Abstract
Semaphorin-3F (SEMA3F) is a member of the class III semaphorin family, and is seen as a candidate tumor suppressor gene. The aims of this study were to evaluate the effect of SEMA3F in colorectal cancer (CRC) patients, and to explore the mechanism for that SEMA3F suppresses tumor progression and metastasis. The expression levels of SEMA3F in the colorectal cancer tissues and corresponding non-tumor colorectal tissues were determined by Western blotting and real-time quantitative PCR (qRT-PCR). In addition, we evaluate the effects of SEMA3F on CRC cell migration and colony formation in vitro. Subsequently, quantitative methylation-specific PCR (qMSP) was used to detect the DNA methylation status in the CpG islands of SEMA3F gene promoter in normal colon and colorectal cancer cell lines, colorectal cancer tissues and corresponding non-tumor colorectal tissues. We found that SEMA3F was downregulated in the protein (P < 0.01) and mRNA (P < 0.001) levels in CRC tissues as compared to matched adjacent non-tumor tissues. Moreover, MSP assay showed high levels of SEMA3F gene promoter methylation in the CpG islands in some CRC cell lines and tissue samples. Furthermore, SEMA3F expression was reactivated in CRC cell lines after treatment with 5-Aza-CdR, demethylation of SW620 cells resulted in cell colony formation and invasion inhibition. These findings suggest DNA methylation of promoter CpG island-mediated silencing of the tumor suppressor SEMA3F gene plays an important role in the carcinogenesis of CRC.
Keywords: Colorectal cancer, SEMA3F, DNA methylation
Introduction
Colorectal cancer (CRC) remains the third most commonly diagnosed cancer and the fourth leading cause of cancer mortality worldwide, the incidence of CRC seems to have rapidly become an epidemic in Asian, especially in China [1,2]. In the last decade, great efforts have been focused on the development of new treatment modalities and diagnostic technologies, however, there is a lack of sensitive and specific biomarkers for early diagnosis, and CRC is complex and poorly understood. Thus, identification of the molecular mechanisms of CRC is critical to improving diagnosis and treatment.
Semaphorins, were first identified as a family of genes encoding guidance molecules for the embryologic development of the nervous system, as negative mediators of axonal guidance in the central nervous system [3]. Although semaphorins play a critical role as axon guidance molecules in the developing nervous system, several semaphorins are expressed in adult nonneuronal tissues, suggesting other functions. Based on structure of the semaphoring family, they are classified into seven subclasses, among of these semaphorin 3F (SEMA3F) belongs to the class 3 secreted type of semaphorin protein [4]. In fact, previous studies reported that SEMA3F was initially identified as a candidate tumor suppressor gene at chromosome 3p21.3 [5], and might play an important repressive role in the growth, angiogenesis, invasion, and metastasis of several cancers [6-8]. For example, recent data from immunohistochemical studies of lung cancers, found that reduction of SEMA3F expression in higher stages of lung cancer [9]. Also Nasarre P and his colleagues previously reported that SEMA3F was able to inhibit the attachment and spreading of breast cancer cell lines as evidenced by loss of lamellipodia extensions [7]. Furthermore, studies still further highlighted the roles of SEMA3F as negative regulators of tumor growth in endometrial cancer [10], and might represent a potent inhibitor of melanoma cell proliferation [11]. SEMA3F has been a competition between SEMA3F and vascular endothelial growth factor (VEGF) for binding to their common neuropilin receptor, which displayed a reduced density of blood vessels and explained for the anti-angiogenic activity during tumor development [12]. Although the tumor suppressor function of SEMA3F may be partly attributed to its anti-angiogenesis activity, it is unclear which of these mechanisms is the primary mechanism used by SEMA3F to inhibit tumor development.
Recent study has reported that DNA methylation play a role in SEMA3F expression [13]. In this study, we confirmed once again that SEMA3F was down-regulated in CRC tissues and associated with progressive phenotypes of CRC. Further methylation analysis of SEMA3F gene promoter indicated that its expression was regulated by methylation of correlated CpG islands in some ways. In the end, we found that reactivation of SEMA3F expression is associated with demethylation of SEMA3F gene by 5-Aza-CdR treated in SW620 cells.
Materials and methods
Patients and samples
Fresh CRC tissues and their matched adjacent non-tumor tissues that located more than 3 cm from the tumor were collected from 36 CRC patients who underwent surgery at the Second Affiliated Hospital of Nantong University between 2013 and 2014, as approved by the institutional ethics committee of the Second Affiliated Hospital of Nantong. No patients had received chemotherapy or radiotherapy before surgery. Tissue samples collected and immediately stored in liquid nitrogen until RNA, DNA or protein extraction.
Cell lines and cell culture
The human CRC cell lines (LoVo, HCT116, SW480 and SW620) were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China), and one normal colon mucosa cell line (FHC) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). HCT116 and FHC cells were grown in DMEM Medium (Gibco) with 10% fetal bovine serum (FBS, HyClone, UT). SW480 and SW620 cells were grown in Leibovitz’s L-15 Medium (Gibco) with 10% FBS. LoVo cells were grown in RPMI-1640 Medium (Gibco) with 10% FBS. All the cell lines were incubated at 37°C in a humidified atmosphere with 5% CO2.
Plasmid construction and transfection
The recombinant plasmid pEGFP-N1-SEMA3F (p-SEMA3F) was transfected into SW620 cells using Lipofectamine 3000 (Invitrogen, Carlsbad, USA) transfection method according to the manufacturer’s instructions. At the 48 h after the transfection, cells were treated with G418 selection (Promega) for 2 weeks. A stable transfectant of the pEGFP-N1 empty vector was used as a control. Then the resistant clones were picked out to culture extensively. The SEMA3F-overexpression SW620/SEMA3F cell line stably expressing SEMA3F gene was established. The level of SEMA3F expression after transfection was assayed by real-time PCR.
Western blot analysis
Protein lysates were prepared from collected tumor, their adjacent non-tumor tissues and cells with RIPA lysis buffer (ProMab Biotechnology), and the supernatant was collected for determination of total protein concentration by DC-protein assay method (Bio-Rad) to maintain the same loads after centrifugation for 20 min at 12000 rpm. Protein was loaded onto SDS-PAGE and transferred onto PVDF membrane. After probed with 1:400 diluted goat anti-SEMA3F (sc-68795, Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight, the blots were subsequently incubated with HRP-conjugated secondary antibody (1:5000). Signals were visualized using ECL Substrates (Millipore, MA, USA). β-actin was used as an endogenous protein for normalization.
Real-time RT-PCR
The mRNA expression of SEMA3F was analyzed by quantitative real-time PCR. Total RNAs were extracted using the Trizol (Gibco) and cDNA was subsequently synthesized from total RNA using an Omniscript RT kit (Qiagen, Valencia, CA) following the supplier’s instructions. Quantitative real-time PCR was performed using 2 × Fast EvaGreenTM qPCR Master Mix on the Mastercycler Ep Realplex (Eppendorf 2S, Hamburg, Germany). According to the protocol, 25 μl reactions were run of with 1 μl cDNA. Cycling parameters were as follows: hot start at 96°C for 2 min; 40 cycles of amplification/quantification at 96°C for 15 s, and 60°C for 1 min during which time fluorescence was measured. The experiment was performed in triplicate and repeated for three times. Primer sequences were as follows: SEMA3F forward: 5’-CGATGACGGTGGTCACTGTTG-3’; SEMA3F reverse: 5’-CAGCGTAAATGACAGGGTTCCT-3’. The β-actin gene served as an internal control. The sequences of the primers for β-actin as follows: β-actin forward 5’-TAGTTGCGTTACACCCTTTCTTG-3’; β-actin reverse 5’-CACCTTCACCGTTCCAGTTTT-3’. Relative expression level for each target gene was normalized by the Ct value of β-actin using a 2-ΔΔCt relative quantification method.
Methylation-specific PCR
The methylation status of CpG island of all samples was initially screened at SEMA3F gene promoter regions by methylation-specific PCR (MSP) as described previously. Genomic DNA extracted from tissues and cells was modified with bisulfite reagents following the manufacturer’s instructions (Zymo Research, CA). This modification converts unmethylated cytosine to thymine, whereas methylated cytosine remained unchanged. A total of 20 ng of bisulfite-modified DNA was subjected to PCR amplification and directly sequenced using the ABI 3700 automated sequencing system (Applied Biosystems, CA). When the CpG sites in the region analyzed by MSP are methylated, the methylated (M) band would show up. However, the unmethylated (U) band would be present when the sites are unmethylated. Occasionally, both bands could be present if the sites are partially methylated. The MSP primers designed for SEMA3F were as follows: methylated: MSP-M-F1: 5’-GGTTTACGTACGGGATTAGGGGTAC-3’, MSP-M-R1: 5’-AAACTACAACGCCAAAAAACAACGA-3’, MSP-U-F1: 5’-TTTATGTATGGGATTAGGGGTATGG-3’, MSP-U-R1: 5’-AACTACAACACCAAAAAACAACAAA-3’.
Treatment of cells with 5-Aza-CdR
CRC cell line was incubated in culture media with the demethylating agent 5-Aza-2’-deoxycytidine (5-Aza-CdR) (Sigma Chemical Co., St. Louis, MO, USA) for 3 days using daily media changes. Cells were harvested and RNA and DNA were extracted on the third day.
MTT assay
Cells were seeded in each well of 96-well plate (1.5 × 103/well) and incubated overnight. After transfection with pEGFP-N1-SEMA3F, cell proliferation was determined at 0, 24, 48, and 72 hours using methylthiazolyl tetrazolium (MTT) assay. The cells were seeded and 20 μL of the MTT solution (5 mg/mL) was then added to each well at the indicated time. The absorbance value (OD) of each well was measured at 490 nm. Cells transfected with pEGFP-N1 and untreated cells were used as the control groups. All experiments were repeated at least three times.
Colony formation assay
Cells with stably transfected pEGFP-N1-SEMA3F were separately seeded into six-well plates. The media were changed twice a week, and after 14 days, cells were fixed with methanol for 10 min, stained with 0.5% crystal violet for 15 min, and rinsed three times with PBS. The number of colonies larger than 50 μm in diameter was counted under a microscope and the average number of colonies was achieved. Scrambled pEGFP-N1 and untreated cells were used as the control groups. The experiment was triplicated independently.
Cell migration assay
The cell migratory capacity was determined using transwell chambers (Corning, Corning, NY, USA). Briefly, cells (1 × 105/well) were suspended in 100 μL serum-free medium and then added to the upper chamber of the inserts, DMEM medium (GIBCO) containing 10% FBS (500 μL) was added to the lower chamber as the chemotactic factor. After culture for 24 hours at 37°C, non-migrated cells on the upper surface were removed gently with a cotton swab and cells that migrated to the lower side of the department were fixed and dyed with 0.1% crystal violet. The numbers of migrated cells were calculated by counting five different views under the microscopy. Independent experiments were repeated three times, with triplicates in each experiment.
Statistical analysis
Statistical analysis was performed using the SPSS 18.0 statistical software (SPSS Inc., Chicago, IL, USA). The paired-samples t-test was used in the analysis of differential SEMA3F expression between tumor and adjacent non-tumor tissues. For experiments in vitro, independent-samples t-test was used for assessing the significance of difference between the treatment and control groups. P-value < 0.05 was considered as a statistically significant difference.
Results
Expression level of SEMA3F is downregulated in CRC tissues
The levels of SEMA3F protein expression in 24 randomly selected pairs of CRC and their matched non-tumor colorectal tissues from CRC patients was estimated by Western blotting analysis. Six representative cases of the Western blotting result are shown in Figure 1A (normalized to the β-actin of the same samples). The SEMA3F protein levels was significantly reduced in 19 (79%) of the CRC tissue samples compared with their adjacent non-tumor colorectal tissues, and the average SEMA3F protein level in 24 CRC tissues was significantly lower than that in their adjacent non-tumor colorectal tissues (Figure 1B, P < 0.01).
Figure 1.
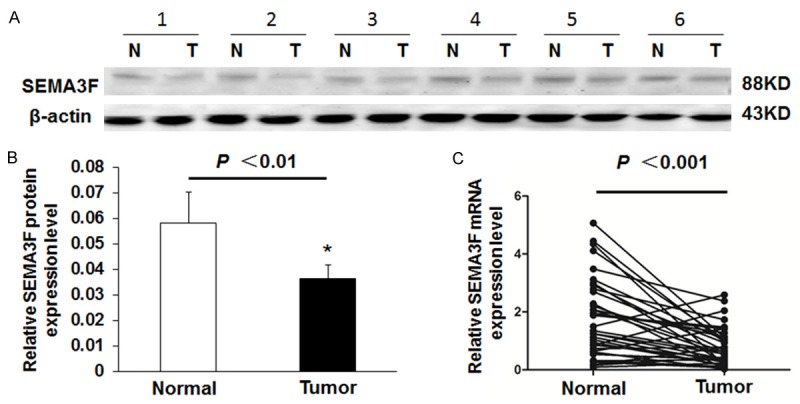
SEMA3F expression in CRC tissues and adjacent non-tumor tissues. A: Representative result of SEMA3F protein expression in six matched samples of CRC tissues (T) and adjacent non-tumors tissues (N). B: Summary of Western blot results from CRC tissues and adjacent normal tissues presented as relative bands density normalized to the β-actin of the same samples. The average SEMA3F expression level in CRC tissues was significantly lower than that in adjacent non-tumor tissues (n = 24, P < 0.01). C: The average relative expression of mRNA level of SEMA3F in CRC tissues compared to paired adjacent non-tumor tissues (n = 36, P < 0.001).
The above results were confirmed to examine SEMA3F mRNA level by real-time RT-PCR analysis in the 36 CRC tissues and their corresponding non-tumor colorectal mucosal samples. We found the average relative expression of SEMA3F in all 36 cases of CRC tissues is lower than that in adjacent non-tumor tissues (Figure 1C, P < 0.001), which was consistent with the results of previous studies by real-time RT-PCR [14].
Reduced expression of SEMA3F is related to DNA methylation
To determine whether an epigenetic mechanism contributes to the downregulation of SEMA3F expression in CRC, DNA methylation status at the promoter region of the SEMA3F gene was examined with MSP assay. Furthermore, we searched for CpG islands in the SEMA3F promoter by using the online accessible software Methprimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) (Figure 2A). The results of the MSP assay showed that SEMA3F gene was hypermethylated in SW620, and partially methylated in SW480 and HCT116, but not methylated in LoVo and FHC cells (Figure 2B). Then we assessed the methylation status of the SEMA3F promoter CpG islands in 36 CRC tissue samples and their matched adjacent non-tumor colorectal tissues by using MSP (Figure 2C). Hypermethylation of the SEMA3F gene was detected in 69.4% (25/36) of CRC tissues, while hypermethylation in non-tumor colorectal tissues was only found in 16.7% (6/36) cases. In addition, SEMA3F gene promoter was partially methylated in 6 cases and non-methylated in 5 cases in CRC tissues, whereas 5 cases of partially methylation and 25 cases of non-methylation in non-tumor colorectal tissues, respectively. The difference in methylation between CRC tissues and non-tumor colorectal tissue specimens was significant (P < 0.001).
Figure 2.
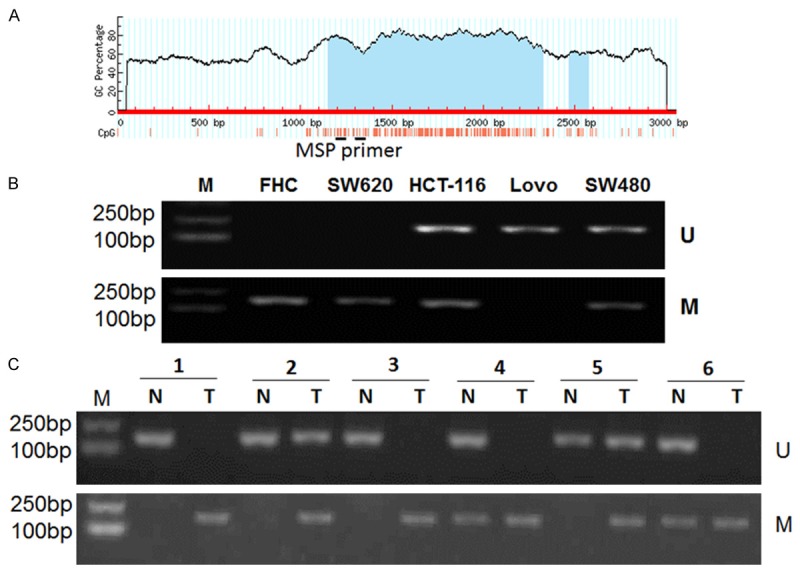
Down-regulation of SEMA3F in CRC cells is associated with DNA methylation of SEMA3F gene promoter region. A: A schematic illustration of the CpG island of the SEMA3F gene, which spans from -965 to + 540 with respect to the transcription start site (+ 1): red vertical bars, the locations of the CpG sites. The regions of MSP were shown. B: DNA methylation of SEMA3F in FHC and CRC cell lines. SEMA3F was hypermethylated in SW620, and partially methylated in SW480 and HCT116, but not methylated in LoVo and FHC. C: MSP showing DNA methylation of SEMA3F in CRC tissue specimens and corresponding non-tumor tissues. M: methylated; U: unmethylated.
Overexpression of SEMA3F inhibited cell migration and cell colony formation in vitro
Some research has recently reported that overexpression of SEMA3F inhibited the proliferation and metastatic potential of CRC cells in vitro and in vivo [14]. In this study, we once again evaluated the potential role of SEMA3F on cellular migration by transwell assays. SW620 cells were transfected with SEMA3F overexpressing or empty vector plasmid and seeded in the chamber, and their migratory abilities were determined 24 hours later. The results showed over-expression of SEMA3F was associated with a significant reduction of migration compared to the empty vector (Figure 3A and 3B, P < 0.05).
Figure 3.
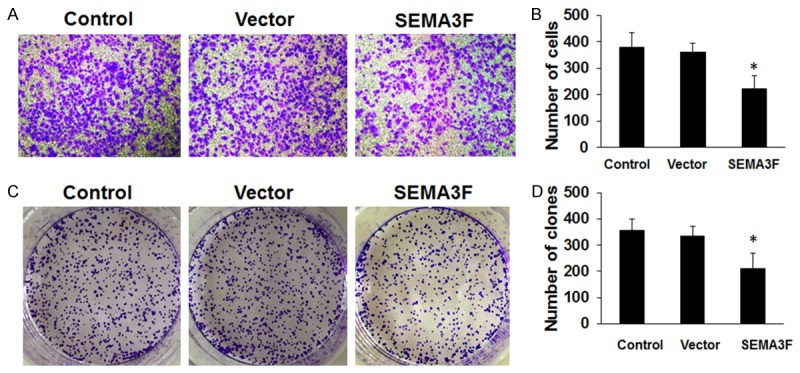
The effect of SEMA3F on cell migration and colony formation. A and B: Cell migration was measured by transwell assay, which indicated that overexpression of SEMA3F inhibited cell migration in SW620 cell line. C and D: About 1 × 1000 cells were seeded on each plate and transfected with pEGFP-N1-SEMA3F or empty vector. After 14 days, cells were stained with crystal violet. Colonies consisting of > 50 cells were counted. The data are presented as the means ± standard deviation (SD) normalized to the negative control (NC) transfected cells. *P < 0.05 (values were compared with untreated SW620 cells or empty vector).
These findings were further confirmed by colony formation assay. The overexpression of SEMA3F significantly inhibited the migration of SW620 cells at 48 h after transfection (Figure 3C and 3D, P < 0.05).
Reactivation of SEMA3F expression after treatment with 5-Aza-CdR
To further examine whether the reactivation of SEMA3F expression can regulate CRC growth and invasion, we analyzed the invasive and colony formation capability of the SEMA3F-silencing cell line, SW620. We treated the SW620 cells with DNA methyltransferase inhibitor 5-Aza-2’-deoxycytidine, 5-Aza-CdR, which was used to determine whether SEMA3F expression can be reactivated. First, SEMA3F expression significantly increased in SW620 cells after treatment with 5-Aza-CdR, with the highest expression occurring at a concentration of 10 μm by qRT-PCR (P < 0.05, Figure 4A) and western blot analysis (P < 0.05, Figure 4B and 4C). Next we found that the colony formation rate of the cells significantly decreased after a 10 μm 5-Aza-CdR treatment, compared to an untreated control after 24 h (P < 0.05; Figure 4F and 4G) by using colony formation assay. Moreover, transwell assay showed that the number of invading cells was significantly decreased when SW620 cells were treated with 5-Aza-CdR, compared with the untreated group that invaded through the membrane (P < 0.05; Figure 4D and 4E). These studies show that colony formation and invasive capacity of SW620 cells treated with 5-Aza-CdR was significantly decreased than SW620 cells without any treatment.
Figure 4.
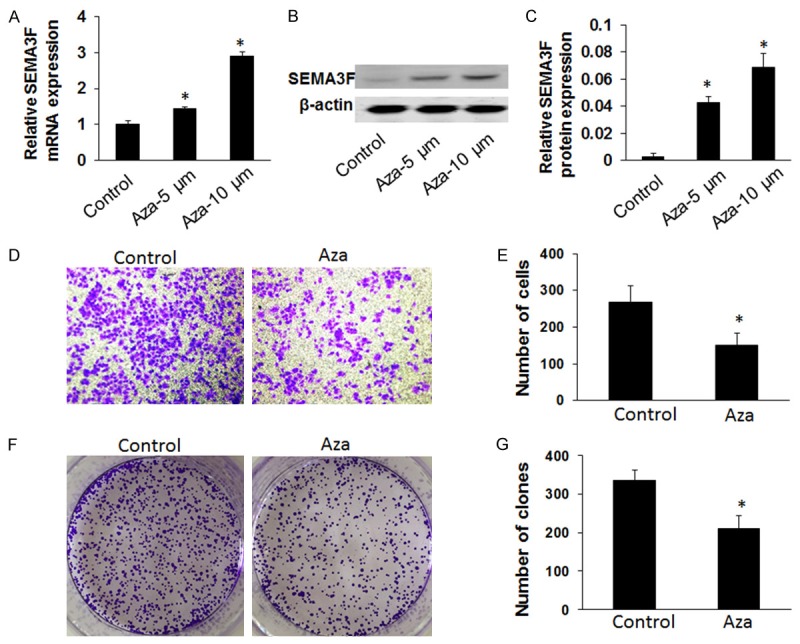
Effect of 5-Aza-CdR on SW620 cell invasion and colony formation. A-C: Comparison of SEMA3F mRNA expression by qRT-PCR and western blot analysis in SW620 cell line after treatment with 5-Aza-CdR (5 or 10 μM). P < 0.05 (compared with control cells). D and E: The transwell assay was performed to assess the effect on cell invasion. Cells were treated or untreated with 5-Aza-CdR (10 μM). Representative photos of treated and untreated cells are presented with the original magnification of 100 ×. The columns indicate the number of cells invaded at the 24 h time point. Results showed the number of SW620 cells was significantly reduced after treatment with5-Aza-CdR compared with the control group. *P < 0.05 (values were compared with untreated SW620 cells). The values represent the mean values ± SD. F and G. Clone formation rate of SW620 cells after treatment with 5-Aza-CdR (10 μM) was assessed by the colony formation assay (*P < 0.05). The columns indicate the number of clones at the 24 h time point. Results showed the clone number of SW620 cells was significantly reduced after treatment with 5-Aza-CdR compared with the control group.
Discussion
Although semaphorins play a critical role as axon guidance molecules in the developing nervous system, they are expressed in a variety of adult and embryonic tissues, suggesting a broader spectrum of the functions for semaphorins. In fact, class 3 semaphorins (SEMA3s) serve as key regulators of cellular processes, such as cell survival, proliferation, apoptosis, and migration [6,15]. Recently, the role of SEMA3s in the pathogenesis of various types of cancer has been investigated in many studies, and SEMA3F previously have been implicated in the inhibition of tumor metastasis in vivo [16,17] and that SEMA3F inhibits in vitro cell attachment and motility [7,18]. In addition, numerous transcription factors, such as ZEB-1, p53, ID-2, and RORα have been reported to regulate SEMA3F expression in lung, melanoma, prostate and breast cancer [19-22]. Moreover, SEMA3F inhibits the stemness of human CRC cells by suppressing Rac1 [23]. Interestingly, some experimental results showed overexpression of endogenous SEMA3F in tumor cells inhibited their proliferation and mobility which is likely via the SEMA3F/NRP2/plexinA complex signaling pathway [14]. In the current study, we estimated the expression of SEMA3F in CRC by Western blotting and qRT-PCR, in addition to analyzing its impact on cell migration and colony formation. We illustrated that SEMA3F is expressed at lower levels with respect to its protein and mRNA levels in CRC tissues compared with corresponding non-cancerous tissues, and found overexpression of this gene protein inhibited the migration and colony formation of cancer cells in vitro, in association with suppression of growth and metastasis of the xenografted tumors in vivo [14], which was in agreement with previous statistics shown in other types of the above-mentioned tumor samples.
Understanding how SEMA3F regulate cell invasion and metastasis could lead to the development of new anticancer therapies for CRC patients. Since the expression of SEMA3F was frequently downregulated epigenetically in a number of cancers, implying that alterations of SEMA3F may be involved in tumorigenesis [13]. Research has proved that downregulation of SEMA3F expression in tumorigenesis of human tumors is attributed to LOH and promoter hypermethylation [24]. Nevertheless, the precise mechanism for that SEMA3F suppresses tumor progression and metastasis still remains largely unclear. DNA methylation plays an important role in tumorigenesis. CpG island methylation of tumor suppressor gene resulted in inactivation of the gene transcription has become an important part of cancer epigenetics research. Aberrant CpG island methylation is associated with genes silencing in CRC, which promotes tumor formation through the deregulation of various cellular processes including proliferation and apoptosis [25]. In this study, we observed SEMA3F gene was hypermethylated in SW620, and partially methylated in SW480 and HCT116. SEMA3F gene promoter hypermethylation was observed in 69.4% CRC tissues, and in non-tumor colorectal tissues hypermethylation was tested in 16.7% cases. The current study shows that the loss of SEMA3F expression is associated with methylation of CpGs in the putative SEMA3F promoter region [26]. Subsequently, we treated the SW620 cell that showed the lowest SEMA3F expression with 5-Aza-CdR, and found the expression of the epigenetic silenced SEMA3F gene reactivate after treatment, revealing that hypermethylation of SEMA3F gene may lead to its silencing. Therefore, the identification of SEMA3F contributing to the development of CRC is critical to the understanding of molecular mechanisms of tumorigenesis and may provide new strategies for clinical therapy.
In conclusion, this study proved that SEMA3F acts as a tumor suppressor in CRC tissues cells once again, and the expression of SEMA3F gene in CRC cells is partly downregulated by DNA hypermethylation. Re-expression of SEMA3F suppresses cell proliferation and invasion in vitro, which offer a new therapeutic way for the treatment of CRC. Moreover, the methylation status and the expression level of SEMA3F may serve as potential biomarkers of CRC.
Therefore, these findings suggest DNA methylation of CpG island of SEMA3F gene promoter -mediated silencing of the tumor suppressor SEMA3F gene plays an important role in the carcinogenesis of CRC.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Moghimi-Dehkordi B, Safaee A. An overview of colorectal cancer survival rates and prognosis in Asia. World J Gastrointest Oncol. 2012;4:71–75. doi: 10.4251/wjgo.v4.i4.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo Y, Raible D, Raper JA. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell. 1993;75:217–227. doi: 10.1016/0092-8674(93)80064-l. [DOI] [PubMed] [Google Scholar]
- 4.Unified nomenclature for the semaphorins/collapsins. Semaphorin Nomenclature Committee. Cell. 1999;97:551–552. doi: 10.1016/s0092-8674(00)80766-7. [DOI] [PubMed] [Google Scholar]
- 5.Xiang RH, Hensel CH, Garcia DK, Carlson HC, Kok K, Daly MC, Kerbacher K, Berg A, Van Den, Veldhuis P, Buys CH. Isolation of the human semaphorin III/F gene (SEMA3F) at chromosome 3p21, a region deleted in lung cancer. Genomics. 1996;32:39–48. doi: 10.1006/geno.1996.0074. [DOI] [PubMed] [Google Scholar]
- 6.Neufeld G, Shraga-Heled N, Lange T, Guttmann-Raviv N, Herzog Y, Kessler O. Semaphorins in cancer. Front Biosci. 2005;10:751–760. doi: 10.2741/1569. [DOI] [PubMed] [Google Scholar]
- 7.Nasarre P, Constantin B, Rouhaud L, Harnois T, Raymond G, Drabkin HA, Bourmeyster N, Roche J. Semaphorin SEMA3F and VEGF Have Opposing Effects on Cell Attachment and Spreading. Neoplasia. 2003;5:83–92. doi: 10.1016/s1476-5586(03)80020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessler O, Shraga-Heled N, Lange T, Gutmann-Raviv N, Sabo E, Baruch L, Machluf M, Neufeld G. Semaphorin-3F Is an Inhibitor of Tumor Angiogenesis. Cancer Res. 2004;64:1008–1015. doi: 10.1158/0008-5472.can-03-3090. [DOI] [PubMed] [Google Scholar]
- 9.Brambilla E, Constantin B, Drabkin H, Roche J. Semaphorin SEMA3F localization in malignant human lung and cell lines: A suggested role in cell adhesion and cell migration. Am J Pathol. 2000;156:939–950. doi: 10.1016/S0002-9440(10)64962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen H, Ivanova VS, Kavandi L, Rodriguez GC, Maxwell GL, Syed V. Progesterone and 1,25-Dihydroxyvitamin D-3 Inhibit Endometrial Cancer Cell Growth by Upregulating Semaphorin 3B and Semaphorin 3F. Mol Cancer Res. 2011;9:1479–1492. doi: 10.1158/1541-7786.MCR-11-0213. [DOI] [PubMed] [Google Scholar]
- 11.Isabelle CDP, Valérie B, Karen L, Martine B, Armand B, Pierre W, Anne MC. Antiproliferative effect of semaphorin 3F on human melanoma cell lines. J Invest Dermatol. 2006;126:2343–2345. doi: 10.1038/sj.jid.5700382. [DOI] [PubMed] [Google Scholar]
- 12.Miao HQ, Soker S, Feiner L, Alonso JL, Raper JA, Klagsbrun M. Neuropilin-1 Mediates Collapsin-1/Semaphorin III Inhibition of Endothelial Cell Motility. J Cell Biol. 1999;146:233–242. doi: 10.1083/jcb.146.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusy S, Potiron V, Chan Z, Franklin W, Brambilla E, Minna J, Bkin HA, Roche Jl. Promoter characterization of Semaphorin SEMA3F, a tumor suppressor gene. Biochim Biophys Acta. 2005;1730:66–76. doi: 10.1016/j.bbaexp.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Wu F, Zhou Q, Yang J, Duan GJ, Ou JJ, Zhang R, Pan F, Peng QP, Tan H, Ping YF. Endogenous Axon Guiding Chemorepulsant Semaphorin-3F Inhibits the Growth and Metastasis of Colorectal Carcinoma. Clin Cancer Res. 2011;17:2702–2711. doi: 10.1158/1078-0432.CCR-10-0839. [DOI] [PubMed] [Google Scholar]
- 15.Gaur P, Bielenberg DR, Samuel S, Bose D, Zhou Y, Gray MJ, Dallas NA, Fan F, Xia L, Lu J. Role of class 3 semaphorins and their receptors in tumor growth and angiogenesis. Clin Cancer Res. 2009;15:6763–6770. doi: 10.1158/1078-0432.CCR-09-1810. [DOI] [PubMed] [Google Scholar]
- 16.Bielenberg DR, Pettaway CA, Seiji T, Michael K. Neuropilins in neoplasms: expression, regulation, and function. Exp Cell Res. 2006;312:584–593. doi: 10.1016/j.yexcr.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 17.Bielenberg DR, Yasuhiro H, Akio S, Arja K, Michael K, Caroline Choi K, Michael K. Semaphorin 3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. J Clin Invest. 2004;114:1260–1271. doi: 10.1172/JCI21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasarre P, Kusy SB, Castellani V, Drabkin HA, Bagnard D, Roche J. Semaphorin SEMA3F has a repulsing activity on breast cancer cells and inhibits E-cadherin-mediated cell adhesion. Neoplasia. 2005;7:180–189. doi: 10.1593/neo.04481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coma S, Amin DN, Shimizu A, Lasorella A, Iavarone A, Klagsbrun M. Id2 promotes tumor cell migration and invasion through transcriptional repression of semaphorin 3F. Cancer Res. 2010;70:3823–3832. doi: 10.1158/0008-5472.CAN-09-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Futamura M, Kamino H, Miyamoto Y, Kitamura N, Nakamura Y, Ohnishi S, Masuda Y, Arakawa H. Possible role of semaphorin 3F, a candidate tumor suppressor gene at 3p21.3, in p53-regulated tumor angiogenesis suppression. Cancer Res. 2007;67:1451–1460. doi: 10.1158/0008-5472.CAN-06-2485. [DOI] [PubMed] [Google Scholar]
- 21.Clarhaut J, Gemmill RM, Potiron VA, Ait-Si-Ali S, Imbert J, Drabkin HA, Roche J. ZEB-1, a Repressor of the Semaphorin 3F Tumor Suppressor Gene in Lung Cancer Cells. Neoplasia. 2009;11:157–166. doi: 10.1593/neo.81074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong G, Wang C, Evers BM, Zhou BP, Xu R. RORα suppresses breast tumor invasion through inducing SEMA3F expression. Cancer Res. 2012;72:1728–1739. doi: 10.1158/0008-5472.CAN-11-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao J, Zhou ZH, Yang J, Shi Y, Xu SL, Wang B, Ping YF, Chen L, Cui YH, Zhang X, Wu F, Bian XW. Semaphorin-3F suppresses the stemness of colorectal cancer cells by inactivating Rac1. Cancer Lett. 2015;358:76–84. doi: 10.1016/j.canlet.2014.12.040. [DOI] [PubMed] [Google Scholar]
- 24.Potiron VA, Sharma G, Nasarre P, Clarhaut JA, Augustin HG, Gemmill RM, Roche J, Drabkin HA. Semaphorin SEMA3F Affects Multiple Signaling Pathways in Lung Cancer Cells. Cancer Res. 2007;67:8708–8715. doi: 10.1158/0008-5472.CAN-06-3612. [DOI] [PubMed] [Google Scholar]
- 25.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe Y, Toyoda R, Nakamura H. Navigation of trochlear motor axons along the midbrain-hindbrain boundary by neuropilin 2. Development. 2004;131:681–692. doi: 10.1242/dev.00970. [DOI] [PubMed] [Google Scholar]


