Abstract
Temozolomide (TMZ) with radiotherapy is the current standard of care for newly diagnosed glioma. However, glioma patients who are treated with the drug often develop resistance to it and some other drugs. Recently studies have shown that microRNAs (miRNAs) play an important role in drug resistance. In present study, we first examined the sensitivity to temozolomide in six glioma cell lines, and established a resistant variant, U251MG/TR cells from TMZ-sensitive glioma cell line, U251MG. We then performed a comprehensive analysis of miRNA expressions in U251MG/TR and parental cells using cancer microRNA PCR Array. Among the downregulated microRNAs was miR-16, members of miR-15/16 family, whose expression was further validated by qRT-PCR in U251MG/TR and U251MG cells. The selective microRNA, miR-16 mimics or inhibitor was respectively transfected into U251MG/TR cells and AM38 cell. We found that treatment with the mimics of miR-16 greatly decreased the sensitivity of U251MG/TR cells to temozolomide, while sensitivity to these drugs was increased by treatment with the miR-16 inhibitor. In addition, the downregulation of miR-16 in temozolomide-sensitive AM38 cells was concurrent with the upregulation of Bcl-2 protein. Conversely, overexpression of miR-16 in temozolomide-resistant cells inhibited Bcl-2 expression and decreased temozolomide resistance. In conclusion, MiR-16 mediated temozolomide-resistance in glioma cells by modulation of apoptosis via targeting Bcl-2, which suggesting that miR-16 and Bcl-2 would be potential therapeutic targets for glioma therapy.
Keywords: Human glioma cells, temozolomide resistance, miR-16, BCL-2
Introduction
Human gliomas are the most common primary tumors of the central nervous system, accounting for 35% of all brain tumors; they are among the most aggressive and lethal malignancies [1,2]. Glioblastoma (GBM) constitutes 55% of all primary gliomas [3,4]. Despite surgical operation combined with radiotherapy and chemotherapy, the prognosis for GBM patients remains poor, with a median survival of about 12-15 months, one of the worst 5-year survival rates of all human cancers [5-7]. The postoperative use of temozolomide (TMZ) concurrently and after radiotherapy, which has been reported to significantly improve the overall survival and has the advantage of wide applicability and minimal additional toxicity, has become the current standard regimen for treatment of GBM [8]. However, little is known about acquired TMZ resistance, which is a serious impediment in the treatment of GBM.
MicroRNAs (miRNAs), a class of dogenous small non-coding RNAs of 19-25 nt in length, are a novel type of post-transcriptional gene regulator by binding to a target site in the 3’-untranslatedregion of target mRNAs [9]. Recent evidence has shown that more than 50% of miRNAs are located in cancer-associated genomic break points, and can function as tumor suppressor genes or oncogenes, depending on their targets [10-12]. In addition, several studies have indicated that some miRNAs likemiR-21, miR-195 and miR-455-3p were implicated in the regulation of the chemosensitivity of glioblastoma cells to TMZ [13-18]. We previously identified miRNA-16 (miR-16) is most strongly correlated with malignancy in nearly all analyzed human tumors. Other authors have shown the downregulation of miR-16 in a wide range of cancers, including breast, prostate, and lung cancers, as well as in chronic lymphocytic leukemia [19-22]. These findings suggest that miR-16 is a possible tumor suppressor that acts in a variety of cancers. However, little is known about whether miR-16 is implicated in the regulation of the chemosensitivity of glioma cells to TMZ.
Previous studies have reported thatmiR-16 can significantly inhibit the in vivo growth of U87MG glioma [23]. And upregulation of miR-16 promoted apoptosis by suppressing BCL2 expression [24]. MiR-16 was also reported to suppress EMT mainly through the downregulation of p-FAK and p-Akt expression, and nuclear factor-κB and Slug transcriptional activity [25]. These results suggested an important role of miRNA-16 in glioma.
Recent studies have shown that Bcl-2 was important for TMZ resistance in cultured glioblastoma cells. However, the underlying molecular mechanism of Bcl-2 action in TMZ resistance of GBM is still needed to be elaborated. It has been reported that the expression of BCL2 protein gradually decreased with the increasing degree of malignancy in glioma. Tian-Quan Yang et al showed that miR-16 directly downregulated BCL2 expression and induced the apoptosis of human glioma cells [24]. Therefore, in this present study, we explored whethermiR-16 was indeed involved in the effects of Bcl-2 action on temozolomide resistance in glioma cells.
In the present study, therefore, we used TMZ-sensitive glioma cell lines to generate TMZ resistant variants by continuous exposure to the drug. We then performed comprehensive analysis of miRNA expression using miRNA microarray to explore the mechanisms of acquired resistance against TMZ.
Materials and methods
Cell culture and treatment
Human glioblastoma cell lines (U-138MG and A172) were obtained from American Type Culture Collection ATCC (Rockville, MD, USA). Other glioblastoma cell lines (LN382, AM-38, U-251MG and KMG4) were obtained from Biofavor company (Wuhan, China). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), 100 units/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen, California, USA) in 5% CO2 atmosphere at 37°C. For miR-16 overexpression, cells were transfected with 100 nmol/L of miR-16 mimics, which are small, chemically modified double-strand RNA molecules that are designed to mimic endogenous mature miRNAs. For inhibition, cells were transfected with miR-16 inhibitors, which are chemically modified, single-strand oligonucleotides designed to specifically bind to and inhibit endogenous miRNAs. The sequences were as follows: for miR-16 mimics, 5’-UAGCAGCACGUAAAUAUUGGCGCCAAUAUUUACGUGCUGCUAUU-3’; for the negative control oligonucleotide, 5’-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3’; for miR-16 inhibitors, 5’-CGCCAAUAUUUACGUGCUGCUA-3’; and for the negative control oligonucleotide, 5’-CAGUACUUUUGUGUAGUACAA-3’.
Cell viability assay (CCK-8 assay)
Cells were seeded into 96-well culture plates and incubated at 37°C. Temozolomide was purchased from Sigma (St. Louis, MO) and dissolved in DMSO. After treatment with different concentrations of temozolomide for 72 h, 20 μL CCK-8 reagent was added to each well and incubated for 2 h at 37 C and the optical density (OD) was measured by a microplate reader (Beckman Coulter, Fullerton, CA, USA) at 490 nm. Each temozolomide concentration was tested in triplicate in 96-well plates, and experiments were repeated independently at least three times. The 50% inhibitory concentration (IC50) was calculated with GraphPad Prism software using the sigmoidal dose-response function.
Establishment of temozolomide-resistant U-251MG cell line
To establish the temozolomide-resistant cell line, U-251MG cells were exposed to a low dose of temozolomide in culture media for 6 months and established temozolomide-resistant cells designated as U-251 MG/TR. IC50 for the growth inhibition of temozolomide to U-251MG and U-251 MG/TR are 25.3 mM and 151.6 mM, respectively.
Cancer microRNA PCR array
The expression profile of 88 cancer-related miRNAs was determined using a 96-well plate cancer RT2 miRNA PCR array from SA Biosciences in an in vitro cell culture model composed of glioma cell line U251MG and U251TMZ-resistant (U251MG/TR) cells. Total RNA was extracted using the TRIzol reagent (Invitrogen) and reverse transcribed using the RT2 miRNA First Strand Kit from SA Biosciences. The resulting cDNA was then diluted, mixed with 2×RT2 SYBR Green PCR Master Mix (SA Biosciences), and loaded into the wells of a PCR array plate to allow real-time PCR amplification and detection. Data analysis was performed with the Web-based software package for the miRNA PCR array system.
Quantitative real-time RT-PCR
Total RNA was extracted using Trizol reagents (Invitrogen) according to the manufacturer’s instructions and diluted to 200 ng/mL. Then, quantitative real-time RT-PCR (qRT-PCR) was performed using One Step SYBR® PrimeScript™ RT-PCR Kit II (TaKaRa, China) according to standard protocol. GAPDH gene was used as an internal control.
Western blot analysis
Cells were harvested and homogenized with cell lysis buffer (Beyotime, China). Then, the homogenates were centrifuged for 30 min at 4°C, 12000 rpm, and the supernatants were collected as protein samples. Protein amounts were measured using BCA Protein Assay Kit (Beyotime, China). Equal amounts of protein samples were separated by denaturing 10% SDS-PAGE and transferred onto polyvinylidenedifluoride (PVDF) membranes. After transfer, membranes were incubated in blocking solution, probed with various antibodies, washed, and visualized using horseradish peroxidase (HRP)-conjugated secondary antibodies (GE Healthcare) and enhanced chemiluminescence reagents (Amersham).
Determination of apoptosis
The extent of apoptosis was determined by the flow cytometric measurement through AnnexinV-FITC apoptosis detection kit (Beyotime, China). Cells treated as above described. After 4 d, cells were harvested and washed twice with cold PBS. Then, cells were stained in 1 mL AnnexinVbinding buffer with 10 μL of PI solution and 5 μL of AnnexinV-FITC for 10 min at RT and analyzed by flow cytometry.
Statistical analysis
Data are reported as mean ± standard deviation (SD). Statistical significance was determined using Double-sided Student’s t test. Multiple groups were analyzed using ANOVA. A P value of less than 0.05 was considered to be significant.
Results
Establishment and characterization of temozolomide-resistant sublines
To determine whether temozolomide was associated with chemoresistance of glioma cells, CCK-8 assay was performed to detect temozolomide sensitivity in six glioma cell lines. Two of them LN382, and U138MG, exhibited a higher IC50 than the other four cell lines A-172, AM-38, U-251MG and KMG4 (Figure 1A). Next, U251MG wild type (U251Wt) cells were exposed to 100 μM TMZ for 2 weeks to generate TMZ-resistant variant. The majority of the cells died, but a small population survived and propagated. We then selected surviving colonies and established U251MG TMZ-resistant cells (U251MG/TR). IC50 for the growth inhibition of temozolomide to U-251MG and U-251MG/TR are 25.3 mM and 151.6 mM, respectively (Figure 1B). Furthermore, flow cytometry was used to detect cells apoptosis of U-251MG and U-251MG/TR cells after temozolomide treatment (50 μM), and showed that apoptosis of U-251MG significantly increased compared with control cells, But the correspondingly converse results was observed in U-251MG/TR cells (Figure 1C).
Figure 1.
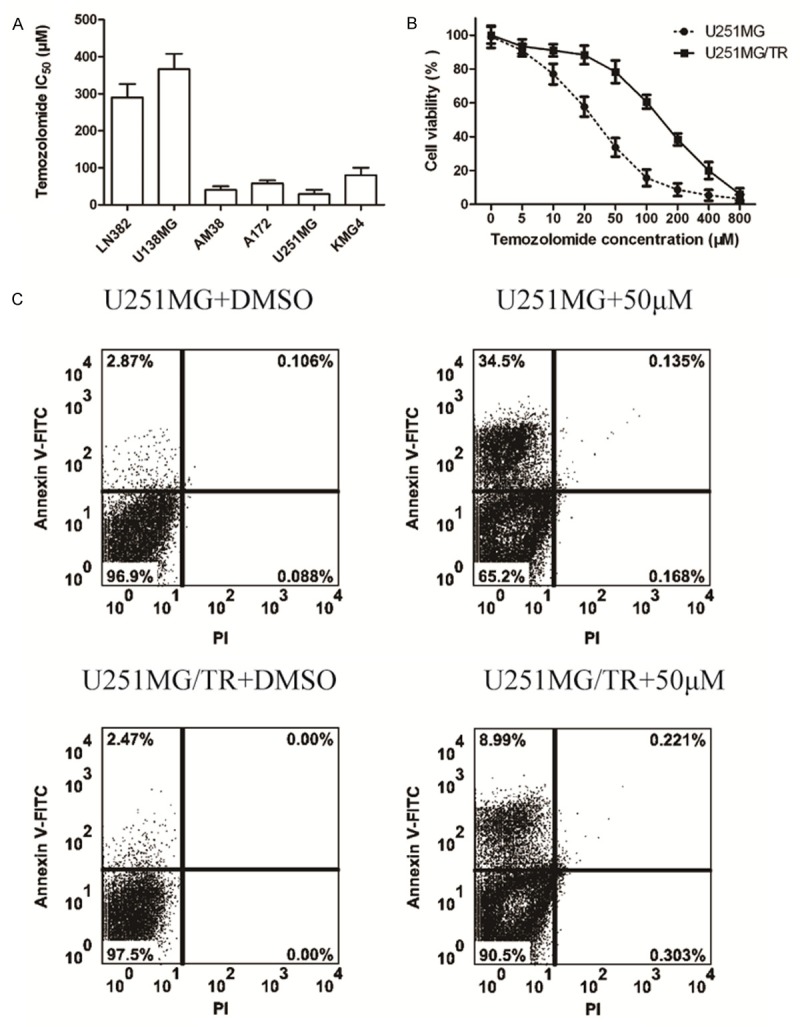
Screen and identify the resistant glioma cell lines to temozolomide. A: The IC50 values of temozolomide in different glioma cell lines. B: Cell viability was assessed by CCK-8 assay. C: Cell apoptosis was evaluated by flow cytometry. All experiments were performed in triplicate.
MiR-16 is significantly downregulated in temozolomide-resistant cell line (U-251MG/TR) compared with parental cell line (U-251MG)
To identify miRNAs specifically deregulated in U251MG/TR cells, we performed a comprehensive analysis of miRNA expression in U251MG and U251MG/TR cells using cancer microRNA PCR Array. Twelve miRNAs were overexpressed (>2.0-fold) and eight were under expressed (<2.0-fold) in U251MG/TR cells compared to U251MG cells. Fold changes of representative miRNAs expression are listed in Figure 2A. To validate the microRNA PCR Array data, we utilized TaqMan real-time RT-PCR assay for miR-150, let-7a, miR-9, miR-16, miR-98 and miR-125b, the three most up-regulated miRNAs and the three most down-regulated miRNAs. As shown in Figure 2B, these miRNAs were certainly up-regulated and down-regulated in U251MG/TR cells, suggesting that deregulation of those miRNAs is involved in the acquisition of TMZ resistance in GBM cells.
Figure 2.
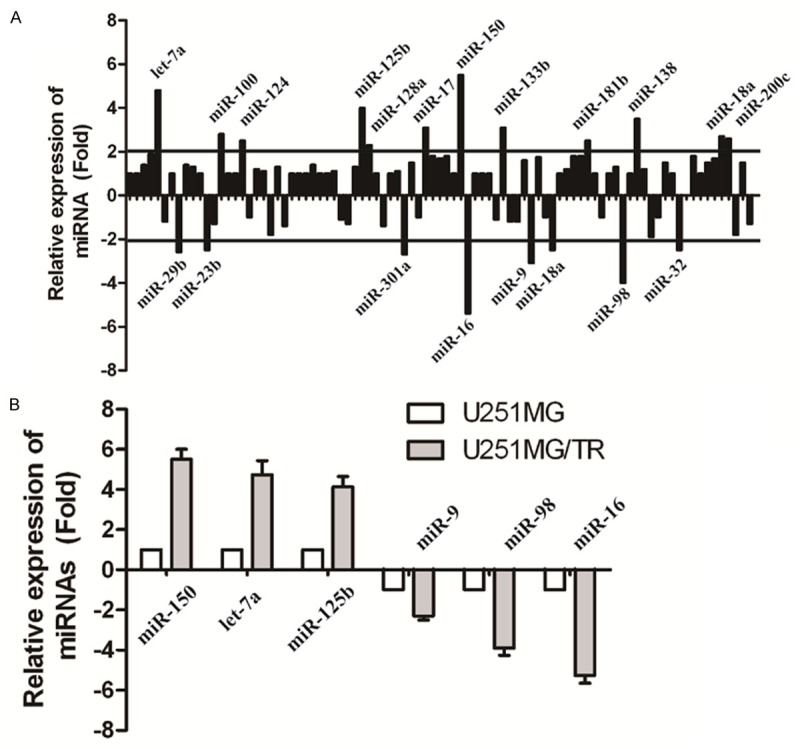
Eighty-eight cancer-related microRNAs were profiled using cancer microRNA PCR Array in U251 MG/TR and U251 MG cells. A: A graphic representation of the 88 microRNA readouts for U251 MG/TR and U251 MG cells. Each vertical line represents a single microRNA value of relative expression in U251 MG/TR and U251 MG cells. B: miR-150, let-7a, miR-9, miR-16, miR-98 and miR-125b were determined to be differentially expressed by cancer microRNA PCR Array in U251 MG/TR and U251 MG cells, and this result was validated by quantitative polymerase chain reaction result (qPCR). miR-150, let-7a and miR-125b, 3 specifically up-regulated microRNAs in U251MG/TR cells, were confirmed to be highly expressed in U251MG/TR cells, while miR-16 was confirmed to be down-regulated in U251MG/TR cells.
From the list of differentially expressed LncRNAs, we focused on miR-16, as previous reports showed that it was related with resistance.
MiR-16 contributed to TMZ resistance in glioma cells
To identify the effect of miR-16 on TMZ sensitivity in U251MG cells, we examined the mRNA expression of miR-16 in these cell lines firstly (Figure 3A). The correlation between the IC50 values and the relative mRNA expression of miR-16 was analyzed. The IC50 values of temozolomide significantly correlated with the expression level of miR-16 in these glioma cells (Figure 3B, Spearman r=-0.9014, P=0.0141). Furthermore, we performed CCK-8 assay to detect the cell viability of AM38 cells transfected with miR-16 inhibitor or U251MG/TR cells transfected with miR-16 mimics for 24 h, respectively, followed by treatment with TMZ for 48 h. Results showed that overexpression of miR-16 rendered U251MG/TR cells more resistant to temozolomide. U251MG/TR cells transfected with vector control had an IC50 value of 38.2 μM; whereas the IC50 value of U251MG/TR cells overexpressed with miR-16 was 93.6 μM (Figure 3C). Instead, transfection of miR-16 inhibitor significantly increased TMZ resistance in AM38cells (IC50Control: 151.6 μM; IC50AM38/miR-16inhibitor: 50.4 μM, P<0.05). In summary, these results suggest that miR-16 contribute to TMZ resistance in glioma cells.
Figure 3.
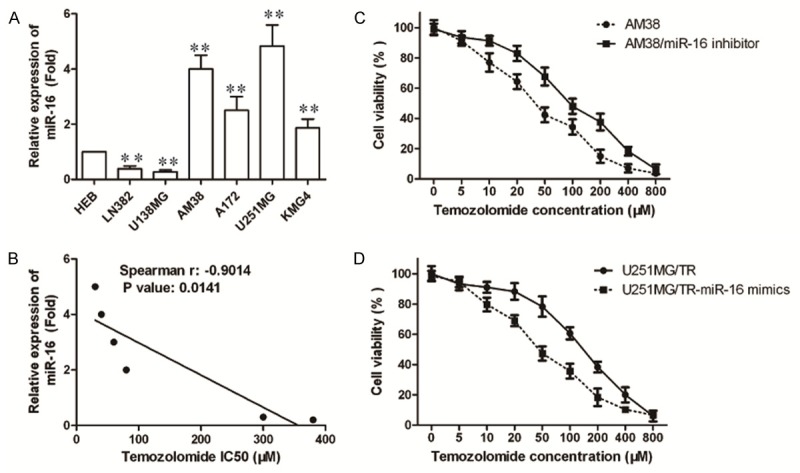
Correlation between miR-16 expression and temozolomide resistance in glioma cells. A: The expression of miR-16 in different glioma cell lines by qRT-PCR. B: The correlation between the relative miR-16 expression and the IC50 values in glioma cells was quantified by Spearman’s rank correlation. C: Temozolomide-sensitive AM38 cells were transfected with specific inhibitor to miR-16. D: Temozolomide-resistant U251MG/TR cells were transfected with miR-16 mimics. After incubation with temozolomide for 48 hr, cell viability was assessed using CCK-8 assay and IC50 value to temozolomide was calculated. All experiments were performed in triplicate.
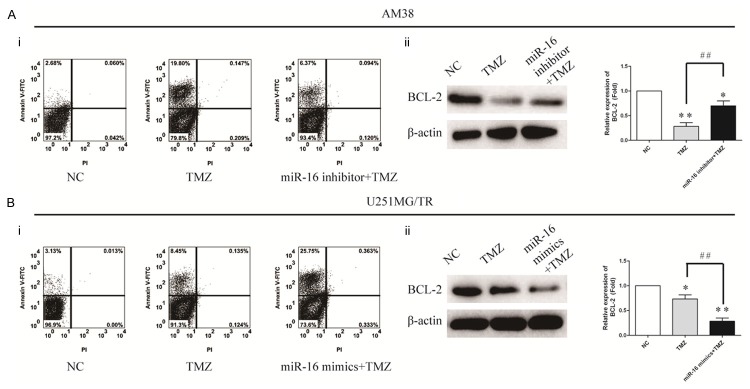
MiR-16 inhibits temozolomide induced apoptosis via upregulation of Bcl-2
To determine the mechanisms by which miR-16 decreased resistance of glioma cells to temozolomide, we analyzed the effect of miR-16 low expression on temozolomide induced apoptosis. AM38 cells stably transduced with miR-16 were treated with 50 μM temozolomide for 6 h, and then cells were washed twice and placed in temozolomide free medium to grow for 48 h. Cell apoptosis was determined by Annexin V/PI flow cytometry assays. The apoptosis rate of AM38 cells group and miR-16 inhibitor group were 3% and 6%, respectively (*P>0.05, Figure 4Ai). It suggests that low expression of miR-16 inhibits temozolomide induced apoptosis. To study underlying molecular mechanisms by which miR-16 inhibits temozolomide induced apoptosis, we analyzed several apoptosis related proteins and found Bcl-2 was significantly upregulated in AM38 cells stably transduced with miR-16 (Figure 4Aii), But the correspondingly converse results was observed in U251MG/TR cells (Figure 4Bi and 4Bii). Taken together, these data suggest that miR-16 inhibits temozolomide-induced apoptosis via upregulation of Bcl-2 in glioma cells.
Figure 4.
MiR-16 inhibits temozolomide induced apoptosis via upregulation of Bcl-2. A and B: AM38 cells transfected with miR-16 inhibitor and U251MG/TR cells were transfected with miR-16 mimics were labeled with FITC-Annexin V/PI, and cell apoptosis was evaluated by flow cytometry. Western blot analysis showed miR-16 upregulated Bcl-2 expression in AM38 cells and downregulated Bcl-2 expression in U251MG/TR cells, *P<0.05, **P<0.01 compared to NC, ##P<0.01. β-actin was used as an internal control. All data represent the means ± SEM of three replications.
Discussion
In the present study, we used TMZ-sensitive glioma cell lines to generate TMZ resistant variants by continuous exposure to the drug. We then found that miR-16 was downregulated in the human U251MG/TR cell lines. Furthermore, our findings demonstrate that the mechanism responsible for resistance of glioma cells to temozolomide is associated with miR-16-mediated downregulation of Bcl-2.
Recently much research effort had been intensely focused on studying the role of altered miRNA expression in tumor progression and drug resistance. There have been several reports regarding the relationship between miRNAs and TMZ resistance in glioma, such as miR-181b, miR-183/96/182, miR-221/222, miR-17 and miR-9 [26-31]. One particularly well-studied example was the ubiquitously expressed and highly conserved miR-16, one of the first miRNAs to be linked to human malignancies [19]. Evidence indicated that miR-16 could modulate the cell cycle, inhibited cell proliferation, promoted cell apoptosis and suppressed tumorigenicity both in vitro and in vivo [32]. Consistently, miR-16 was frequently deleted and/or downregulated in many types of cancer, such as chronic lymphocytic leukemia [19,33], prostate cancer [34] and lung cancer [35]. However, the correlation of miR-16 with chemosensitivity of tumor cells is unclear and remains to be elucidated.
Here, we demonstrated an altered miR-16 expression in TMZ resistant U251MG cells. To further determine whether the miR-16 could modulate the chemosensitivity of U251MG/TR cells, the miR-16 mimics or inhibitor were respectively transfected into U251MG/TR cells. We found that treatment with the mimics of miR-16 greatly decreased the sensitivity of U251MG/TR cells to temozolomide, while sensitivity to temozolomide was increased by treatment with the miR-16 inhibitor. All these results indicated that miR-16 may be an important modulator of TMZ resistance in glioma cells.
Dysregulation of apoptosis-regulating genes and proteins was one of the most common mechanisms of temozolomide resistance [36]. Bcl-2 translation was repressed by binding of miR-16 to a seed sequence in Bcl-2 mRNA 3’-untranslated regions, and loss of miR-16 in several cancer cell lines and tumors was associated with BCL-2 upregulation [20,22,34]. Therefore, we measured the expression of the Bcl-2 through miRNA overexpression or knockout experiment under in vitro condition to verify if miR-16 could regulate Bcl-2 expression by Western blot. We found that miR-16 knockdown led to up-regulation of Bcl-2 protein levels in AM38 cells, while overexpression of miR-16 resulted in decreased expression of Bcl-2 protein levels in U251MG/TR cells. Previous studies reported that Bcl-2 was essential for temozolomide induced cell death in human glioma, and thus may be a target to overcome therapeutic resistance toward temozolomide [22,37]. Consistent with previous studies, our results also showed that miR-16 inhibited temozolomide induced apoptosis via downregulation of Bcl-2.
In conclusion, our findings demonstrate that the mechanism responsible for resistance of glioma cells to temozolomide was associated with miR-16-mediated downregulation of Bcl-2. MiR-16 may function as an important modifier of the response of glioma cells to temozolomide. A new strategy combining current regimens with compounds targeting miR-16 may significantly improve the therapeutic outcome of temozolomide-resistant glioma.
Disclosure of conflict of interest
None.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins VP. Gliomas. Cancer Surv. 1998;32:37–51. [PubMed] [Google Scholar]
- 4.Gurney JG, Kadan-Lottick N. Brain and other central nervous system tumors: rates, trends, and epidemiology. Curr Opin Oncol. 2001;13:160–166. doi: 10.1097/00001622-200105000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Krex D, Klink B, Hartmann C, von Deimling A, Pietsch T, Simon M, Sabel M, Steinbach JP, Heese O, Reifenberger G, Weller M, Schackert G German Glioma Network. Long-term survival with glioblastoma multiforme. Brain. 2007;130:2596–2606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- 6.Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, Baeza N, Pisani P, Yonekawa Y, Yasargil MG, Lutolf UM, Kleihues P. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 7.Smith JS, Jenkins RB. Genetic alterations in adult diffuse glioma: occurrence, significance, and prognostic implications. Front Biosci. 2000;5:D213–231. doi: 10.2741/smith. [DOI] [PubMed] [Google Scholar]
- 8.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Asuthkar S, Velpula KK, Chetty C, Gorantla B, Rao JS. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 2012;3:1439–1454. doi: 10.18632/oncotarget.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010;1352:255–264. doi: 10.1016/j.brainres.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Shi L, Zhang S, Feng K, Wu F, Wan Y, Wang Z, Zhang J, Wang Y, Yan W, Fu Z, You Y. MicroRNA-125b-2 confers human glioblastoma stem cells resistance to temozolomide through the mitochondrial pathway of apoptosis. Int J Oncol. 2012;40:119–129. doi: 10.3892/ijo.2011.1179. [DOI] [PubMed] [Google Scholar]
- 16.Ujifuku K, Mitsutake N, Takakura S, Matsuse M, Saenko V, Suzuki K, Hayashi K, Matsuo T, Kamada K, Nagata I, Yamashita S. miR-195, miR-455-3p and miR-10a (*) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 2010;296:241–248. doi: 10.1016/j.canlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Wong ST, Zhang XQ, Zhuang JT, Chan HL, Li CH, Leung GK. MicroRNA-21 inhibition enhances in vitro chemosensitivity of temozolomide-resistant glioblastoma cells. Anticancer Res. 2012;32:2835–2841. [PubMed] [Google Scholar]
- 18.Yang YP, Chien Y, Chiou GY, Cherng JY, Wang ML, Lo WL, Chang YL, Huang PI, Chen YW, Shih YH, Chen MT, Chiou SH. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials. 2012;33:1462–1476. doi: 10.1016/j.biomaterials.2011.10.071. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo CJ, Pan Q, Li DG, Sun H, Liu BW. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J Hepatol. 2009;50:766–778. doi: 10.1016/j.jhep.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 22.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 23.Zhang N, Zhou H, Yu L, Lin J, Sheng H, Wang M. [The influence of miR-16 on proliferation and angiogenesis of U87MG in vivo] . Zhonghua Yi Xue Za Zhi. 2014;94:2618–2621. [PubMed] [Google Scholar]
- 24.Yang TQ, Lu XJ, Wu TF, Ding DD, Zhao ZH, Chen GL, Xie XS, Li B, Wei YX, Guo LC, Zhang Y, Huang YL, Zhou YX, Du ZW. MicroRNA-16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor-kappaB1/MMP9 signaling pathway. Cancer Sci. 2014;105:265–271. doi: 10.1111/cas.12351. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Chen J, Wang J, Zhang Y, Chen D, Yang C, Kai C, Wang X, Shi F, Dou J. Observation of ovarian cancer stem cell behavior and investigation of potential mechanisms of drug resistance in three-dimensional cell culture. J Biosci Bioeng. 2014;118:214–222. doi: 10.1016/j.jbiosc.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Zhang J, Han L, Zhang A, Zhang C, Zheng Y, Jiang T, Pu P, Jiang C, Kang C. Downregulation of miR-221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncol Rep. 2012;27:854–860. doi: 10.3892/or.2011.1535. [DOI] [PubMed] [Google Scholar]
- 27.Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F, Pirtoli L, Miracco C. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 2013;14:574–586. doi: 10.4161/cbt.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li P, Lu X, Wang Y, Sun L, Qian C, Yan W, Liu N, You Y, Fu Z. MiR-181b suppresses proliferation of and reduces chemoresistance to temozolomide in U87 glioma stem cells. J Biomed Res. 2010;24:436–443. doi: 10.1016/S1674-8301(10)60058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol Ther Nucleic Acids. 2013;2:e126. doi: 10.1038/mtna.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang H, Bian Y, Tu C, Wang Z, Yu Z, Liu Q, Xu G, Wu M, Li G. The miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Curr Cancer Drug Targets. 2013;13:221–231. doi: 10.2174/1568009611313020010. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Sai K, Chen FR, Chen ZP. miR-181b modulates glioma cell sensitivity to temozolomide by targeting MEK1. Cancer Chemother Pharmacol. 2013;72:147–158. doi: 10.1007/s00280-013-2180-3. [DOI] [PubMed] [Google Scholar]
- 32.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 33.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 34.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 35.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 36.Johannessen TC, Bjerkvig R. Molecular mechanisms of temozolomide resistance in glioblastoma multiforme. Expert Rev Anticancer Ther. 2012;12:635–642. doi: 10.1586/era.12.37. [DOI] [PubMed] [Google Scholar]
- 37.Cittelly DM, Das PM, Salvo VA, Fonseca JP, Burow ME, Jones FE. Oncogenic HER2{Delta}16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31:2049–2057. doi: 10.1093/carcin/bgq192. [DOI] [PMC free article] [PubMed] [Google Scholar]