Abstract
Hepatoma is a tumor with high degree of malignancy. A number of oncogenes and tumor suppressor genes play certain roles in tumorigenesis and progression. Among which, miRNA, as an important class of gene regulators, play important roles in regulating tumorigenesis and development of hepatoma. So know well the unique molecular pathway is very important. Here, we showed that there is a different miR-143 expression patterns in different hepatoma tissues, and that miR-143 expressions contribute disease progress. By contrast, we down-regulated the expression of miR-143 with miR-143 mimics in HepG2 cells resulting in decreased proliferation. And the decreased proliferations of HepG2 cells were due to a G0/G1 arrest of cell cycle. During this progress, the increased apoptosis may be another major cause for decreased proliferation of HepG2 cells. And then, we found miR-143 down-regulation induced decreased mRNA and protein expressions of TLR2 and NF-κB. These results show that HepG2 cells depend to a greater extent on miR-143 for proliferation, and miR-143 down-regulation may induce a cell cycle arrest though TLR and NF-κB pathway. miR-143 blockade may be beneficial in therapy of Hepatoma.
Keywords: miR-143, hepatoma, TLR2, proliferation, invasion
Introduction
Primary hepatocellular carcinoma (HCC) is one of the tumors with higher mortality due to its high degree of malignancy and difficulty with early detection. And surgical operation is an option for only a small portion of these patients [1]. Besides early-stage surgical resection, chemotherapy and radiotherapy often cannot obtain satisfactory results; accordingly, breakthrough in hepatoma treatment may take place with biotherapy [2]. The MicroRNA (MiRNA) as an important gene regulation mechanism in epigenetics has attracted widespread attention. The MiRNA is a class of endogenous small non-coding single-stranded RNA molecule comprising approximately 18-25 nucleotides; this evolutionarily highly-conserved MiRNA is widely present in eukaryotic cells and plays important regulatory roles in transcription and translation. As a regulator negatively regulating the target gene, miRNA can bind to the 3’-untranslated regions (3’-UTR) of a target gene in complete or incomplete complementary manner, leading to direct degradation or translational block of the target gene [3]. Therefore, miRNA plays an important role in tumorigenesis and development.
The miR-143 is localized to human No. 5 chromosome; its mature form miR-143-3p is under-expressed in a variety of tumors such as colorectal cancer [4], pancreatic cancer [5] and lung cancer [6], in which miR-143 plays the role of a tumor suppressor gene. A study found that miR-143 was under-expressed in hepatoma [7], while Zhang et al [8] found that miR-143 was over-expressed in hepatoma and behaved like an oncogene to promote the invasion and metastasis of hepatoma cells. Currently, the relationship between MiR-143 and pathogenesis and development of hepatoma is still unknown; neither is its mechanism of action. Therefore, this study determined miR-143 expression in hepatoma using quantitative real-time PCR (qRT-PCR); meanwhile, the hepatoma HepG2 cells were transfected with synthetic miR-143 mimics to observe its effects on the biological behavior of hepatoma cells and to explore its possible mechanisms of action.
Materials and methods
Materials
Hepatoma tissue and its corresponding peritumor tissue from 32 pairs of surgically resected specimens (24 males and 8 females, mean age was 53.8 years) in the Department of Hepatobiliary Surgery, the First Hospital of Xi’an Jiaotong University; all hepatoma tissue were confirmed through postoperative pathological sectioning; none of these patients received radiotherapy and chemotherapy postoperatively. The specimens were collected intraoperatively; the peritumor tissues came from normal liver tissue >3 cm away from the tumor margin; the specimens were stored in liquid nitrogen immediately after resection and then placed in a -80°C refrigerator for long-term preservation. This study was approved by the Medical Ethics Committee of the First hospital of Xi’an Jiaotong University; all subjects were informed of the related matters prior to participation in the experiments and all subjects signed the informed consent form. Human hepatocellular carcinoma HepG2 cell line was provided and stored by the Experimental Center for Biomedical Research, Xi’an Jiaotong University School of Medicine; fetal bovine serum (Evergreen) was purchased from Zhejiang Tianhang Biotech Limited; DMEM/high glucose (1×) culture medium was purchased from HyClone; TRIzol reagent and liposomes (LipofectamineTM2000) were purchased from Invitrogen; the design and synthesis of all primers were entrusted to Beijing Dingguo Changsheng Biotechnology (miR-143SL: GTC GTA TCC AGT GCG TGT CGT GGA GTC GGC AAT TGC ACT GGA TA CG AC GAG CTA C; upstream primer was CAG TGC GTG TCG TGG AG, downstream primer was GCG GTG AGA TGA AGC ACT; U6 upstream primer was CTC GCT TCG GCA GCA CA, downstream primer was AAC GCT TCA CGA ATT TGC GT); miR-143 mimics and negative control were purchased from Shanghai GenePharma (miR-143 mimics: 5’-UGA GAU GAA GCA CUG UAG CUC-3’; negative control: 5’-UUC UCC GAA CGU GUC ACG UTT-3’).
Determination of miR-143 expression in hepatoma tissue using qRT-PCR assay
The total RNA was extracted from hepatoma tissue stored in a -80°C freezer according to TRIzol reagent instructions; total RNA concentration and A260/280 ratio were measured with Nanodrop-2000, RNA within a ratio range of 1.8-2.0 was used for reverse transcription. Measure 1 μl total RNA; then, add 2 μl of 5× PrimeScript buffer, 0.5 μl PrimeScript RT Enzyme Mix I, 0.5 μl specific stem-loop (SL) primer and 6 μL RNase-free H2O; follow TaKaRa reverse transcription kit instructions, reversely transcribe miR-143 into cDNA, reaction temperature: 42°C for 15 min, 85°C for 5 s; then, store at 4°C for later use. The iQ5 Multicolor Real-Time PCR Detection System was used to detect the expression of miR-143 in hepatoma and the transfection efficiency of miR-143 mimics in HepG2 (using U6 internal reference). Reaction condition: denaturation at 95°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 30 s, a total of 40 cycles, then extension at 72°C for 5 min before terminating the reaction. Three duplicate wells were set for all reactions; the relative expression level of tumor versus peritumor tissue and transfection efficiency was calculated using 2-ΔΔCt (Table 1).
Table 1.
The gene primer sequences
| Gene | Primer Sequences (5’-3’) |
|---|---|
| TLR2 | Forward: GCCACCATTTCCACGGACT |
| Reverse: GGCTTCCTCTTGGCCTGG | |
| NF-κB | Forward: TGCTGTGCGGCTCTGCTTCC |
| Reverse: AGGCTCGGGTCTGCGTAGGG | |
| MMP-2 | Forward: GGCCCTGTCACTCCTGAGAT |
| Reverse: GGCATCCAGGTTATCGGGGA | |
| MMP-9 | Forward: CAACATCACCTATTGGATCC |
| Reverse: CGGGTGTAGAGTCTCTCGCT | |
| CD44 | Forward: TCCAACACCTCCCAGTATGACA |
| Reverse: GGCAGGTCTGTGACTGATGTACA | |
| MMP14 | Forward: GGATACCCAATGCCCATTGGCCA |
| Reverse: CCTCGGTGCACCATGTTTGGC | |
| Integrin β1 | Forward: GAGATGTGTCAGACCTGCCTTG |
| Reverse: ATTTGTCCCGACTTTCTACCTTGG | |
| Integrin β4 | Forward: TGACGATCTGGACAACCTCAAGCA |
| Reverse: ATCCAATGGTGTAGTCGCTGGTGA | |
| E-cadherin | Forward: GGTTATTCCTCCCATCAGCT |
| Reverse: CTTGGCTGAGGATGGTGTA | |
| GAPDH | Forward: CGGAGTCAACGGATTTGGTCGTAT |
| Reverse: AGCCTTCTCCATGGTTGGTGAAGAC |
Cell culture and transfection
Hepatoma HepG2 cells were cultured in DMEM medium 100 mL/L containing fetal bovine serum and double antibiotics in an incubator under 37°C and 50 mL/L CO2 condition. According to the instructions, LipofectamineTM 2000 was used to transiently transfect hepatoma HepG2 cell line with miR-143 mimics; the experiment consisted of mimics group (i.e. miR-143 mimics transfection group) and NC group (meaningless oligonucleotide transfection group).
Determination of cell proliferation using MTT assay
Cells were prepared into single-cell suspension, 5×105 cells/mL; these cells were seeded in 96-well plates, 100 μL/well; at 24, 48 and 72 h after the intervention, 20 μL MTT (final concentration was 5 mg/mL) was added in the dark and incubated in a 37°C incubator for 4 h; then, the supernatant was discarded and 150 μl DMSO was added; after shaking on a shaker until the crystalline was completely dissolved in DMSO, the A value was determined at 490 nm with a microplate reader; the mean of 5 wells was calculated.
Apoptosis assay
Cells were seeded in 6-well plates; transfection intervention was carried out after the attached cells entered in logarithmic growth phase. In 48 h after the intervention, the 2 groups of cells were digested with trypsin; the cells were collected and washed with cold PBS for 3 times; 5×105 cells/mL cell suspension was obtained and centrifuged; then, 500 μL binding buffer, 5 μl annexin V-FITC and 10 μL propidiumiodide (PI) were added sequentially, followed by repeated gentle pipetting to mix evenly. After standing at room temperature away from light for 10 min, the apoptosis was detected with flow cytometry. The experiment was repeated 3 times.
Cell cycle assay
For cell cycle analysis, HepG2 cells in exponential growth phase were permeated with 75% ethanol for overnight at 4°C and stained with Propidium Iodide (PI) in the presence of 5 µg/ml RNase (Sigma) for 10 min. And cell cycle distribution (G0-G1, S and G2-M) was analyzed with DNA cell cycle analysis software (ModFit, Becton Dickinson).
Cell apoptosis assay
Apoptosis was measured with a commercial kit (Tianjin Sungene Biotech, China) as recommended by the manufacturer. Approximately, 105 HepG2 cells were stained for 15 minutes with Annexin V- allophycocyanin (APC) and PI at room temperature in the dark. After analysis, the apoptotic HepG2 cells were analyzed with software of Flowjo 7.6. HepG2 cells that not stained with the dyes are the viable cells (Annexin V-APC negative, PI negative). HepG2 cells will stain with Annexin V-APC but not a PI dye, thus distinguishing cells in early apoptosis. However, in late stage apoptosis, the cell membrane loses integrity thereby allowing Annexin V-APC and PI to stain the cells (Annexin V-APC positive, PI positive).
Cell invasion assay
The melted Matrigel was diluted with DMEM medium and 100 μl of which was added to the upper chamber of TranswellTM; the chamber was incubated at 37°C for 4 h to allow Matrigel to solidify; after intervention for 48 h, the cells were digested with trypsin and re-suspended in DMEM medium containing 10 mL/L FBS; 200 μl of the cell suspension, 1×105 cells/mL, were seeded in the upper chamber while 600 μl DMEM medium containing 200 mL/L FBS were added to the lower chamber. After incubation in an incubator under 37°C and 50 mL/L CO2 condition for 24 h, cotton swabs were used to gently wipe off the cells remaining on the membrane surface without passing through. After fixation in 40 g/L paraformaldehyde, staining in 1 g/L crystal violet and wash with PBS, the cells were counted under high-power fields microscopically, 5 randomly selected fields for each chamber; the cell counts in each visual field was recorded. The cell counts passing through the membrane were used to indirectly reflect tumor cell invasion ability.
Western blot assay
After intervention for 48 h, the grouped HepG2 cells were washed with PBS to remove the residual culture medium; 200 μl RIPA lysis buffer was added, followed by repetitive pipetting and lysis on the ice for 1 h; the contents were collected into centrifuge tubes and centrifuged at 12000 r/min; after loading buffer was added to the supernatant, the samples were treated at 100°C for 5 min to denature the proteins; 10 μl sample was loaded into each well for SDS-PAGE and then the proteins were transferred to a PVDF membrane. After blocking in 50 g/L skim milk, rabbit anti-human GAPDH antibody (1:2000), mouse anti-human TLR2 antibody (1:1500), mouse anti-human NF-κB antibody (1:1500), rabbit anti-human MMP-2 antibody (1:1500), rabbit anti-human MMP-9 antibody (1:1500), rabbit anti-human CD44 antibody (1:1500), rabbit anti-human MMP14 antibody (1:1500), rabbit anti-human E-cadherin antibody (1:1500), rabbit anti-human integrin β1 antibody (1:1500) and rabbit anti-human integrin β4 antibody (1:1500) were added to incubate at 4°C overnight. After washing with PBST, the membrane was incubated with HRP-conjugated goat anti-rabbit IgG (1:2000) or HRP-conjugated rabbit anti-mouse IgG (1:2000) for 2 h; after the membrane was washed, chemiluminescence detection and exposure in a dark room were carried out.
Statistical analysis
SPSS13.0 statistical software was used to analyze the above experimental data. Measurement data were represented as x ± s; ANOVA and paired t-test was used to analyze the inter-group differences. When P<0.05, the difference was considered to have statistical significance.
Results
MiR-143 under-expression in hepatoma tissue
The qRT-PCR assay found that miR-143 was under-expressed in hepatoma tissue. In the 32 pairs of hepatoma tissue and peri-tumor tissue, miR-143 was under-expressed in 78.13% (25/32) hepatoma tissue (down-regulation exceeded 2 folds); the mean down-regulation was 4.36 folds (Figure 1A). After transfection with miR-143 mimics (the final concentration was 100 nmol/L), miR-143 expression level in HepG2 was significantly higher than that in control group (Figure 1B), suggesting successful transfection of mimics (P<0.05).
Figure 1.
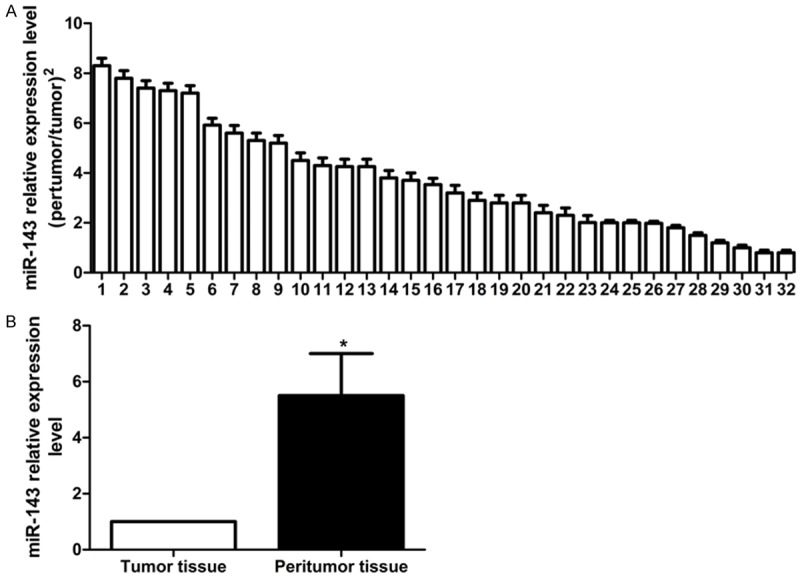
The miR-143 expressions in the 32 pairs of peritumor tissue and the corresponding tumor tissue. A. Relative expression of miR-143 in 32 pairs of hepatoma tissue (peritumor tissue/tumor tissue. B. Down-regulation of miR-143 expression in hepatoma tissue after treatment with miR-143 mimics.
MiR-143 inhibits proliferation and enhances apoptosis of hepatoma cells
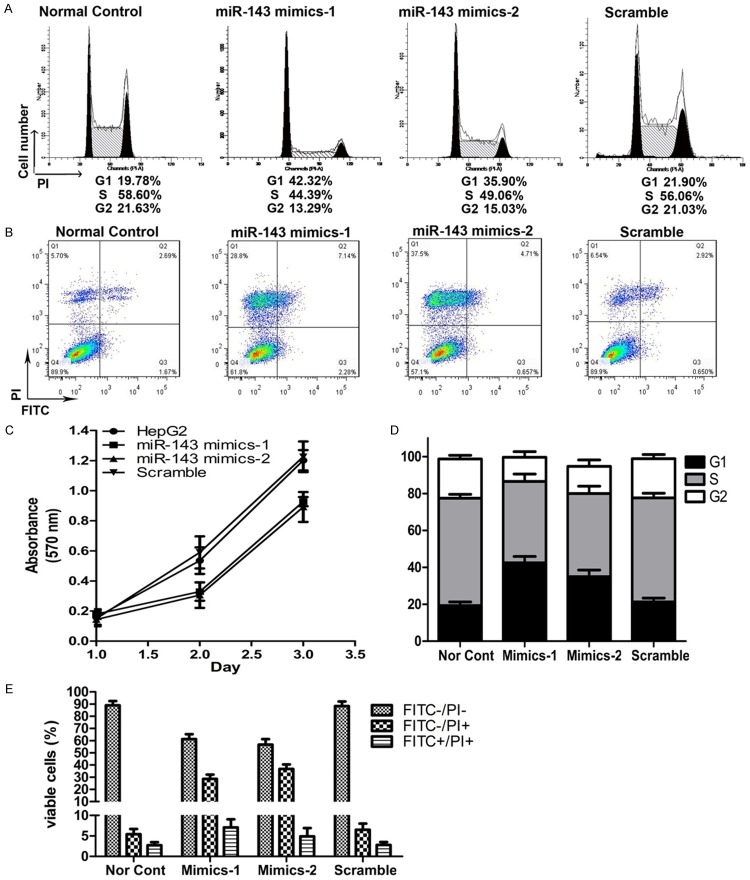
MTT assay results showed that, in 24, 48 and 72 h of intervention after hepatoma HepG2 cell line was transfected with mimics, the proliferation rate was significantly lower in intervention group than in control group; the difference had statistical significance (P<0.05, Figure 2C). In order to verify whether miR-143 up-regulation inhibited tumor cell proliferation by promoting apoptosis, flow cytometry was used to determine the cell cycle and early-phase apoptosis rate in the 2 groups of cells in 48 h after intervention (Figure 2A and 2B). The HepG2 cells after miR-143 silence were selected for cell cycle assay. The results showed that the proportion of cells in G0/G1 phase increased in miR-143 silenced HepG2 cells, as shown in Figure 4. In addition, the apoptosis experimental results showed that the apoptosis of HepG2 was significantly enhanced in mimics intervention group than in control group (Figure 2D and 2E).
Figure 2.
The miRNA-143 silence attenuates the proliferation of HepG2 cell. A. Cell cycle distribution of HepG2 cells before and after treatment with miR-143 mimics. B. Cell death distribution of HepG2 cells after treatment with different concentration of halofuginone. C. Effect of miR-143 silence on cellular proliferation in HepG2 cells assessed by MTT assay. D. The relative distribution of cell cycle of HepG2 cells after treatment with miR-143 mimics. E. The relative distribution of cell death of HepG2 cells after treatment with miR-143 mimics.
Figure 4.
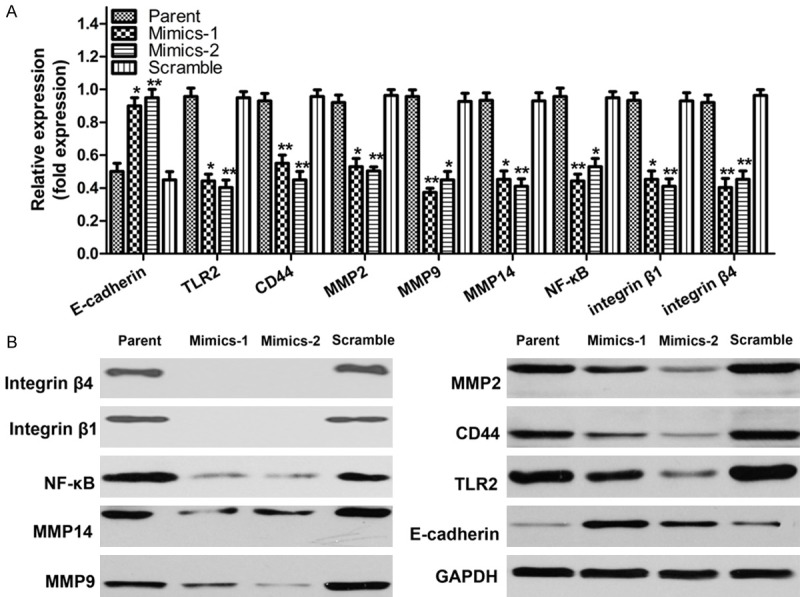
The miR-143 regulates tumor gene expression in hepatoma cells. A, B. Detection of the mRNA and protein expression levels of TLR2, NF-κB, MMP-2, MMP-9, CD44, MMP14, E-cadherin, integrin β1 and integrin β4 in HepG2 cells after treatment with miR-143 mimics.
MiR-143 inhibits hepatoma cell invasion
A key feature of HepG2 cells that have higher miR-143 expression is their increased migration and invasion capacity. The migration and invasion abilities of HepG2 cells after miR-143 silence were detected with wound-healing assay and transwell experiment. The results of the cell invasion (Figure 3A) and the wound-healing assay (Figure 3B) showed that the metastatic capacity of HepG2 cells was inhibited by down-regulation of miR-143. The amount of cells that migrated to the lower side of the collagen-coated membrane was significantly reduced and the migration of HepG2 cells was also prominently decreased after transfected with miR-143 mimics. But the migration and invasion abilities of scramble control cells were unaffected when compared to the parental cells (Figure 3C).
Figure 3.
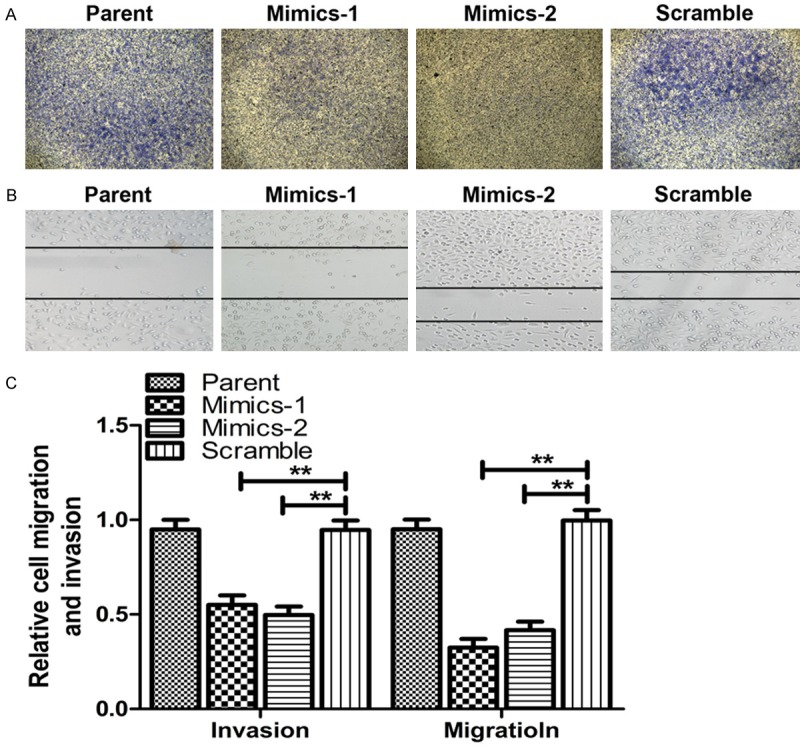
MiR-143 silence attenuates the metastasis of HepG2 cells. A. Representative images of transwell assay for cell invasion after miR-143 silence. B. Representative images of wounding-heal assay for cell migration after miR-143 silence. C. The relative cells migration and invasion. *P<0.05, **P<0.01, compared with the control
.
The miR-143 regulates tumor gene expression in hepatoma cells
TLR2 may play a role in tumorigenesis and development. But it is still unknown whether miR-143 affects hepatoma progression through TLR2. Therefore, real-time PCR and western blot assay were used to determine the changes in expression of TLR2 and related pathways in HepG2 cells after intervention with miR-143 mimics. The experimental results showed that, in 48 h after intervention with the mimics, the mRNA and protein expression levels of TLR2, NF-κB, MMP-2, MMP-9, CD44, MMP14, integrin β1 and integrin β4 all were decreased significantly, and the expressions of E-cadherin were significantly increased (Figure 4).
Discussion
Hepatoma is a tumor with high degree of malignancy. Because most of the cases are already in advanced stage at the time of diagnosis, surgical treatment is greatly limited. In recent years, hepatoma biotherapy has become an important breakthrough point for the treatment of hepatoma; searching for therapeutic targets has important significance for the treatment of hepatoma. A number of oncogenes and tumor suppressor genes play certain roles in tumorigenesis and progression. Among which, miRNA, as an important class of gene regulators, play important roles in regulating tumorigenesis and development of hepatoma. Accordance to domestic and foreign study reports, the role of miR-143 expression in hepatoma progression is still unclear. Therefore, we further studied the mechanisms of miR-143 action on hepatoma progression.
The miR-143 is under-expressed in a variety of tumors and can be used as a prognostic marker [9]. Its target genes have been identified in many tumors successively, including COX-2, MMP-13, GLI3 [10], which function as tumor suppressor genes in tumorigenesis and development. Bauer et al [11] found that miR-143 was under-expressed in colon cancer. In 2009, Chen et al [12] observed similar phenomenon and pointed out that the under-expression of miR-143 resulted in the loss of inhibition on KRAS expression, leading to enhanced tumor cell proliferation. Currently, there are only a few studies on miR-143 and hepatoma. Two reports on miR-143 expression in hepatoma and the relationship between them are still controversial. In order to explore the relationship between them, we first use real-time quantitative PCR to detect miR-143 expression in hepatoma tissue in this experiment and found that miR-143 was under-expressed in hepatoma.
In a study of prostate cancer, the investigators found that, after miR-143 expression, prostate cancer cell proliferation was suppressed by inhibiting target gene ERK5 [13]. In this study, we found with MTT assay and apoptosis assay that hepatoma HepG2 cell proliferation could be inhibited through increasing the apoptosis rate after miR-143 expression had been restored. Kapoor et al [14] found in their study that miR-143 as a tumor suppressor gene was able to suppress the invasion ability of prostate cancer by inhibiting GOLM1. While, we confirmed with TranswellTM assay that miR-143 could inhibit the proliferation and invasion of hepatoma HepG2 cell line. To further investigate the mechanism of miR-143 action in enhancing hepatoma apoptosis and inhibiting its invasion, we examined the possible target gene TLR2 [15] and its downstream pathway NF-κB.
The toll-like receptor (TLR) is mainly expressed on the surface of immune cells and is able to pass on the pathogen-caused damages to T or B cells. In a study conducted by Soares et al [16], TLR2 was over-expressed in hepatoma; we speculate that this may be related to the loss of miR-143 inhibition, but further study is needed to determine the relationship between them. NF-κB signaling pathway is presently considered to be frequently involved in the progression of tumor cells, including liver cancer [17]. Therefore, we determined NF-κB expression activity in mimics intervention group. Western blot results showed that, after miR-143 expression was up-regulated in HepG2 cells, the expression of its potential target gene TLR2 was down-regulated and subsequently, NF-κB in the downstream pathway was also down-regulated. The studies in recent years showed that the abnormal activation of NF-κB as a transcription factor during tumorigenesis can inhibit tumor apoptosis and directly affect the tumorigenesis and development of malignant tumors [18]. In addition, we also examined the invasion-related factor MMP-2 and MMP-9; the results showed that the expression of MMP-2 and MMP-9 were also significantly down-regulated after intervention with mimics; accordingly, we believe that miR-143 can also inhibit hepatoma cell invasion by down-regulating MMP-2 and MMP-9 expression.
This study demonstrated that miR-143 was under-expressed in hepatoma; up-regulating its expression level could inhibit its potential target gene TLR2 and NF-κB in the downstream pathway, leading to inhibition of hepatoma proliferation. In addition, down-regulating the expression of invasion-related factor MMP-2 and MMP-9 also play an important role in promoting hepatoma cell invasion. However, given the numerous miRNA target genes present, miR-143 may inhibit hepatoma proliferation and invasion through other target genes. Therefore, further studies are needed to elucidate the mechanisms of miR-143 mediated inhibition on hepatoma.
Disclosure of conflict of interest
None.
References
- 1.Dong J, An W, Ma W, Guo X, Gao Y, Liu C, Li Y. Primary hepatic malignant fibrous histiocytoma mimicking hepatocellular carcinoma: A report of two cases. Oncol Lett. 2014;5:2150–2154. doi: 10.3892/ol.2014.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu M, Chen Y, Zheng S, Duan Z, Zhang JY. Biotherapy for autoimmune liver diseases. Curr Pharm Biotechnol. 2014;6:510–5. doi: 10.2174/138920101506140910145539. [DOI] [PubMed] [Google Scholar]
- 3.Nadal E, Truini A, Nakata A, Lin J, Reddy RM, Chang AC, Ramnath N, Gotoh N, Beer DG, Chen G. A Novel Serum 4-microRNA Signature for Lung Cancer Detection. Sci Rep. 2015;5:12464. doi: 10.1038/srep12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su J, Liang H, Yao W, Wang N, Zhang S, Yan X, Feng H, Pang W, Wang Y, Wang X, Fu Z, Liu Y, Zhao C, Zhang J, Zhang CY, Zen K, Chen X, Wang Y. MiR-143 and MiR-145 regulate IGF1R to suppress cell proliferation in colorectal cancer. PLoS One. 2014;12:e114420. doi: 10.1371/journal.pone.0114420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masamune A, Nakano E, Hamada S, Takikawa T, Yoshida N, Shimosegawa T. Alteration of the microRNA expression profile during the activation of pancreatic stellate cells. Scand J Gastroenterol. 2014;3:323–31. doi: 10.3109/00365521.2013.876447. [DOI] [PubMed] [Google Scholar]
- 6.Wei J, Ma Z, Li Y, Zhao B, Wang D, Jin Y, Jin Y. miR-143 inhibits cell proliferation by targeting autophagy-related 2B in non-small cell lung cancer H1299 cells. Mol Med Rep. 2015;1:571–6. doi: 10.3892/mmr.2014.2675. [DOI] [PubMed] [Google Scholar]
- 7.Zeng XL, Zhang SY, Zheng JF, Yuan H, Wang Y. Altered miR-143 and miR-150 expressions in peripheral blood mononuclear cells for diagnosis of non-small cell lung cancer. Chin Med J (Engl) 2013;126:4510–6. [PubMed] [Google Scholar]
- 8.Zhang N, Su Y, Xu L. Targeting PKCε by miR-143 regulates cell apoptosis in lung cancer. FEBS Lett. 2013;22:3661–7. doi: 10.1016/j.febslet.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Wu P, Guo XH, Hu Y, Li YL, Shi J, Wang KZ, Chu WY, Zhang JS. miR-143: a novel regulator of MyoD expression in fast and slow muscles of Siniperca chuatsi. Curr Mol Med. 2014;3:370–5. doi: 10.2174/1566524014666140228100250. [DOI] [PubMed] [Google Scholar]
- 10.Xu B, Niu X, Zhang X, Tao J, Wu D, Wang Z, Li P, Zhang W, Wu H, Feng N, Wang Z, Hua L, Wang X. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem. 2011;350:207–13. doi: 10.1007/s11010-010-0700-6. [DOI] [PubMed] [Google Scholar]
- 11.Bauer KM, Hummon AB. Effects of the miR-143/-145 microRNA cluster on the colon cancer proteome and transcriptome. J Proteome Res. 2012;11:4744–54. doi: 10.1021/pr300600r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Ma C, Zhang W, Chen Z, Ma L. Down regulation of miR-143 is related with tumor size, lymph node metastasis and HPV16 infection in cervical squamous cancer. Diagn Pathol. 2014;9:88. doi: 10.1186/1746-1596-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pekow J, Meckel K, Dougherty U, Butun F, Mustafi R, Lim J, Crofton C, Chen X, Joseph L, Bissonnette M. Tumor suppressors miR-143 and miR-145 and predicted target proteins API5, ERK5, K-RAS, and IRS-1 are differentially expressed in proximal and distal colon. Am J Physiol Gastrointest Liver Physiol. 2015;3:G179–87. doi: 10.1152/ajpgi.00208.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapoor S. mir-143 and it’s emerging role as a modulator of systemic carcinogenesis. Cell Biochem Biophys. 2014;3:639–40. doi: 10.1007/s12013-013-9751-0. [DOI] [PubMed] [Google Scholar]
- 15.Guo H, Chen Y, Hu X, Qian G, Ge S, Zhang J. The regulation of Toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol Cancer. 2013;12:77. doi: 10.1186/1476-4598-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X, Liu D, Gong W, Zhao G, Liu L, Yang L, Hou Y. The toll-like receptor 3 ligand, poly (I:C), improves immunosuppressive function and therapeutic effect of mesenchymal stem cells on sepsis via inhibiting MiR-143. Stem Cells. 2014;2:521–33. doi: 10.1002/stem.1543. [DOI] [PubMed] [Google Scholar]
- 17.He G, Karin M. NF-kappaB and STAT3-key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thapa D, Meng P, Bedolla RG, Reddick RL, Kumar AP, Ghosh R. NQO1 suppresses NF-κB-p300 interaction to regulate inflammatory mediators associated with prostate tumorigenesis. Cancer Res. 2014;19:5644–55. doi: 10.1158/0008-5472.CAN-14-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]