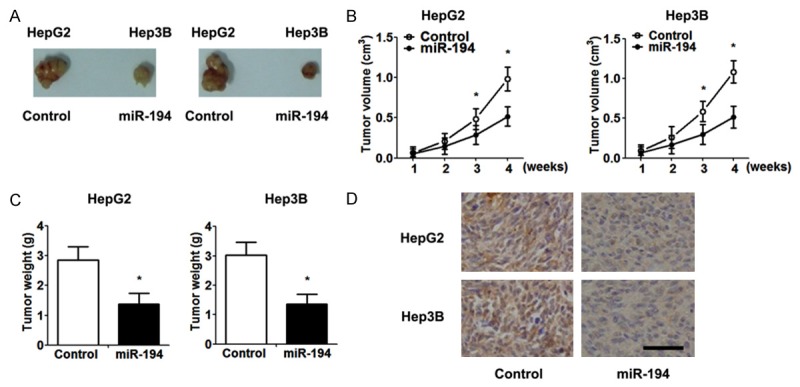
The overexpression of miR-194 suppresses tumor growth in vivo. A. Macroscopic xenograft tumor in nude mice with injection of HepG2 and Hep3B cells. B. The growth curves of xenografts. Tumor volume was determined using the following formula: volume = (length×width2)/2. C. Xenograft weight was measured when the mice were sacrificed. D. Representative images of tumor xenografts stained by anti-MAP4K4 antibody (scale bar = 50 μm). Data are expressed as the mean ± SD. *, P<0.05 vs. control group, n = 5.
miR-194 directly inhibits MAP4K4 gene expression via targeting its 3’UTR
miRNAs exert biological function via their target gene. To determinate whether MAP4K4 was the target gene of miR-194, we first examined the correlation between MAP4K4 and miR-194 expression in HCC tissue specimens. Immunohistochemistry analysis showed that the expression of MAP4K4 in tumors with high miR-194 expression was significantly lower than those in tumors with low miR-194 expression (Figure 4A). Western blot analysis demonstrated that overexpression of miR-194 significantly down-regulated MAP4K4 expression (Figure 4B). Then, we employed TargetScan 5.1. algorithms to search the potential binding sequence of miR-194 in 3’UTR of MAP4K4 and cloned the wild type and mutant 3’UTR fragment into a luciferase reporter gene system (Figure 4C). As expected, miR-194 in both HCC cell lines led to significantly reduced luciferase activity of MAP4K4 containing a wild type 3’UTR but did not suppress activity of MAP4K4 with a mutant (Figure 4D).
Figure 4.
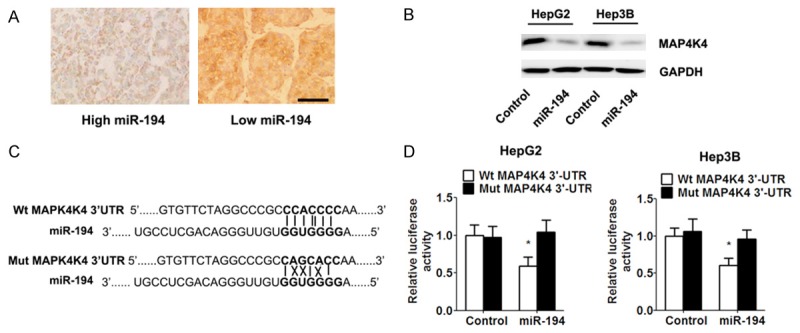
miR-194 directly targeted MAP4K4 by binding toits 3’UTR. A. Immunohistochemistry showed an inverse relationship between the expression of miR-194 and MAP4K4 in HCC specimens (scale bar = 50 μm). B. Western blot analysis of the expression of MAP4K4 in indicated cells. C. Predicted miR-194 target sequences in 3’UTR of MAP4K4 is shown (solid lines indicate matching base pairs and crosses represent non-matching base pairs). D. miR-194-overexpressing and control cells were transfected with the luciferase reporter plasmid carrying the wild type/mutant 3’UTR sequences of MAP4K4. A dual-luciferase reporter system analysis was performed. Data are expressed as the mean ± SD. *, P<0.05 vs. control group, n = 5.
Reintroduction of MAP4K4 reverses miR-194-induced inhibitory effects
To determine whether MAP4K4 is a functional target of miR-194, we restored the MAP4K4 expression in HCC cells by infecting MAP4K4 adenovirus (lacking the 3’UTR region). Western blot analysis determinate increased expression of MAP4K4 protein in the HCC cells after MAP4K4 adenovirus infection (Figure 5A). Functionally, restoration of MAP4K4 expression reversed the inhibitory effects of exogenous miR-194 on proliferation, resulting in significant increase of cell viability and DNA synthesis (Figure 5B and 5C). Similarly, re-expression of MAP4K4 exhibited an apparent decreased apoptosis rate and rescued S phase in HCC cells (Figure 5D and 5E).
Figure 5.
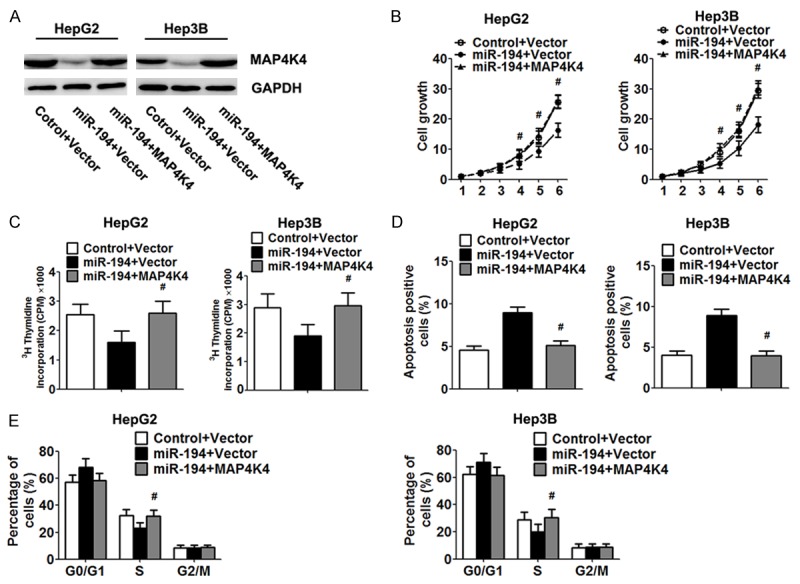
Re-expression of MAP4K4 abrogated the effect of miR-194. HepG2 and Hep3B cells were infected with MAP4K4 and control adenovirus for 48 h. A. Expression of MAP4K4 protein was analysed by Western blot. B. Cell viability was measured by the MTT assay. C. Proliferation capacity was examined by 3H thymidine incorporation assay. D, E. The cell apoptosis and cycle progression was analysed by flow cytometry. Data are expressed as the mean ± SD. *, P<0.05 vs. control group, n = 5.
Discussion
miRNAs can act as either oncogenes or tumor suppressors on the basis of the roles of their target genes. Increasing evidences strongly suggest that that the deregulation or dysfunction of miRNAs contributes to human carcinogenesis and cancer progression [12,13]. Recent studies have shown that miR-194 is associated with various malignancies, such as pancreatic cancer [14], osteosarcoma [15], gastric [16] and colon cancers [17]. The high expression of miR-194 in the liver has been known for a long time, but its function has not been clearly characterized.
Our results found that the expression of miR-194 was reduced in HCC tissues, and the level of miR-194 was associated with tumour size, histologic grade, TNM stage, intrahepatic metastasis, suggesting that miR-194 might be closely related with tumorigenesis and differentiation of HCC. Moreover, patients with low expression of miR-194 had shorter overall and disease-free survival. We demonstrated that miR-194 was an independent prognostic marker for predicting overall and disease-free survival of HCC patients. Altogether, these data indicated that the status of miR-194 was critical for prognosis determination in HCC patients. Overexpression of miR-194 in HCC cells suppressed proliferation in vitro and in vivo through blocking G1-S transition and inducing apoptosis, suggesting that miR-194 acted as a novel tumor suppressor and played a critical role in hepatocarcinogenesis. Recent studies have reported that miR-194 is involved in epithelial mesenchymal transition, and overexpressing miR-194 suppresses HCC cell migration and invasiveness in culture and metastatic seeding in mice [18,19]. Our results were consistent with previous studies demonstrating that miR-608 inhibits biological features of HCC.
Furthermore, current study demonstrated that an inverse correlation between the expression of miR-194 and MAP4K4 protein was observed in xenograft tumor and HCC tissues. Up-regulation of miR-194 decreased the expression of MAP4K4 protein and the luciferase reporter activity in HCC cells. This phenomenon coincides with previous study that MAP4K4 is the direct target of miR-194 in colorectal cancer [20]. MAP4K4, a germinal center protein kinase belonging to the mammalian STE20/MAP4K family, is implicated in the activation of the c-jun N-terminal kinase (JNK) pathway [21]. Increasing studies have documented that MAP4K4 participates in many types of human cancer, and targeted reduction of MAP4K4 by small interfering RNA inhibits tumorigenesis in a variety of cancers, such as ovarian cancer, breast cancer, prostate cancer, and malignant melanoma [22]. Moreover, elevated MAP4K4 expression has an independent prognostic value in predicting overall survival for patients with lung adenocarcinoma [23]. Recent study provided direct evidence that MAP4K4 was aberrantly overexpressed in HCCs relative to adjacent non-tumor liver tissues, and knockdown of the MAP4K4 expression reduced cell proliferation, blocked cell cycle at S phase, and increased apoptosis [24]. These findings imply an oncogenic role for MAP4K4. In this study, we demonstrated that restoration of MAP4K4 expression abrogated the functional effect of miR-194 on HCC cell viability, proliferation and apoptosis, supporting that MAP4K4 overexpression is a notable feature and may be one of the many critical events that occur in HCC tumourigenesis. Taken together, these data provide solid evidences to support that miR-194 exerts it inhibitory effect on HCC, at least in part, through inhibiting MAP4K4.
In conclusion, our results indicate that miR-194 over expression is significantly associated with tumour size, histologic grade, TNM stage, and intrahepatic metastasis and serves as an independent prognostic factor for HCC. Our data reveal the important molecular mechanism by which miR-194 exerts its negative effects on cell viability and proliferation in HCC cells by suppressing of MAP4K4. This newly identified target of miR-194 may provide a potential prognostic marker and therapeutic target for HCC patients.
Disclosure of conflict of interest
None.
References
- 1.Fang F, Chang RM, Yu L, Lei X, Xiao S, Yang H, Yang LY. MicroRNA-188-5p Suppresses Tumor Cell Proliferation and Metastasis by Directly Targeting FGF5 in Hepatocellular Carcinoma. J Hepatol. 2015;63:874–85. doi: 10.1016/j.jhep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Huang Z, Ye Q, Ming Y, Zhang S, Zhao Y, Liu L, Wang Q, Cheng K. Prognostic significance and anti-proliferation effect of microRNA-365 in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:1705–1711. [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, Qin J, Lu S. Up-regulation of miR-877 induced by paclitaxel inhibits hepatocellular carcinoma cell proliferation though targeting FOXM1. Int J Clin Exp Pathol. 2015;8:1515–1524. [PMC free article] [PubMed] [Google Scholar]
- 4.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 6.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dou C, Wang Y, Li C, Liu Z, Jia Y, Li Q, Yang W, Yao Y, Liu Q, Tu K. MicroRNA-212 suppresses tumor growth of human hepatocellular carcinoma by targeting FOXA1. Oncotarget. 2015;6:13216–13228. doi: 10.18632/oncotarget.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G, Shen Q, Li C, Li D, Chen J, He M. Identification of circulating MicroRNAs as novel potential biomarkers for hepatocellular carcinoma detection: a systematic review and meta-analysis. Clin Transl Oncol. 2015;17:684–93. doi: 10.1007/s12094-015-1294-y. [DOI] [PubMed] [Google Scholar]
- 9.Cai J, Xu L, Cai Z, Wang J, Zhou B, Hu H. MicroRNA-146b-5p inhibits the growth of gallbladder carcinoma by targeting epidermal growth factor receptor. Mol Med Rep. 2015;12:1549–1555. doi: 10.3892/mmr.2015.3461. [DOI] [PubMed] [Google Scholar]
- 10.Qi J, Zhao P, Li F, Guo Y, Cui H, Liu A, Mao H, Zhao Y, Zhang X. Down-regulation of Rab17 promotes tumourigenic properties of hepatocellular carcinoma cells via Erk pathway. Int J Clin Exp Pathol. 2015;8:4963–4971. [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S, Xie Y, Yang P, Chen P, Zhang L. HCV core protein-induced down-regulation of microRNA-152 promoted aberrant proliferation by regulating Wnt1 in HepG2 cells. PLoS One. 2014;9:e81730. doi: 10.1371/journal.pone.0081730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 13.Han G, Wang Y, Bi W, Jia J, Wang W. MicroRNA-124 functions as a tumor suppressor and indicates prognosis in human osteosarcoma. Exp Ther Med. 2015;9:679–684. doi: 10.3892/etm.2014.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KH, Lee JK, Choi DW, Do IG, Sohn I, Jang KT, Jung SH, Heo JS, Choi SH, Lee KT. Postoperative Prognosis Prediction of Pancreatic Cancer With Seven MicroRNAs. Pancreas. 2015;44:764–768. doi: 10.1097/MPA.0000000000000346. [DOI] [PubMed] [Google Scholar]
- 15.Han K, Zhao T, Chen X, Bian N, Yang T, Ma Q, Cai C, Fan Q, Zhou Y, Ma B. microRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. Int J Oncol. 2014;45:1437–1449. doi: 10.3892/ijo.2014.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Wang Y, Zang W, Du Y, Li M, Zhao G. miR-194 targets RBX1 gene to modulate proliferation and migration of gastric cancer cells. Tumour Biol. 2015;36:2393–2401. doi: 10.1007/s13277-014-2849-1. [DOI] [PubMed] [Google Scholar]
- 17.Zhao HJ, Ren LL, Wang ZH, Sun TT, Yu YN, Wang YC, Yan TT, Zou W, He J, Zhang Y, Hong J, Fang JY. MiR-194 deregulation contributes to colorectal carcinogenesis via targeting AKT2 pathway. Theranostics. 2014;4:1193–1208. doi: 10.7150/thno.8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove R, Xu R, Huang W. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148–2157. doi: 10.1002/hep.23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao C, Li Y, Huan L, Zhang Y, Zhao F, Wang Q, Liang L, Ding J, Liu L, Chen T, Li J, Yao M, Huang S, He X. NF-kappaB signaling relieves negative regulation by miR-194 in hepatocellular carcinoma by suppressing the transcription factor HNF-1alpha. Sci Signal. 2015;8:ra75. doi: 10.1126/scisignal.aaa8441. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Shen ZL, Gao ZD, Zhao G, Wang CY, Yang Y, Zhang JZ, Yan YC, Shen C, Jiang KW, Ye YJ, Wang S. MiR-194, commonly repressed in colorectal cancer, suppresses tumor growth by regulating the MAP4K4/c-Jun/MDM2 signaling pathway. Cell Cycle. 2015;14:1046–1058. doi: 10.1080/15384101.2015.1007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang N, Wang Y, Hui L, Li X, Jiang X. Silencing SOX2 Expression by RNA Interference Inhibits Proliferation, Invasion and Metastasis, and Induces Apoptosis through MAP4K4/JNK Signaling Pathway in Human Laryngeal Cancer TU212 Cells. J Histochem Cytochem. 2015;63:721–33. doi: 10.1369/0022155415590829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins CS, Hong J, Sapinoso L, Zhou Y, Liu Z, Micklash K, Schultz PG, Hampton GM. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc Natl Acad Sci U S A. 2006;103:3775–3780. doi: 10.1073/pnas.0600040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu MH, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH. Expression and prognostic significance of MAP4K4 in lung adenocarcinoma. Pathol Res Pract. 2012;208:541–548. doi: 10.1016/j.prp.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Liu AW, Cai J, Zhao XL, Jiang TH, He TF, Fu HQ, Zhu MH, Zhang SH. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin Cancer Res. 2011;17:710–720. doi: 10.1158/1078-0432.CCR-10-0331. [DOI] [PubMed] [Google Scholar]


