Abstract
Early in prostate cancer development, tumor cells express vascular endothelial growth factor C (VEGF-C), a secreted molecule that is important in angiogenesis progression. CC-chemokine receptor 7 (CCR7), another protein involved in angiogenesis, is strongly expressed in most human cancers, where it activated promotes tumor growth as well as favoring tumor cell invasion and migration. The present study aimed to investigate the effect of down-regulating CCR7 expression on the growth of human prostate cancer cells stimulated by VEGFC. The CCR7-specific small interfering RNA (siRNA) plasmid vector was constructed and then transfected into prostate cancer cells. The expression of CCR7 mRNA and protein was detected by quantitative polymerase chain reaction and western blot analysis, respectively. Cell proliferation, apoptosis, cell cycle distribution and cell migration were assessed following knockdown of CCR7 by RNA interference (RNAi). Western blot analysis was used to identify differentially expressed angiogenesis- and cell cycle-associated proteins in cells with silenced CCR7. The expression levels of CCR7 in prostate cancer cells transfected with siRNA were decreased, leading to a significant inhibition of prostate cancer cell proliferation, migration and invasion induced by VEGFC. Western blot analysis revealed that silencing of CCR7 may inhibit vascular endothelial growth factor, matrix metalloproteinase (MMP)-2 and MMP-9 protein expression. In conclusion, the present study demonstrated that RNAi can effectively silence CCR7 gene expression and inhibit the growth of prostate cancer cells, which indicates that there is a potential of targeting CCR7 as a novel gene therapy approach for the treatment of prostate cancer.
Keywords: Prostate cancer, VEGFC, CCR7
Introduction
Prostate cancer is one of the most common types of fatal tumors of the male and also a major cause of cancer mortality [1]. There is a lack of effective screening and early detection strategies; therefore, the majority of males are diagnosed with advanced-stage metastatic cancer for which surgical and pharmaceutical treatment options are significantly less effective [2]. Standard treatment options include debulking followed by chemotherapy with platinum agents. Although there is a good response to primary surgery and chemotherapy treatments, the recurrence rates are high and salvage therapies available are not curative [3]. Therefore, it is important to understand the molecular mechanisms underlying this disease in order to develop novel treatment strategies to improve the clinical outcomes for these patients.
In tumor biology, angiogenesis is required to permit increased delivery of oxygen and nutrients to the tumor cells [4]. This pathological process involves several steps, including release of extracellular factors, tumor cells migration, proliferation and formation of new vessels. Amongst all the molecules participating in these events, vascular endothelial growth factor C (VEGFC) is particularly well known master of angiogenesis [5], and promotes every step of angiogenesis. CC-chemokine receptor 7 (CCR7) activity is inducible by inflammatory stimuli, including cytokines, growth factors and tumor promoters, and is up-regulated in a variety of malignancies. CCR7 up-regulation favors the growth of malignant cells by stimulating proliferation and angiogenesis [6]. A large number of previous studies demonstrated that CCR7 is over-expressed in prostate cancer [7]. Furthermore, previous research had found that CCR7 can induce angiogenesis via vascular endothelial growth factor (VEGF) and can also inhibit apoptosis by inducing the anti-apoptotic factor B-cell lymphoma 2 as well as activating anti-apoptotic signaling through Akt/protein kinase B (one of the serine/threonine kinases). These results suggest that CCR7 has a significant role in the generation and progression of solid tumors and the inhibition of CCR7 may inhibit the growth of a variety of solid malignancies. Therefore, down-regulation of CCR7 in cancer cells may prove useful in improving clinical outcomes in cancer patients.
RNA interference (RNAi) is a powerful method for gene inactivationand cancer gene therapy [8]. The advantage of RNAi technology is that it can be used to target a large number of different genes, which are involved in a number of distinct cellular pathways. The technology of RNA silencing is poised to have a major impact on the treatment of human disease, particularly cancer [9]. The aim of the present study was to investigate the role of CCR7 in the growth of human prostate cancer cells. The effect of RNAi-induced CCR7 suppression on the proliferation, invasion and migration of prostate cancer cells was also evaluated.
Materials and methods
Cell culture and transfection
PC-3 cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in 1640 Medium (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL, Carlsbad, CA, USA). The cells were maintained in a humidified 37°C incubator with 5% CO2. The short small interfering RNA (siRNA) was constructed by Nanjing Genscript Biotechnology Co., LTD with sequence specifically targeted to CCR7 gene: (5’-GAAGUGCAUACACCGAGAC-3’). Transient transfection was performed using the Lipofectamine RNAi MAX reagent (Invitrogen) and following the manufacturer’s instructions. After transfection, cell proliferation was measured by MTT assay.
Cell viability assay
At 48 and 72 h post-transfection with CCR7 siRNAs, the cells were seeded in quadruplicate into 96-well plates (5,000 cells/well in 100 μl of medium). At the indicated times, the cells were incubated with 1 mg/ml MTT in normal culture medium for 6 h at 37°C. The medium was then aspirated and the formazan was dissolved in 200 μl dimethyl sulfoxide. The absorbance was measured at 570 nm by a STAKMAX™ microplate reader (Molecular Devices, Sunnyvale, CA, USA) and the experiments were repeated five times.
Cell cycle analysis
The PC-3 cells were treated with siRNA for 48 h or 72 h in 1640 containing 5% FBS. All the cells were collected, and 1×106 cells were centrifuged, resuspended in ice-cold 70% ethanol and stored at -20°C until further analysis. Washed cells were stained by 0.1% Triton X-100 in 0.01 M phosphate-buffered saline (pH 7.2) with 50 μg/ml propidium iodide (Sigma-Aldrich) and 1 mg/ml RNase A (Invitrogen), and incubated at 37°C for 30 min in the dark. Samples of the cells were then analyzed for their DNA content using FACScan flow cytometry (Beckman, Miami, FL, USA), and cell cycle phase distributions were analyzed by the Cell Quest acquisition software (BD Biosciences, Franklin Lanes, NJ, USA). All experiments were performed in duplicate and repeated twice.
Cell migration assay
The migration assay was performed using a 12-well Boyden Chamber (Neuro Probe, Inc., Gaithersburg, MD, USA) with an 8 μm pore size. In total, ~1×105 cells were seeded into upper wells of the Boyden Chamber and incubated for 6 h at 37°C in medium containing 1% FBS. 1640 containing with 30 ng/ml VEGFC was used as a chemoattractant in the bottom wells. The cells that did not migrate through the pores of the Boyden Chamber were manually removed with a rubber swab. The cells that migrated to the lower side of the membrane were stained with hematoxylin and eosin and photographed using an inverted microscope.
Transwell invasion assay
The invasiveness of PC-3 cancer cells was assessed using 24-well Transwell plates (Corning, Lowell, MA, USA). In brief, 2×105 cells in 1640 with 0.5% FBS were added to the upper chamber containing a 8-mm pore polycarbonate coated with 1 mg/ml Matrigel; the lower chamber was filled with media containing 30 ng/ml VEGFC. Subsequent to 16 h incubation, the upper surface of the membrane was scrubbed with a cotton-tipped swab. The invading cells on the lower surface of the membrane were fixed and stained with 0.5% crystal violet dye. A total of five random fields per membrane were photographed and cells were quantified. All experiments were performed in triplicate and repeated four times.
Quantitative polymerase chain reaction (qPCR)
To evaluate CCR7 mRNA expression, the cells were harvested following transfection. The total RNA was extracted from the cells using TRIzol reagent (Invitrogen Life Technologies) for reverse transcription. The RNA was transcribed to cDNA using the Superscript First-Strand Synthesis kit (Takara, Dalian, China) following the manufacturer’s instructions. qPCR assays were performed using SYBR-Green Real-Time PCR Master Mix (Toyobo, Osaka, Japan) and RT-PCR amplification equipment (7300 Real-Time PCR System; Applied Biosystems, Foster City, CA, USA) using specific primers: CCR7 sense, 5’-CCCTTGGGTGTCAAAGGTAAA-3’ and antisense, 5’-AAACTGATGCGTGAAGTGCTG-3’; and β-actin sense, 5’-GCGAGCACAGAGCCTCGCCTTTG-3’ and antisense, 5’-GATGCCGTGCTCGATGGGGTAC-3’. The PCR conditions were as follows: Pre-denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 10 sec and annealing/extension at 58°C for 20 sec. The amplification specificity was checked by melting curve analysis. The expression of the genes of interest was determined by normalization of the threshold cycle (Ct) of these genes to that of the β-actin control.
Matrix metalloproteins (MMPs) activity assay
The activity of MMP-9 and MMP-2 were determined by QuickZyme MMPs activity assay (QucikZyme BioSciences) according to the manufacturer’s instructions. Briefly, after transfection, cells were washed with fresh medium and replaced with serum-free medium. After additional 24 h, the medium was collected and centrifuged at 10000 g for 10 min. Respective supernatant was added to the 96-well strip coated with MMP-9 antibody or MMP-2 antibody and incubated at 4°C overnight. After washing with wash buffer for 3 times, 50 µl assay buffer was added into the well, followed by adding 50 µl detection reagent. After incubation at 37°C for 1 h, OD405 was measured with Microplate Reader (Bio-Tek).
Western blot analysis
The cells were harvested and lysed in Triton X-100 in Hepes buffer [150 mM NaCl, 50 mM Hepes, 1.5 mM MgCl2, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS) and a protease inhibitor cocktail (Sigma)]. Western blot analysis was performed using conventional protocols. Briefly, the protein concentration of the extracts was determined using a bicinchoninic acid kit (Sigma) with bovine serum albumin used as the standard. The total protein samples (40 μg) were separated by 8% SDS-PAGE, proteins were transferred to nitrocellulose membranes (Millipore, Bedford, MA, USA), immunoblotted with specific primary antibodies and incubated with corresponding horseradish peroxidase (HRP)-conjugated secondary antibody. The other primary antibodies used in the western blots were as follows: Antibodies against CCR7, β-actin, VEGFC, CD31 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA); MMP-9 and MMP-2 (Sigma Aldrich, St. Louis, MO, USA); secondary antibodies used for immunodetection were as follows: HRP-conjugated goat anti-mouse immunoglobulin (Ig) G and goat anti-rabbit IgG (Amersham Biosciences, Uppsala, Sweden). All the immunoblots were visualized by enhanced chemiluminescence (Pierce Biotechnology, Inc., Rockford, IL, USA). All the assays using CCR7 knockdown PC-3 cells were performed after the third day of siRNA transfection.
Mouse xenografts
All animal procedures were approved by the ethical commission of the Third Military Medical University. Cells were injected s.c. into the right posterior flanks of 7-wk-old immunodeficient NOD/SCID female mice (6 mice per group). Tumor volume was measured every 3 days. Tumor volume was calculated as mm3 =0.5× length (mm)3 width (mm)2. All animal experiments were carried out in compliance with the Guidelines for the Third Military Medical University.
Statistical analysis
The values were expressed as the mean ± standard deviation. The data were analyzed by one-way analysis of variance and P < 0.05 was considered to be statistically significant.
Results
Down-regulation of CCR7 mRNA and protein expression levels by CCR7-siRNA
A recombinant vector was designed and constructed, which expresses siRNA targeting the CCR7 gene, and transfected it into PC-3 cells. Subsequent to 48 h or 72 h transfection, the cells were harvested and the CCR7 mRNA levels were analyzed by qPCR. CCR7 expression was significantly down-regulated in PC-3 cells transfected with siRNA compared with the cells transfected with the control vector and un-transfected cells (P<0.01). There was no significant difference between the un-transfected and empty vector groups (Figure 1A). The results indicated that most of the CCR7 mRNA was degraded by CCR7 siRNA in the PC-3 cells.
Figure 1.
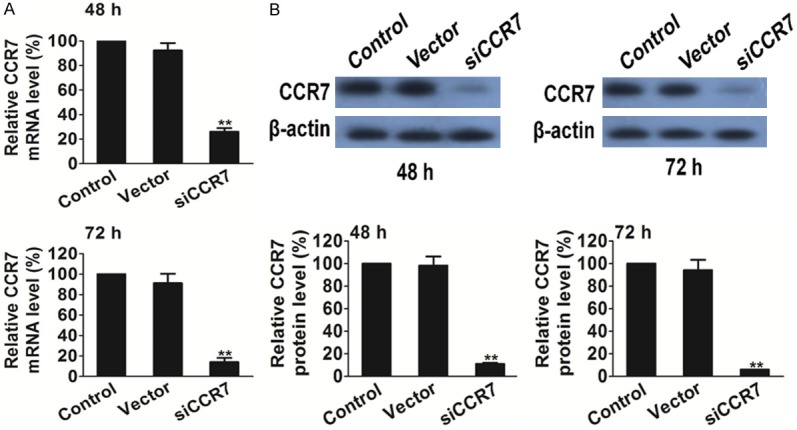
Silencing of CCR7 decreases CCR7 expression in PC-3 tumor cells. A. Quantitative polymerase chain reaction analysis of CCR7 after RNAi silencing. B. Western blot analysis of CCR7 after RNAi silencing. **P<0.01 vs. the control.
The effect of CCR7 siRNA treatment on protein expression was assessed by western blot analysis. As shown in Figure 1B, there was no difference between the un-transfected and empty vector group, while the band density clearly decreased in the CCR7 siRNA group as compared with the un-transfected and empty vector group. These results demonstrated that siRNA targeting CCR7 significantly silenced CCR7 protein expression in PC-3 prostate cancer cells (P<0.01).
CCR7 silencing affects PC-3 cell proliferation and the cell cycle
To elucidate the effects of the CCR7 siRNA on PC-3 prostate cancer cell proliferation in the presence of VEGFC, the MTT assay was used and cell proliferation was determined by counting the number of viable cells. PC-3 prostate cancer cells transfected with CCR7-siRNA proliferated at a much lower rate compared with the control cells at 48 and 72 h post-transfection (Figure 2A). These data demonstrate that the inhibition of CCR7 by RNAi can inhibit the proliferation of PC-3 cells induced by VEGFC. In addition, to determine the effects of PC-3 cell cycle progression following CCR7 silencing, flow cytometry was performed in the present study. In the siRNA therapy group, the percentage of cells in G0/G1 phase was significantly increased as compared with the scrambled-treated and control cells. While, VEGFC had no effect of phase arrest in PC-3 cells. These results indicated that CCR7 silencing can induce cell cycle arrest in G0/G1 phase in PC-3 cells (Figure 2B).
Figure 2.
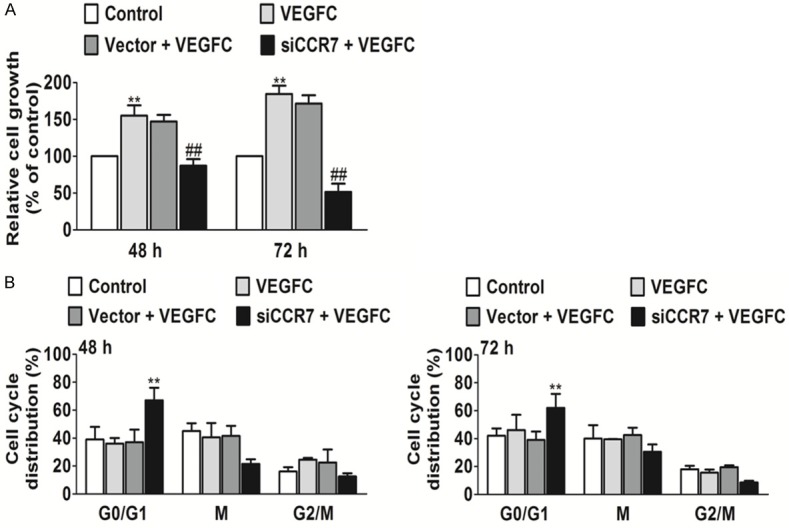
Silencing of CCR7 inhibits cell proliferation and the cell cycle in vitro. A. Down-regulation of CCR7 by siRNA significantly suppressed the proliferation of PC-3 cells. **P<0.01 vs. the control and ##P<0.01 vs. VEGFC treated group. B. Down-regulation of CCR7 by siRNA significantly induced G0/G1 phase arrest of PC-3 cells. **P<0.01 vs. the control.
CCR7 silencing inhibits PC-3 cell migration and invasion induces by VEGFC
To analyze whether CCR7 siRNA affects PC-3 cell migration induced by VEGFC, migration assays were performed using Boyden chambers. RNAi-mediated CCR7 silencing significantly inhibited PC-3 cell migration as compared with the control (un-transfected) and negative control-transfected (Vector) groups (Figure 3A). On the other hand, CCR7 knockdown in PC-3 cells markedly inhibited invasion in vitro as compared with the vector-treated and control groups (Figure 3B). The control cells and vector cells remained invasive and no statistically significant differences were observed.
Figure 3.
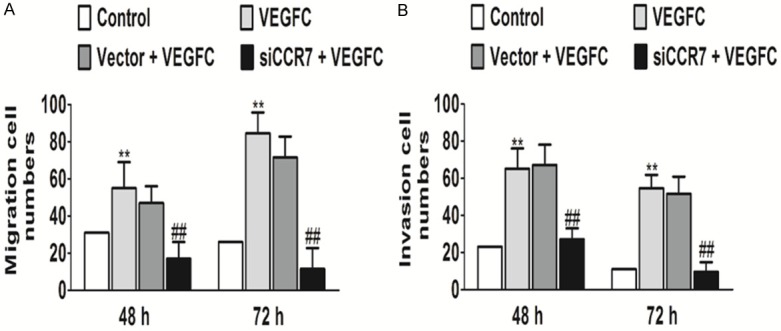
Silencing of CCR7 affects cell migration and invasion in vitro. Down-regulation of the CCR7 gene by siRNA caused significantly suppressed (A) cell migration and (B) cell invasion of PC-3 cells. **P<0.01 vs. the control and ##P<0.01 vs. the VEGFC treatment group.
In order to investigate the mechanisms involved in the inhibition of the invasion and migration ability by down-regulation of CCR7 silencing of PC-3 cells, MMP-2/9 levels in PC-3 cells were determined by ELISA analysis. As shown in Figure 4A, the MMP-2/9 levels in the siRNA therapy groups were significantly lower compared with those in the un-transfected and empty vector groups. In addition, MMP-2/9 protein expression levels were determined following CCR7 silencing. It was found that CCR7 silencing was found to significantly inhibit MMP-2/9 secretion and expression in the PC-3 tumor cells compared with the control and empty vector cells (Figure 4B).
Figure 4.
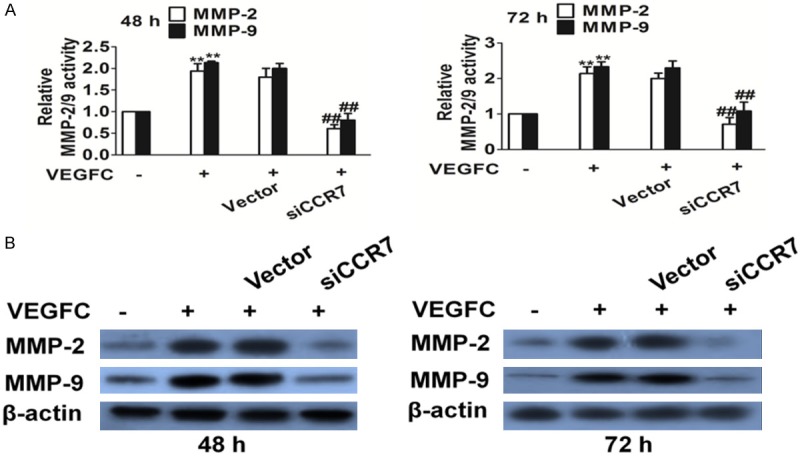
Silencing of CCR7 affects MMPs activity expression by ELISA, and protein expression by western blot analysis. A. MMP-2 and MMP-9 levels were significantly decreased after silencing CCR7 as compared with the control cells. **P<0.05 vs. the control and ##P<0.01 vs. the VEGFC treated group. B. Protein expression levels of MMP-9 and MMP-2 were detected by western blot analysis after CCR7 silencing.
CCR7 knock-down inhibits growth of PC-3 cells in vivo
To evaluate the effects of CCR7 knockdown on prostate cancer cells growth in vivo, we further constructed an experiment using PC-3 cells xenograft mouse model. It was found that CCR7 knockdown dramatically suppressed tumor volumes compared with control group (Figure 5A). To further examine whether CCR7 knockdown suppress PC-3 cells growth in vivo via inhibit angiogenesis, VEGFC, CD31 and MMPs expression activity in tumor tissues were assayed by western blot analysis with specific antibody. Mice implantation with CCR7 knockdown cells showed a significant reduction of protein expression in tumors (Figure 5B). All the results demonstrated that CCR7 knockdown inhibited growth of PC-3 cells via suppressed tumor growth and angiogenesis.
Figure 5.
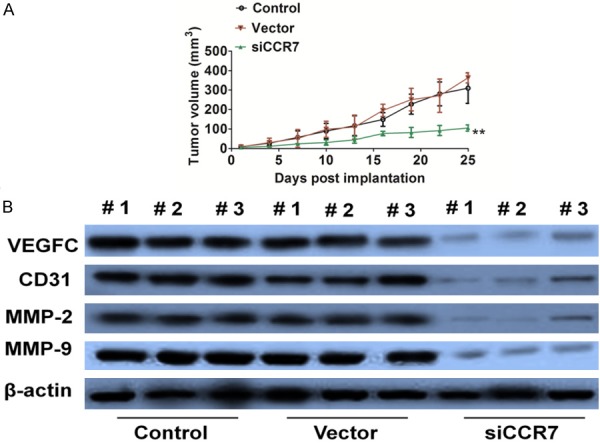
CCR7 siRNA inhibits prostate cancer cells growth in vivo. A. PC-3 siCCR7 or control cells were subcutaneously implanted into mice to build prostate cancer xenograft. On day 25 post injection, tumors were removed and photographed. CCR7 siRNA resulted in significantly tumor growth inhibition versus control mice. Data were from three independent experiments and were average ± S.E. values. n=6, **P<0.01, compared to mice injected with control cells. B. VEGFC, CD31, and MMP-2/9 expression in tumor sections from different mice group was detected by western blot with the antibodies.
Discussion
RNAi is a fundamental cellular mechanism used for silencing gene expression. RNAi has been widely used in cancer therapy to silence the expression of oncogenes and growth factors or their receptors, which may result in inhibition of the cell cycle, cell proliferation and tumor angiogenesis as well as induction of cell apoptosis [10]. To date, RNAi has been regarded as an effective and useful approach for therapeutic applications of cancer [11]. In the present study, RNAi strategies were used to reduce the expression of CCR7 in the prostate cancer cell line PC-3 and to evaluate the role of CCR7 in PC-3 cells. The present study demonstrated that down-regulation of CCR7 reduced proliferation, cell cycle, cell migration and invasion in PC-3 cells.
CCR7 is known to be involved in multiple pathophysiological processes, including inflammation and tumorigenesis [12]. CCR7 up-expression is undetectable in numerous normal tissues, yet it is commonly over-expressed in various human cancers, including prostate cancer [13], with its downstream of chemokine (C-X-C motif) ligand 7 (CXCL7), which is linked to more aggressive behavior of tumors and thus contributing to prostate cancer progression [13]. Tumors require a vascular supply to grow and can achieve this via the expression of pro-angiogenic growth factors. Angiogenesis has been understood to be an important therapeutic target, and drugs targeting vascular endothelial growth factor A (VEGFA) such as a bevacizumab has been developed and approved for clinical use, however, few other angiogenic factors switch on during cancer progression. VEGFC is also a significant mediator in tumor-promoting angiogenesis [14]. The present study revealed that down-regulation of the CCR7 gene by silencing decreased VEGFC induces tumor cell proliferation. The poor prognosis of prostate cancer is mainly due to dissemination caused by the aggressive migration activity of the cancer cells [15]. In the present study, siRNA-mediated down-regulation of CCR7 expression in human prostate cancer cells lead to a significant decrease in PC-3 cell invasion and migration. These results were consistent with previous studies [16], demonstrating that CCR7 mediates the invasive and metastatic potential of prostate cancer cells.
In conclusion, the vector expressing three siRNAs targeting the CCR7 gene in tandem was able to silence the expression of CCR7 in prostate cancer cells. The CCR7 knock-down not only resulted in a decrease of cell proliferation in prostate tumor cells both in vitro and in vivo, but it also suppressed cell migration and invasion of prostate cancer cells induced by VEGFC. These results indicated that CCR7 is a potential therapeutic target for the prevention or treatment of prostate cancer.
Disclosure of conflict of interest
None.
References
- 1.Zhang HM, Yan Y, Wang F, Gu WY, Hu GH, Zheng JH. Ratio of prostate specific antigen to the outer gland volume of prostrate as a predictor for prostate cancer. Int J Clin Exp Pathol. 2014;7:6079–84. [PMC free article] [PubMed] [Google Scholar]
- 2.Heresi GA, Wang J, Taichman R, Chirinos JA, Regalado JJ, Lichtstein DM, Rosenblatt JD. Expression of the chemokine receptor CCR7 in prostate cancer presenting with generalized lymphadenopathy: report of a case, review of the literature, and analysis of chemokine receptor expression. Urol Oncol. 2005;23:261–7. doi: 10.1016/j.urolonc.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Huang X, Su K, Zhou L, Shen G, Dong Q, Lou Y, Zheng S. Hypoxia preconditioning of mesenchymal stromal cells enhances PC3 cell lymphatic metastasis accompanied by VEGFR-3/CCR7 activation. J Cell Biochem. 2013;114:2834–41. doi: 10.1002/jcb.24629. [DOI] [PubMed] [Google Scholar]
- 4.Lu Y, Cai Z, Xiao G, Liu Y, Keller ET, Yao Z, Zhang J. CCR2 expression correlates with prostate cancer progression. J Cell Biochem. 2007;101:676–85. doi: 10.1002/jcb.21220. [DOI] [PubMed] [Google Scholar]
- 5.Malietzis G, Lee GH, Bernardo D, Blakemore AI, Knight SC, Moorghen M, Al-Hassi HO, Jenkins JT. The prognostic significance and relationship with body composition of CCR7-positive cells in colorectal cancer. J Surg Oncol. 2015;112:86–92. doi: 10.1002/jso.23959. [DOI] [PubMed] [Google Scholar]
- 6.Li F, Zou Z, Suo N, Zhang Z, Wan F, Zhong G, Qu Y, Ntaka KS, Tian H. CCL21/CCR7 axis activating chemotaxis accompanied with epithelial-mesenchymal transition in human breast carcinoma. Med Oncol. 2014;31:180. doi: 10.1007/s12032-014-0180-8. [DOI] [PubMed] [Google Scholar]
- 7.Mo M, Zhou M, Wang L, Qi L, Zhou K, Liu LF, Chen Z, Zu XB. CCL21/CCR7 enhances the proliferation, migration, and invasion of human bladder cancer T24 cells. PLoS One. 2015;10:e0119506. doi: 10.1371/journal.pone.0119506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z, Zheng X, Yang L, Liu F, Zhang E, Duan W, Bai S, Safdar J, Li Z, Sun C. Chemokine receptor 7 promotes tumor migration and invasiveness via the RhoA/ROCK pathway in metastatic squamous cell carcinoma of the head and neck. Oncol Rep. 2015;33:849–55. doi: 10.3892/or.2014.3631. [DOI] [PubMed] [Google Scholar]
- 9.Tutunea-Fatan E, Majumder M, Xin X, Lala PK. The role of CCL21/CCR7 chemokine axis in breast cancer-induced lymphangiogenesis. Mol Cancer. 2015;14:35. doi: 10.1186/s12943-015-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng S, Guo J, Yang Q, Yang X. Crk-like adapter protein regulates CCL19/CCR7-mediated epithelial-to-mesenchymal transition via ERK signaling pathway in epithelial ovarian carcinomas. Med Oncol. 2015;32:47. doi: 10.1007/s12032-015-0494-1. [DOI] [PubMed] [Google Scholar]
- 11.Goranova TE, Bozhanov SS, Lozanov VS, Mitev VI, Kaneva RP, Georgieva EI. Changes in gene expression of CXCR4, CCR7 and BCL2 after treatment of breast cancer cells with saponin extract from Tribulus terrestris. Neoplasma. 2015;62:27–33. doi: 10.4149/neo_2015_004. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Zhou Y, Yang Y. CCR7 pathway induces epithelial-mesenchymal transition through up-regulation of Snail signaling in gastric cancer. Med Oncol. 2015;32:467. doi: 10.1007/s12032-014-0467-9. [DOI] [PubMed] [Google Scholar]
- 13.Liu FY, Safdar J, Li ZN, Fang QG, Zhang X, Xu ZF, Sun CF. CCR7 regulates cell migration and invasion through JAK2/STAT3 in metastatic squamous cell carcinoma of the head and neck. Biomed Res Int. 2014;2014:415375. doi: 10.1155/2014/415375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Ghonaimy EA, El-Shinawi M, Ibrahim SA, El-Ghazaly H, Abd-El-Tawab R, Nouh MA, El-Mamlouk T, Mohamed MM. Positive lymph-node breast cancer patients-activation of NF-κB in tumor-associated leukocytes stimulates cytokine secretion that promotes metastasis via C-C chemokine receptor CCR7. FEBS J. 2015;282:271–82. doi: 10.1111/febs.13124. [DOI] [PubMed] [Google Scholar]
- 15.Liu FY, Safdar J, Li ZN, Fang QG, Zhang X, Xu ZF, Sun CF. CCR7 regulates cell migration and invasion through MAPKs in metastatic squamous cell carcinoma of head and neck. Int J Oncol. 2014;45:2502–10. doi: 10.3892/ijo.2014.2674. [DOI] [PubMed] [Google Scholar]
- 16.Shi JY, Yang LX, Wang ZC, Wang LY, Zhou J, Wang XY, Shi GM, Ding ZB, Ke AW, Dai Z, Qiu SJ, Tang QQ, Gao Q, Fan J. CC chemokine receptor-like 1 functions as a tumour suppressor by impairing CCR7-related chemotaxis in hepatocellular carcinoma. J Pathol. 2015;235:546–58. doi: 10.1002/path.4450. [DOI] [PubMed] [Google Scholar]


