Abstract
The aim of this study was to identify differently expressed proteins in the presence and absence of EPHX2 gene in mouse hypothalamus using proteomics profiling and bioinformatics analysis. This study was performed on 3 wild type (WT) and 3 EPHX2 gene global knockout (KO) mice (EPHX2 -/-). Using the nano- electrospray ionization (ESI)-LC-MS/MS detector, we identified 31 over-expressed proteins in WT mouse hypothalamus compared to the KO counterparts. Gene Ontology (GO) annotation in terms of the protein-protein interaction network indicated that cellular metabolic process, protein metabolic process, signaling transduction and protein post-translation biological processes involved in EPHX2 -/- regulatory network. In addition, signaling pathway enrichment analysis also highlighted chronic neurodegenerative diseases and some other signaling pathways, such as TGF-beta signaling pathway, T cell receptor signaling pathway, ErbB signaling pathway, Neurotrophin signaling pathway and MAPK signaling pathway, were strongly coupled with EPHX2 gene knockout. Further studies into the molecular functions of EPHX2 gene in hypothalamus will help to provide new perspective in neurogenesis.
Keywords: EPHX2, hypothalamic neurogenesis, proteomics profiling, protein network
Introduction
Soluble epoxide hydrolase (sEH), a ubiquitously expressed predominantly cytosolic enzyme that encoded by EPHX2 gene, was found to be over expressed in liver, kidney, heart and ovary tissues [1]. Meanwhile, the epoxyeicosatrienoic acids or EETs have been reported to be important endogenous substrates for sEH which produced by cytochrome P-450 epoxygenases [2]. As a ubiquitously hydrolytic enzyme, sEH can catalyze EETs into dihydroxyeicosatrienoic acids (DHETs), which are biologically less active [3]. By far, a growing body of evidence indicates that EETs play a series of benignant roles in ischemia/reperfusion [4], inflammatory response [5], fibrinolysis [6], tube formation [7] etc.
In brain, EETs have been identified to promote vasodilatation, resulting in a protective effect against ischemia-induced tissue damage [8-10]. Besides, the stabilized EETs were also found to antagonize inflammation via negative regulation of nuclear factor-κB (NF-κB) [5]. Given the salutary effects of sEH deficiency, it global expression of proteins in tissues and fluids [11]. The proteins or peptides that are preferentially expressed and identified pathological state are well suited for diagnostic assays and medical treatment. In addition, with the rapid development of computational biology, rapid advances in network biology indicate that cellular is an attractive therapeutic target for several disorders [12-14]. Thus, insights into the physiological functions of sEH have emerged from studies in mice with global EPHX2 gene deficiency or pharmacological inhibition of sEH, which in turn intrigues curiosities of neuroscientists.
Recent advances in analytic technique present a new opportunity to examine the networks are governed by universal laws and offer a new conceptual framework that could potentially revolutionize our view of biology and disease pathologies in the twenty-first century [15]. Systematic mapping of protein-protein interactions networks was initiated in model organisms, starting with defined biological processes to proteome [16]. However, detailed network perspective associated with sEH in hypothalamic neurogenesis is still unclear. Given the role of sEH deficiency in neural systems and the lack of proteomics based network biological studies related to sEH, the aim of our present study is to extract the significantly expressed proteins with or without EPHX2 gene knockout in mouse and explored the potential signaling pathways involved in EPHX2 gene. Detailed elucidations are as follows.
Materials and methods
Ethics statement
This experiment was conducted according to the Guide for the Care and Use of Laboratory Animals of the US National Institutes of Health (NIH Publication, 8th Edition, 2011) and approved by the Peking University Committee on Animal Care and Use. All surgeries were performed under sodium pentobarbital anesthesia, and all efforts were made to ameliorate animal suffering and euthanasia.
Animal models
Male C57BL/6 background mice (8-10 weeks old, 20 ± 4 g body weight) with and without targeted disruption of EPHX2 gene (EPHX2-/-) were provided by Professor Yi Zhu at the department of physiology and pathophysiology in Peking University [17]. Mice were housed individually in air-conditioned facilities at room temperature with 55 ± 5% humidity under 12:12 h light/dark artificial cycle conditions, and supplied with food and water ad libitum.
Protein sample processing
Mouse hypothalamic tissues were washed with chilled phosphate buffered saline (PBS) and homogenized using a motor-driven glass-teflon homogenizer. After centrifuging and boiling at 100°C for 5 min, proteins from hypothalamus were extracted. The concentration of whole proteins was determined at 570 nm using the bicinchoninic acid (BCA, Pierce, Rockford, IL, USA) assay kit. Proteins from three tubes of EPHX2-/- and the control groups were pooled to minimize individual variation. Protein samples (200 µg) from each group were processed per the manufacturer’s protocol for FASP (Filter Assisted Sample Preparation, Mann). Briefly, to Vivacon 500 filtrate tube (Cat No. VNO1HO2, Sartorius Stedim Biotech) containing protein concentrates, 100 µL of 8 M urea in 0.1 M Tris/HCL, pH 8.5 (UA) was added and samples were centrifuged at 14,000 g for 15 min at room temperature. Then 10 µL of 0.05 M TCEP in water was added to the filters and incubated at 37°C for 1 h. 10 µL of 0.1 M IAA in UA was added to the filters, and the samples were incubated for 30 min in darkness. Filters were washed twice with 200 µL of 50 mM NH4HCO3. Finally, 4 µg trypsin (Promega, Madison, WI) was added in 100 µL of 50 mM NH4HCO3 to each filter. The protein to enzyme ratio was 50:1. Samples were incubated over night at 37°C and released peptides were collected by centrifugation.
Protein separation
The digested peptide mixture was reconstituted with 600 µL buffer A (20 mM ammonium formate in water, pH = 10) and loaded onto a 2.1 × 150 mm Waters XBridge BEH130 C18 column containing 3.5 µm particles (Waters, Milford, MA). The peptides were eluted at a flow rate of 230 µL/min with a gradient of 5% buffer B (20 mM ammonium formate in 80% acetonitrile, pH = 10) for 5 min, 1-15% buffer B for 15 min, 15-25% buffer B for 10 min, 25-55% buffer B for 10 min, and finally 55-95% buffer B for 5 min. The system was then maintained in 95% buffer B for 5 min before equilibrating with 1% buffer B for 8 min prior to the next injection. Elution was monitored by measuring absorbance at 214 nm, and fractions were collected every 2 min. The eluted peptides were pooled as 15 fractions and vacuum-dried. Then samples were ready for nano-ESI-LC-MS/MS analysis.
Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis
The MS analysis experiments were performed on a nano-flow HPLC system (Easy-nLC II, Thermo Fisher Scientific, USA) connected to a LTQ-OrbitrapVelos Pro (Linear quadrupole ion trap-Orbitrap mass analyzer) mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA), equipped with a Nanospray Flex Ion Source (Thermo Fisher Scientific, USA). The peptide mixtures were injected (5 μL) at a flow rate of 5 μL/min onto a pre-column (Easy-column C18-A1, 100 μm I.D. × 20 mm, 5 μm, Thermo Fisher Scientific). The chromatographic separation was performed on a reversed phase C18 column (Easy-column C18-A2, 75 μm I.D. × 100 mm, 3 μm, Thermo Fisher Scientific) at a flow rate of 300 nL/min with a 60 min gradient of 2% to 40% acetonitrile in 0.1% formic acid. The electrospray voltage was maintained at 2.2 kV, and the capillary temperature was set at 250°C. The LTQ-Orbitrap was operated in the data dependent mode to simultaneously measure full scan MS spectra (m/z 350-2000) in the Orbitrap with a mass resolution of 60,000 at m/z 400. After the completion of the full-scan survey, the 15 most abundant ions detected in the full-MS scan were measured in the LTQ part by collision induced dissociation (CID), respectively.
Protein identification and quantization
As to the protein identification, data analysis was performed using MaxQuant software (version 1.4.1.2, http://www.maxquant.org/). Briefly, the raw MS/MS data were submitted to the Uniprot human protein database (http://www.uniprot.org/, release 3.43, 72,340 sequences) using the Andromeda search engine with the following settings: trypsin cleavage; fixed modification of carbamidomethylation of cysteine; variable modifications of oxidation of methionine; a maximum of two missed cleavages; the false discovery rate was calculated by decoy data base searching. Label-free quantization was also performed in MaxQuant. The Min. ratio count for LFQ was set 2, and the match-between-runs option was enabled. Other parameters were set as default.
Raw data analytical procedures
All spectra were exported as plain-text files and the *.txt files were read using Perseus software (version 1.5.0.3, http://www.perseus-framework.org/). Perseus, an architecture based on Windows. NET framework, was designed for the statistical analysis of omics data, such as mRNA microarray [18]. Briefly, raw data were firstly processed to remove random noises, subtract the low-frequency baseline, and detect and quantify individual sample peaks using the filter function button. Log 2 based normalized expression data was performed before differently expressed proteins identification.
Differentially expressed proteins identification
The differentially expressed proteins between the 3 couples of EPHX2-/- mice and the matched normal ones were assessed using two-sample t test method as previously described [18]. The parameter p-value was adjusted for multiple testing corrections with p value cutoff of 0.0001. No further false discovery rate was used for multiple testing correction in this experiment.
K-means clustering and principal component analysis
A nonsupervised analysis of global gene expression was performed using k-mean hierarchical clustering. Hierarchical clustering, also known as hierarchical cluster analysis or HCA, is a cluster analysis method which focuses on building a hierarchy of clusters using Agglomerative and Divisive strategies , such as Cluster 3.0 [19], NCSS statistical software, SPSS etc. In this study, cluster proteins with aberrant expression were agglomerated using k-mean average linkage hierarchical clustering rule and delineated based on the Z score method.
Besides, an orthogonal transformation to the linearly uncorrelated variables of differently expressed proteins was also visualized using principal component analysis (PCA). PCA, a variance-focused approach seeking to reproduce the total variable variance, creates variables that are linear combinations of the original variables and reflect both common and unique variance of the variable using correspondence analysis. To better merit the heterogeneity of protein candidate between EPHX2 gene knock and the normal group, we performed a PCA analysis using SPSS for Windows (SPSS 18.0; SPSS, Chicago, IL, USA).
Protein-protein interaction network construction
The protein to protein interaction network associated with EPHX2 was constructed as previously described [20]. To infer a reliable network from sEH to the possible proteins identified in our proteomics analysis, proteins found in proteomic analysis were used as seed to fish out other partners with direct interactions. Nodes relations were retrieved from Database of Interaction Proteins (DIP, http://dip.doe-mbi.ucla.edu/dip/Main.cgi) [21], BIOGRID (Biological General Repository for Interaction Datasets, http://thebiogrid.org/) [22], HPRD (Human Protein Reference Database, http://www.hprd.org/) [23], BOND (Bimolecular Object Network Database, http://bind.ca), MINT (Molecular Interaction database, http://mint.bio.uniroma2.it/mint/Welcome.do) [24] and IntAct (http://www.ebi.ac.uk/intact/) [25]. The optimized network based on Steiner minimal tree algorithm was visualized in the Cytoscape environment [26]. Besides, nodes without connections were removed from the integrated network and only the largest component was regarded as the protein-protein interaction network associated with sEH.
Gene ontology and pathway enrichment analysis
The BiNGO plugin [27] in Cytoscape environment and DAVID web-server [28] (http://david.abcc.ncifcrf.gov/) were used to retrieve the Gene Ontology Consortium (GOC, http://geneontology.org/) [29] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [30] annotations for the protein to protein interaction network as previously described [31].
Results
31 proteins were found to be differently expressed in mouse hypothalamic tissue with or without EPHX2 gene deletion
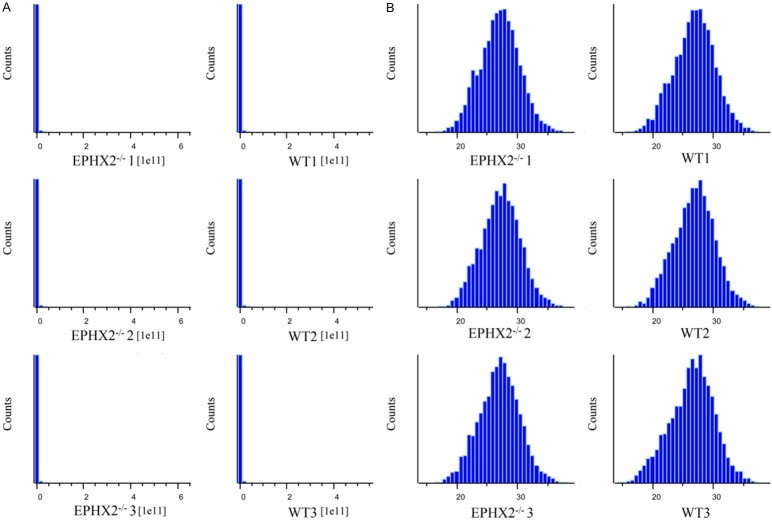
As a result, a total of 6,158 proteins were identified using shotgun method. A logarithm transformation (base 2) of the raw abundance of proteins was pre-performed before differently expressed proteins identification (Figure 1). As shown in Figure 2A and Table 1, we found a total of 31 differently expressed proteins. 2D and 3D PCA (Figure 2B) also indicated that those 31 proteins had an excellent ability to discriminate mouse models with or without EPHX2 gene deletion and could be used as phenotypic discriminators.
Figure 1.
Histogram results of the raw abundance of proteins before and after logarithm transformation.
Figure 2.
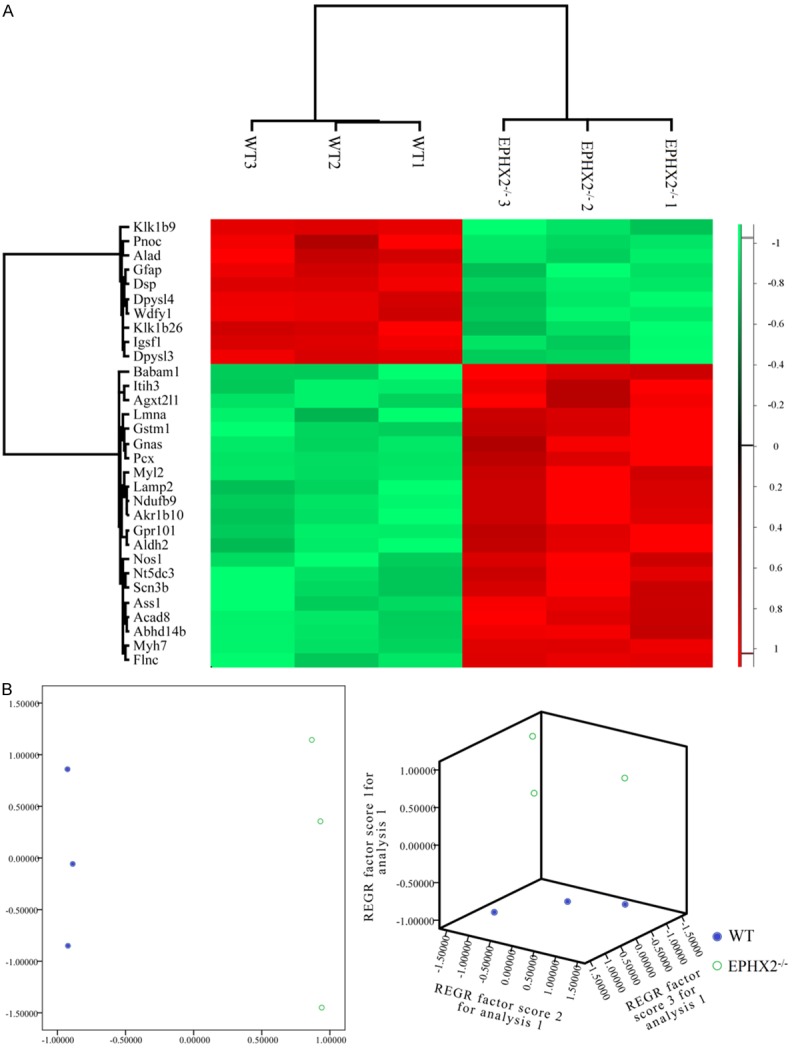
K-means clustering (A) and principal component analysis (B) of the 31 proteins with different expression in EPHX2 -/- (n = 3) compared with the vehicle groups (n = 3). Color key in the hierarchical clustering represents the different expression levels. Red represents up-regulation, while green represents down-regulated proteins.
Table 1.
Identification of proteins associated in EPHX2(+/+) and EPHX2(-/-) mouse hypothalamus using ESI-LC-MS/MS
| Protein ID | Protein names | Official gene symbol | -Log t-test p value | t-test Difference |
|---|---|---|---|---|
| D3YW87 | Filamin C, gamma | Flnc | 5.09635 | 1.91472 |
| E9PWE8 | Dihydropyrimidinase-related protein 3 | Dpysl3 | 4.61218 | -1.14479 |
| E9Q4P1 | WD repeat and FYVE domain containing 1 | Wdfy1 | 4.89347 | -1.80857 |
| E9Q557 | desmoplakin | Dsp | 5.72609 | -1.37034 |
| E9QN99 | Abhd14b abhydrolase domain containing 14b | Abhd14b | 4.75775 | 0.5864 |
| E9QPD7 | pyruvate carboxylase | Pcx | 4.2864 | 0.408389 |
| Q9Z0J4 | nitric oxide synthase 1, neuronal | Nos1 | 4.51849 | 0.471614 |
| G5E895 | aldo-keto reductase family 1, member B10 (aldose reductase) | Akr1b10 | 4.43471 | 0.750383 |
| H3BJR6 | sodium channel, voltage-gated, type III, beta | Scn3b | 4.11826 | 0.797281 |
| Q3TMU8 | dihydropyrimidinase-like 4 | Dpysl4 | 4.79247 | -0.763332 |
| P03995 | glial fibrillary acidic protein | Gfap | 4.4619 | -1.61872 |
| P10518 | aminolevulinate, delta-, dehydratase | Alad | 4.28579 | -0.703655 |
| P10649 | glutathione S-transferase, mu 1 | Gstm1 | 4.17015 | 0.840785 |
| P15949 | kallikrein 1-related peptidase b9 | Klk1b9 | 4.80132 | -5.13527 |
| P16460 | argininosuccinate synthetase 1 | Ass1 | 4.56585 | 0.339808 |
| P17047 | lysosomal-associated membrane protein 2 | Lamp2 | 4.01747 | 2.33493 |
| P36369 | kallikrein 1-related petidase b26 | Klk1b26 | 4.05746 | -4.61297 |
| P47738 | aldehyde dehydrogenase 2, mitochondrial | Aldh2 | 4.10676 | 0.884048 |
| P48678 | lamin A | Lmna | 4.01616 | 1.05971 |
| P51667 | myosin, light polypeptide 2, regulatory, cardiac, slow | Myl2 | 4.48729 | 3.80926 |
| Q3UHB1 | 5’-nucleotidase domain containing 3 | Nt5dc3 | 4.49016 | 1.8812 |
| Q3UI43 | BRISC and BRCA1 A complex member 1 | Babam1 | 4.1279 | 1.24296 |
| Q61704 | inter-alpha trypsin inhibitor, heavy chain 3 | Itih3 | 4.22979 | 0.659157 |
| Q64387 | prepronociceptin | Pnoc | 4.184 | -1.47249 |
| Q6R0H7 | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus | Gnas | 4.16739 | 0.81181 |
| Q7TQA1 | immunoglobulin superfamily, member 1 | Igsf1 | 5.0341 | -0.607182 |
| Q80T62 | G protein-coupled receptor 101 | Gpr101 | 4.27175 | 2.75136 |
| Q8BWU8 | ethanolamine phosphate phospholyase | Agxt2l1 | 4.35029 | 0.943425 |
| Q91Z83 | myosin, heavy polypeptide 7, cardiac muscle, beta | Myh7 | 5.75811 | 3.22376 |
| Q9CQJ8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 | Ndufb9 | 4.62392 | 0.991632 |
| Q9D7B6 | acyl-Coenzyme A dehydrogenase family, member 8 | Acad8 | 4.66179 | 0.494231 |
Protein-protein interaction regulatory network construction
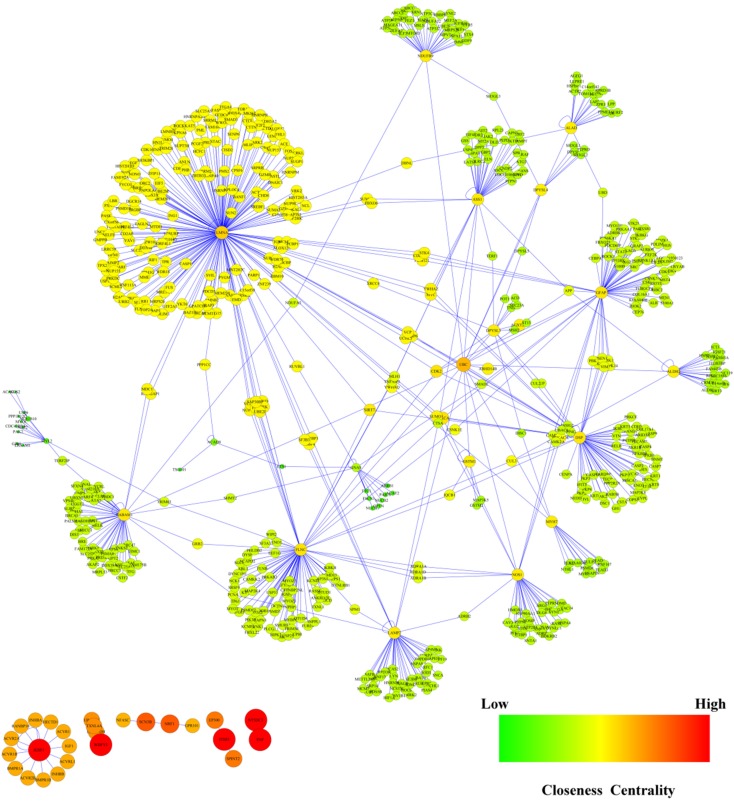
To infer a protein-protein interaction network associated with sEH, we matched the 31 differently expressed genes with 6 public available warehouses to link the known regulatory data between transcriptional factors and the target genes. Totally, we obtained a cohort of 685 nodes and 768 relationships, and the integrated regulatory network was visualized using Cytoscape 2.8.3 (Figure 3).
Figure 3.
Protein-protein interaction network in terms of EPHX2 gene was constructed using DIP, BIOGRID, HPRD, BOND, MINT and IntAct databases. In this network, the size of each node is proportionate to its closeness centrality.
Enrichment of the biological processes
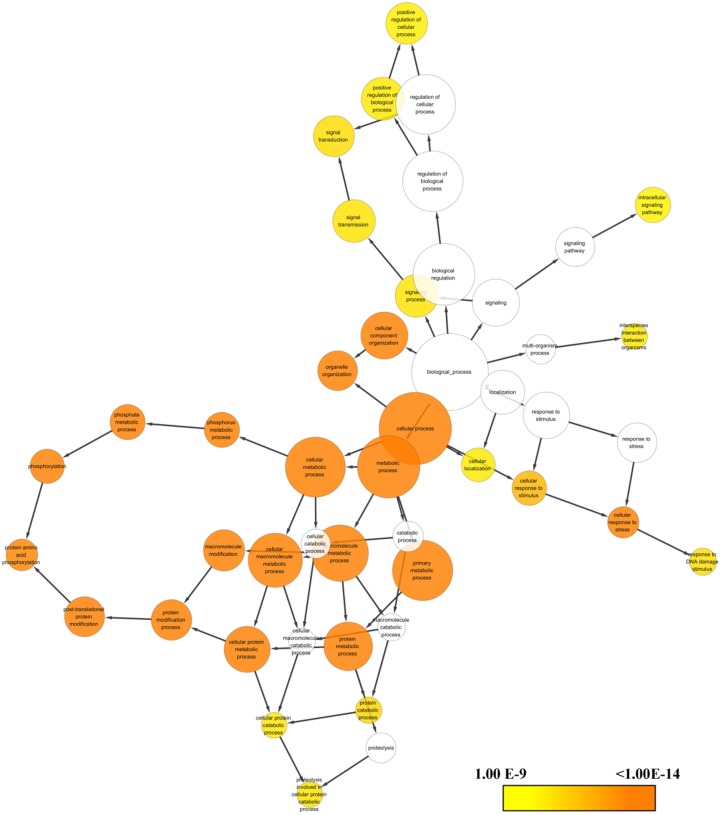
To further extend our knowledge about the regulatory network associated with sEH, we enriched the large list of proteins for functional annotation using BiNGO plugin. As demonstrated in the Figure 4, the biological processes in term of Gene Ontology suggested that EPHX2 gene was highly correlated with cellular metabolic process, protein metabolic process, signaling transduction and protein post-translation.
Figure 4.
Gene Ontology (GO) terms associated with EPHX2 gene. The color gradient shows the proportion associated with the biological processes.
Signaling pathways enrichment analysis
To analyze the whole lists of proteins and better understand the functional annotation involved in EPHX2, differently expressed proteins were submitted to the DAVID Bioinformatics Resources 6.7 for signaling pathway enrichment analysis in terms of KEGG. In this study, we chose the p-value less than 0.001 as the cut-off criterion for canonical pathways. As a result, TGF-beta signaling pathway, T cell receptor signaling pathway, ErbB signaling pathway, Neurotrophin signaling pathway, MAPK signaling pathway, Parkinson’s disease and Alzheimer’s disease seem to be aberrant in EPHX2-/- mouse. All the detailed pathways were listed in Table 2.
Table 2.
The enriched KEGG signaling pathways in DAVID Bioinformatics Resource (P ≤ 0.001)
| KEGG ID | Term | Count | % | P Value | Pop Hits | Fold Enrichment |
|---|---|---|---|---|---|---|
| hsa04350 | TGF-beta signaling pathway | 17 | 0.199157 | 3.73E-05 | 87 | 3.301065 |
| hsa04660 | T cell receptor signaling pathway | 19 | 0.222587 | 4.88E-05 | 108 | 2.972038 |
| hsa04012 | ErbB signaling pathway | 16 | 0.187441 | 1.41E-04 | 87 | 3.106885 |
| hsa04722 | Neurotrophin signaling pathway | 19 | 0.222587 | 3.03E-04 | 124 | 2.588549 |
| hsa04010 | MAPK signaling pathway | 31 | 0.363168 | 4.07E-04 | 267 | 1.961439 |
| hsa05012 | Parkinson’s disease | 19 | 0.222587 | 4.51E-04 | 128 | 2.507657 |
| hsa05010 | Alzheimer’s disease | 22 | 0.257732 | 5.20E-04 | 163 | 2.28013 |
Discussion
High throughput proteomics, in terms of vast amount of proteins, offers highly information towards biology [32]. However, owing to the complexity of biological system, it is difficult to identify and understand the entire proteins well. With the development of sophisticated separation techniques, mass spectrometry (MS)-based high throughput proteomics becomes a core instrumentation for proteins characterization due to its high sensitivity [33]. In the present study, shotgun proteomics method was introduced for protein analysis in a high-throughput way [34]. Interactome network basing on high throughput data, as well as its integration with disease phenotype, has been reported to be a conventional technology for the identification of disease-specific biomarkers, such as cancer [35]. Recently, Diederick et al. [36] discovered FASN, XPO1, ENO1 and PDCD61P were novel biomarkers for prostate cancer progression using a nanoLC and LTQ-Orbitrap-MS/MS mode. Similarly, Shen and colleagues [37] also revealed SSP411 was a band-new biomarker for cholangiocarcinoma, suggesting the high throughput proteomics profiling was a valuable tool for cancer biomarkers identification and diagnosis. Besides, proteomics analysis was also considered to be a powerful for neurodegeneration diagnosis [38]. In 2014, Liguori et al. detected the cerebrospinal fluid proteomic profiles in Multiple Sclerosis (MS) patients basing on the Matrix Assisted Laser Desorption Ionization Time of Flight (MALDI-TOP) mass spectrometer. They concluded Secretograin II and Protein 7B2 were highly expressed in clinically definite MS patients compared to the progressive ones. In addition, Tymosin β4 was also found to be aberrant in clinically isolated syndrome and relapsing remitting (RR) MS patients, suggesting the proteomic profiling technique in combination with the mass spectrometry evaluation provided useful and important information to improve our understanding of the complex pathogenesis of MS.
Previous animal studies have indicated that EPHX2 gene deletion or treatment with sEH inhibitors results in increased levels of EETs and protection against stroke-induced brain damage [39]. Oxidative stress is hypothesized to play a major role in Alzheimer’s disease. Epoxides are potentially reactive intermediates formed that contribute to cytotoxic damage mediated by oxidative stress [40]. In chronic neurodegenerative diseases, epoxide hydrolase were found to be significantly elevated in the hippocampus and associated cortex in Alzheimer’s disease patients [41]. In agreement with the results of previous studies, KEGG based pathway enrichment analysis also confirmed that Neurotrophin signaling pathway, Parkinson’s disease and Alzheimer’s disease highly are strongly coupled with sEH. As an indispensable section of the brain, hypothalamus governed hormone production and regulated homeostasis. Hypothalamic hormones include thyrotropin-releasing, gonadotropin-releasing, growth hormone-releasing, corticotrophin-releasing, somatostatin, and dopamine hormones, which are released into the blood and link the nervous system to the endocrine system via hypophysis.
A number of documents proved that sEH inhibitors possessed protective effects in ischemic or cardiovascular disease. In 2010, Simpkins and colleagues discovered 12-(3-adamantan-1-yl-ure-ido)-dodecanoic acid, an inhibitor of sEH, could significantly improve the increment of inward remodeling. In addition, mice with EPHX2 deletion enhanced inward vascular remodeling induced by carotid ligation [42]. Besides, using sEH-knockout mice, Xiaocui et al. also discovered sEH deficiency and inhibition could decrease 1-methyl-4-phenyl-1,2,3,6-tetrahydro pyridine (MPTP)-treated mice via activating AKT signaling pathway to protect dopamine neurons (Xiaocui et al., 2014). In view of this, insights into the physiological functions of sEH have emerged from studies in mice with global EPHX2 gene deficiency or pharmacological inhibition of sEH, which in turn provides beneficial effects in blood pressure [43], cardiovascular [44], renal [45] in murine models.
Many studies have shown that alternations of neuropeptides from hypothalamic pituitary axis resulted in neurodegenerative diseases, such as Huntington’s disease [46]. Besides, postmortem studies pointed out that dopamine concentrations in the hypothalamus were involved in Parkinson’s disease [47]. More recently, 18F-dopa based positron emission tomography (PET) also discovered the hypothalamic monoamine storage capacity decreased in patients with idiopathic Parkinson’s disease, which was in agreement with the postmortem observations [48].
Pathway enrichment analysis also indicated that TGF-beta signaling pathway, T cell receptor signaling pathway, ErbB signaling pathway and MAPK signaling pathway were engaged in EPHX2 gene deficiency. However, detailed relations associated with EPHX2 gene and these regulatory pathways still lacks and undefined.
In summary, we identified 31 significantly expressed proteins in mouse hippocampus using proteomics analysis and constructed a protein-protein interaction network associated with EPHX2 gene knockout. Our study may shed new lights for sEH in the chronic neurodegenerative diseases, like Parkinson’s disease and Alzheimer’s disease.
Acknowledgements
We wish to thank Prof. Yi Zhu for the generous gift of the wild type and EPHX2 -/- mouse.
Disclosure of conflict of interest
None.
References
- 1.Wu C, Macleod I, Su AI. BioGPS and MyGene. info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karara A, Dishman E, Blair I, Falck JR, Capdevila JH. Endogenous epoxyeicosatrienoic acids. Cytochrome P-450 controlled stereoselectivity of the hepatic arachidonic acid epoxygenase. J Biol Chem. 1989;264:19822–19827. [PubMed] [Google Scholar]
- 3.Spector AA, Norris AW. Action of epoxyeicosatrienoic acids on cellular function. Am J Physiol Cell Physiol. 2007;292:C996–1012. doi: 10.1152/ajpcell.00402.2006. [DOI] [PubMed] [Google Scholar]
- 4.Nithipatikom K, Moore JM, Isbell MA, Falck JR, Gross GJ. Epoxyeicosatrienoic acids in cardioprotection: ischemic versus reperfusion injury. Am J Physiol Heart Circ Physiol. 2006;291:H537–542. doi: 10.1152/ajpheart.00071.2006. [DOI] [PubMed] [Google Scholar]
- 5.Node K, Huo Y, Ruan X, Yang B, Spiecker M, Ley K, Zeldin DC, Liao JK. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science. 1999;285:1276–1279. doi: 10.1126/science.285.5431.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Node K, Ruan XL, Dai J, Yang SX, Graham L, Zeldin DC, Liao JK. Activation of Galpha s mediates induction of tissue-type plasminogen activator gene transcription by epoxyeicosatrienoic acids. J Biol Chem. 2001;276:15983–15989. doi: 10.1074/jbc.M100439200. [DOI] [PubMed] [Google Scholar]
- 7.Munzenmaier DH, Harder DR. Cerebral microvascular endothelial cell tube formation: role of astrocytic epoxyeicosatrienoic acid release. Am J Physiol Heart Circ Physiol. 2000;278:H1163–1167. doi: 10.1152/ajpheart.2000.278.4.H1163. [DOI] [PubMed] [Google Scholar]
- 8.Amruthesh SC, Falck JR, Ellis EF. Brain synthesis and cerebrovascular action of epoxygenase metabolites of arachidonic acid. J Neurochem. 1992;58:503–510. doi: 10.1111/j.1471-4159.1992.tb09749.x. [DOI] [PubMed] [Google Scholar]
- 9.Gebremedhin D, Gopalakrishnan S, Harder DR. Endogenous events modulating myogenic regulation of cerebrovascular function. Curr Vasc Pharmacol. 2014;12:810–817. doi: 10.2174/15701611113116660153. [DOI] [PubMed] [Google Scholar]
- 10.Gebremedhin D, Ma YH, Falck JR, Roman RJ, VanRollins M, Harder DR. Mechanism of action of cerebral epoxyeicosatrienoic acids on cerebral arterial smooth muscle. Am J Physiol. 1992;263:H519–525. doi: 10.1152/ajpheart.1992.263.2.H519. [DOI] [PubMed] [Google Scholar]
- 11.Harry JL, Wilkins MR, Herbert BR, Packer NH, Gooley AA, Williams KL. Proteomics: capacity versus utility. Electrophoresis. 2000;21:1071–1081. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1071::AID-ELPS1071>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Chiamvimonvat N, Ho CM, Tsai HJ, Hammock BD. The soluble epoxide hydrolase as a pharmaceutical target for hypertension. J Cardiovasc Pharmacol. 2007;50:225–237. doi: 10.1097/FJC.0b013e3181506445. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Koerner IP, Noppens R, Grafe M, Tsai HJ, Morisseau C, Luria A, Hammock BD, Falck JR, Alkayed NJ. Soluble epoxide hydrolase: a novel therapeutic target in stroke. J Cereb Blood Flow Metab. 2007;27:1931–1940. doi: 10.1038/sj.jcbfm.9600494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imig JD, Hammock BD. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat Rev Drug Discov. 2009;8:794–805. doi: 10.1038/nrd2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 16.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Li N, Pang W, Zhang X, Hammock BD, Ai D, Zhu Y. Opposite effects of gene deficiency and pharmacological inhibition of soluble epoxide hydrolase on cardiac fibrosis. PLoS One. 2014;9:e94092. doi: 10.1371/journal.pone.0094092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Fan S, Han D, Xie J, Kuang H, Ge P. Microarray gene expression profiling and bioinformatics analysis of premature ovarian failure in a rat model. Exp Mol Pathol. 2014;97:535–541. doi: 10.1016/j.yexmp.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 19.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 20.Feng LX, Jing CJ, Tang KL, Tao L, Cao ZW, Wu WY, Guan SH, Jiang BH, Yang M, Liu X, Guo DA. Clarifying the signal network of salvianolic acid B using proteomic assay and bioinformatic analysis. Proteomics. 2011;11:1473–1485. doi: 10.1002/pmic.201000482. [DOI] [PubMed] [Google Scholar]
- 21.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, Roechert B, van Eijk K, Hermjakob H. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan S, Pan Z, Geng Q, Li X, Wang Y, An Y, Xu Y, Tie L, Pan Y, Li X. Layered signaling regulatory networks analysis of gene expression involved in malignant tumorigenesis of non-resolving ulcerative colitis via integration of cross-study microarray profiles. PLoS One. 2013;8:e67142. doi: 10.1371/journal.pone.0067142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesley SA. High-throughput proteomics: protein expression and purification in the postgenomic world. Protein Expr Purif. 2001;22:159–164. doi: 10.1006/prep.2001.1465. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Wu S, Stenoien DL, Pasa-Tolic L. High-throughput proteomics. Annu Rev Anal Chem (Palo Alto Calif) 2014;7:427–454. doi: 10.1146/annurev-anchem-071213-020216. [DOI] [PubMed] [Google Scholar]
- 34.Nogueira FC, Domont GB. Survey of shotgun proteomics. Methods Mol Biol. 2014;1156:3–23. doi: 10.1007/978-1-4939-0685-7_1. [DOI] [PubMed] [Google Scholar]
- 35.Srinivas PR, Srivastava S, Hanash S, Wright GL Jr. Proteomics in early detection of cancer. Clin Chem. 2001;47:1901–1911. [PubMed] [Google Scholar]
- 36.Duijvesz D, Burnum-Johnson KE, Gritsenko MA, Hoogland AM, Vredenbregt-van den Berg MS, Willemsen R, Luider T, Pasa-Tolic L, Jenster G. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS One. 2013;8:e82589. doi: 10.1371/journal.pone.0082589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen J, Wang W, Wu J, Feng B, Chen W, Wang M, Tang J, Wang F, Cheng F, Pu L, Tang Q, Wang X, Li X. Comparative proteomic profiling of human bile reveals SSP411 as a novel biomarker of cholangiocarcinoma. PLoS One. 2012;7:e47476. doi: 10.1371/journal.pone.0047476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liguori M, Qualtieri A, Tortorella C, Direnzo V, Bagala A, Mastrapasqua M, Spadafora P, Trojano M. Proteomic profiling in multiple sclerosis clinical courses reveals potential biomarkers of neurodegeneration. PLoS One. 2014;9:e103984. doi: 10.1371/journal.pone.0103984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sura P, Sura R, Enayetallah AE, Grant DF. Distribution and expression of soluble epoxide hydrolase in human brain. J Histochem Cytochem. 2008;56:551–559. doi: 10.1369/jhc.2008.950659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pohanka M. Alzheimer s disease and oxidative stress: a review. Curr Med Chem. 2014;21:356–364. doi: 10.2174/09298673113206660258. [DOI] [PubMed] [Google Scholar]
- 41.Liu M, Sun A, Shin EJ, Liu X, Kim SG, Runyons CR, Markesbery W, Kim HC, Bing G. Expression of microsomal epoxide hydrolase is elevated in Alzheimer’s hippocampus and induced by exogenous beta-amyloid and trimethyl-tin. Eur J Neurosci. 2006;23:2027–2034. doi: 10.1111/j.1460-9568.2006.04724.x. [DOI] [PubMed] [Google Scholar]
- 42.Simpkins AN, Rudic RD, Roy S, Tsai HJ, Hammock BD, Imig JD. Soluble epoxide hydrolase inhibition modulates vascular remodeling. Am J Physiol Heart Circ Physiol. 2010;298:H795–806. doi: 10.1152/ajpheart.00543.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imig JD, Zhao X, Capdevila JH, Morisseau C, Hammock BD. Soluble epoxide hydrolase inhibition lowers arterial blood pressure in angiotensin II hypertension. Hypertension. 2002;39:690–694. doi: 10.1161/hy0202.103788. [DOI] [PubMed] [Google Scholar]
- 44.Motoki A, Merkel MJ, Packwood WH, Cao Z, Liu L, Iliff J, Alkayed NJ, Van Winkle DM. Soluble epoxide hydrolase inhibition and gene deletion are protective against myocardial ischemia-reperfusion injury in vivo. Am J Physiol Heart Circ Physiol. 2008;295:H2128–2134. doi: 10.1152/ajpheart.00428.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao X, Yamamoto T, Newman JW, Kim IH, Watanabe T, Hammock BD, Stewart J, Pollock JS, Pollock DM, Imig JD. Soluble epoxide hydrolase inhibition protects the kidney from hypertension-induced damage. J Am Soc Nephrol. 2004;15:1244–1253. [PubMed] [Google Scholar]
- 46.Politis M, Pavese N, Tai YF, Tabrizi SJ, Barker RA, Piccini P. Hypothalamic involvement in Huntington’s disease: an in vivo PET study. Brain. 2008;131:2860–2869. doi: 10.1093/brain/awn244. [DOI] [PubMed] [Google Scholar]
- 47.Javoy-Agid F, Ruberg M, Pique L, Bertagna X, Taquet H, Studler JM, Cesselin F, Epelbaum J, Agid Y. Biochemistry of the hypothalamus in Parkinson’s disease. Neurology. 1984;34:672–675. doi: 10.1212/wnl.34.5.672. [DOI] [PubMed] [Google Scholar]
- 48.Moore RY, Whone AL, Brooks DJ. Extrastriatal monoamine neuron function in Parkinson’s disease: an 18F-dopa PET study. Neurobiol Dis. 2008;29:381–390. doi: 10.1016/j.nbd.2007.09.004. [DOI] [PubMed] [Google Scholar]