Abstract
Background: ITGB1 is a heterodimeric cell-surface receptor involved in cell functions such as proliferation, migration, invasion and survival. The aim of this study was to assess ITGB1 expression in colorectal cancer and correlate it with clinicopathological features, as well as to evaluate its potential prognostic significance. Materials and methods: In this study, we examined the expression of ITGB1 using tissue microarrays containing analyzed specimens by immunohistochemistry. ITGB1 expression was further correlated with clinicopathological and prognostic data. The prognostic significance was assessed using Kaplan-Meier survival estimates and log-rank tests. A multivariate study with the Cox’s proportional hazard model was used to evaluate the prognostic aspects. Results: ITGB1 expression was present in 88.5% of the analyzed specimens. Significant differences in ITGB1 expression were found between normal mucosa and carcinomas (P<0.001). High ITGB1 expression was associated with poor prognosis, and it independently correlated with shortened overall survival and disease-free survival in colorectal cancer patients (P<0.001). More so, ITGB1 expression, bowel wall invasion, lymph node metastasis and distant metastasis were independent prognostic factors for overall survival. Additionally, significant differences in ITGB1 expression were observed in adenomas and tumors from patients with familial adenomatous polyposis compared to normal colon mucosa (P<0.05) Conclusion: The results of this study indicate that ITGB1 overexpression in colorectal tumors is associated with poor prognosis, as well as aggressive clinicopathological features. Therefore, ITGB1 expression could be used as potential prognostic predictor in colorectal cancer patients.
Keywords: Colorectal cancer, prognosis, tissue microarray, ITGB1
Introduction
Colorectal cancer (CRC) is the third most common malignant cancer worldwide [1]. Surgery is the primary method of treatment for CRC, but the high rate of recurrence and/or metastasis after surgery hinders a patient’s recovery, even with postoperative chemotherapy and/or radiation therapy [2,3]. Currently, the gold standard for determining postoperative treatment and prognostication for CRC patients is clinicopathological TNM staging [4]. Nevertheless, the TNM stage offers little help in the treatment of an individual patient. In addition, it is not understood why only some patients respond to therapy or have a good clinical outcome [5]. Therefore, understanding treatment failure and determining a good prognostic marker still remain an important goal in the management of CRC patients. Some genetic changes may have a prognostic role in CRC, but thus far only the prognostic value of K-ras mutations in response to cetuximab therapy of metastatic CRC has been translated into the clinic [6-8]. In addition, chromosomal and microsatellite instability have been associated with the clinical outcome of CRC patients [9]. Nevertheless, despite the efforts to determine molecular markers for individualized CRC treatments, none have been introduced into clinical practice. Therefore, the aim of this study was to find a potential diagnostic marker that could be used to predict the prognosis of CRC patients after curative surgery.
ITGB1 (Integrin β1, CD29) is a member of the integrin family and is comprised of 18 α and 8 β transmembrane subunits that form at least 24 different heterodimeric receptors for cell adhesion to extracellular matrix (ECM) proteins [10]. Integrins activate various protein tyrosine kinases through focal adhesion kinase (FAK), Src-family kinases and serine-threonine kinases, in order to regulate cell functions, such as proliferation, migration, invasion and survival [10-12]. ITGB1 functions as mediator of cell and extracellular matrix signaling in cell proliferation, apoptosis and survival [13]. It has been reported that high ITGB1 expression is associated with poor prognosis of patients with invasive breast [14], lung [15] and pancreatic [16] cancer. However, its value as a prognostic marker, as well as its correlation with clinical significance, has been rarely studied in CRC patients.
Hence, we explored the expression of ITGB1, as well as examined its relationship with clinicopathological features and prognosis, in CRC patients.
Material and methods
Patient’s selection
A total of 726 patients from Changhai Hospital (Second Military Medical University), including 56 normal patients without a tumor, 51 colon polyps patients, 582 CRC patients (stage I-IV), 16 familial adenomatous polyposis patients and 21 colorectal liver metastases patients, were recruited between 2001 and 2013 and included in this study. Patients were included/excluded according to the following the criteria: (a) definitive pathological diagnosis of CRC or normal controls; (b) no anticancer treatment prior to surgery; (c) curative resection with the cut surface being free of cancer as confirmed by a pathologist; (d) availability of suitable paraffin-embedded tissues; and (e) complete clinicopathological and follow-up data. The study was approved by the medical ethical boards of the Changhai Hospital (Second Military Medical University) and patients’ informed consent was obtained. Clinicopathological characteristics, including sex, age, tumor stage, bowel wall invasion, lymph node metastasis, distant metastasis, tumor differentiation, survival, postoperative therapy, carcinoembryonic antigen (CEA) and carbohydrate antigen-199 (CA-199), were included in the records. Tumor stage was determined by the American Joint Committee Against Cancer (AJCC) tumor-node-metastasis (TNM) classification system for primary colon cancer. In our follow-up study, data of all patients were censored from the date of surgery to the date of the last follow-up visit (September 30, 2013) or death. Disease-free survival (DFS) was defined as the probability that patients remained free of tumor recurrence (in situ or metastasis). Overall survival (OS) time was defined as the time from the date of surgery to the confirmed death date for dead patients or from the date of surgery to the date of the last follow-up for surviving patients [17].
Follow-up of patients after surgery
Selected patients were evaluated every 3 months during the first postoperative year, every 6 months during the following year, and afterwards once a year until September 30, 2013, which was the ending date of our study. Follow-up was completed by phone or mail. Dates of death/recurrence, cause of death and postoperative treatment were recorded simultaneously. During the follow-up period all patients were monitored by CEA, CA-199, colonoscopy, and chest X-ray for possible recurrence. If recurrence was suspected, a computed tomography scan (CT) of the abdomen or magnetic resonance imaging (MRI) or positron emission tomography (PET) was performed for further confirmation.
Implementation of tissue microarray and immunohistochemistry
Paraffin sections used in this study were obtained from the Department of pathology, Changhai Hospital. Samples were initially reviewed by hematoxylin and eosin (H&E) staining and representative areas were pre-marked in the paraffin blocks, avoiding necrotic and hemorrhagic areas. The Formaldehyde Fixed-Paraffin Embedded tissues (FFPE) corresponding to the selected histological sections were sampled from these marked regions using a specialized manual arraying instrument (Model MTA1, Beecher Instruments, Sun Prairie, Wis). With this device, 1.5 mm diameter cylinders were obtained for each sample to ensure reproducibility and homogenous staining of the slides. The slides were then aligned in pre-arranged sequences according to TNM stages (Shanghai Biochip). The core samples were then placed into an empty paraffin block. After the arraying was complete, the tissue microarray (TMA) blocks were completely sectioned at a 4 μm thickness, yielding more than 80 slides from each block. Thus, five different tissue microarray blocks were constructed, with each block containing a total of 160 specimens. Finally, 800 samples were aligned in five different tissue microarray blocks. If patients suffered from familial adenomatous polyposis (FAP) or presented with liver metastasis before surgery, three specimens (tumor, matching noncancerous mucosa and adenoma or liver metastatic tissue, respectively) were obtained from each patient.
Immunohistochemistry analysis was performed using a mouse anti-human ITGB1 (1:25 dilution) monoclonal antibody (Abcam, ab3167, USA). Briefly, immunohistochemistry of tissue microarrays was carried out as follows: sections were deparaffinized in xylene, rehydrated, and washed in phosphate buffered saline (PBS) for 10, 5 and 10 min, respectively. After application of endogenous peroxidase for 10 min and antigen retrieval at 98° for 25 min, slides were pre-incubated with blocking serum for 30 min, and then incubated with the ITGB1 monoclonal antibody at 4° overnight. Subsequently, the sections were thoroughly rinsed with PBS, incubated with secondary antibodies, and treated with horseradish peroxidase-conjugated streptavidin. The immunohistochemical reaction was visualized with 3,3’-diaminobenzidine tetrahydrochloride and counterstained with hematoxylin.
Quantification of ITGB1 expression by immunohistochemistry
The density of ITGB1-positive staining was evaluated by two independent pathologists, without prior knowledge of the patient characteristics, using a Leica DMI3000 microscope (magnification of ×200). Positive ITGB1 staining in each photograph was evaluated in the cytosol as follows: the staining intensity was first scored (0 point, negative staining; 1 point, weak staining, light yellow; 2 points, moderate staining, yellowish brown; 3 points, strong staining, brown) and then the proportion of positive cells was scored (0 point, 0-5% positive cells; 1 point, 5-25% positive cells; 2 points, 26-50% positive cells; 3 points, 51-75% positive cells; 4 points, 76-100% positive cells). The final score was obtained by multiplying the scores of staining intensity and percentage of positive cells for each specimen.
For statistical purposes, specimens were divided into four grades according to their overall scores: absent expression (-), 0 points; weak expression (+), a total of 1-4 points; moderate expression (++), 5-8 points; and strong expression (+++), 9-12 points. All samples were anonymized and independently scored by two investigators. In case of disagreement, the slides were re-examined until the final consensus was reached.
Statistical analysis
The associations of ITGB1 expression with clinicopathological features were tested with the Kruskal-Wallis test. For the analysis of the training set, the survival curves were estimated by the Kaplan-Meier method and compared by with the log-rank test. To determine the independence of our classifier to clinicopathological variables in predicting an individual’s risk of survival, we analyzed the validation set using univariate analysis followed by multivariate analysis in a Cox proportional-hazards model for prognostic predictors. All calculations were performed with SPSS statistical package version 17.0 (SPSS, Chicago, IL). P<0.05 (two-sided) was considered statistically significant.
Results
ITGB1 expression in mucosa and carcinoma tissues of the patients at different stages
ITGB1 expression was observed in 610 out of the 689 (88.5%) analyzed specimens including normal mucosa, adenomas and I-IV stages. Examples of different ITGB1 expression in normal mucosa and carcinomas are presented in Figure 1. Positive ITGB1 expression was present in the cell cytoplasm. ITGB1 expression was scored on a scale of (-) to (+++). As shown in Figure 1, expression of TIGB1 in patients with stage II and above carcinomas was significantly higher than those in stage I carcinoma, adenoma and normal mucosa.
Figure 1.

Representative cases of ITGB1 expression in normal mucosa, adenomas and different CRC tumor stages. Positive immunohistochemical staining for ITGB1 was present mainly in the cytoplasm of tumor cells. A. Absence of expression in normal mucosa scored as “-”; B. Absence of expression in adenoma scored as “-”; C. Low intensity expression in tumor stage I scored as “+”; D. Low intensity expression in stage II scored as “+”; E. Moderate intensity expression in stage III scored as “++”; F. High intensity expression in stage IV scored as “+++”. Magnification, ×200.
The Kruskal-Wallis test was used to analyze the relationship between the ITGB1 expression in normal mucosa and adenomas and different tumor stages. Significant differences in ITGB1 expression were observed between carcinomas of all stages compared to ITGB1 expression in normal mucosa and adenomas (P<0.001). The ITGB1 expression was not statistically significant between adenomas and stage I cancer patients, between stage II and II cancer patients, and between stage III and stage IV cancer patients (P>0.05) (Table 1).
Table 1.
Expression of ITGB1 in normal mucosa, adenomas and different tumor stages
| Characteristic | ITGB1 immunostaining (n) | P a | P b | P c | P d | P e | |||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| - | + | ++ | +++ | ||||||
| Normal mucosa | 28 | 26 | 2 | 0 | - | - | - | - | - |
| Adenoma | 5 | 38 | 8 | 0 | <0.001 | - | - | - | - |
| Stage I cancer | 4 | 35 | 14 | 0 | <0.001 | 0.205 | - | - | - |
| Stage II cancer | 22 | 117 | 78 | 16 | <0.001 | 0.003 | 0.094 | - | - |
| Stage III cancer | 12 | 56 | 161 | 12 | <0.001 | <0.001 | <0.001 | <0.001 | - |
| Stage IV cancer | 8 | 10 | 25 | 12 | <0.001 | <0.001 | <0.001 | 0.002 | 0.474 |
Compared with the “Normal mucosa” group;
Compared with the “Adenoma” group;
Compared with the “Stage I cancer” group;
Compared with the “Stage II cancer” group;
Compared with the “Stage III cancer” group.
ITGB1 expression in FAP patients
Among patients selected for this study, 16 FAP patients were included to examine the expression of ITGB1 in their tumors, adenomas and matching non-cancerous mucosa. A significant difference in ITGB1 expression was found in tumors and adenomas of FAP patients compared to ITGB1 expression in matching normal mucosa (P<0.05), while no difference was found between ITGB1 expression in adenomas compared tumors in these patients (P>0.05) (Table 2; Figure 2).
Table 2.
Expression of ITGB1 in normal mucosa, adenomas and tumors of FAP patients
| Characteristic | ITGB1 immunostaining (n) | P a | P b | |||
|---|---|---|---|---|---|---|
|
| ||||||
| - | + | ++ | +++ | |||
| Normal mucosa | 2 | 9 | 5 | 0 | - | - |
| Adenoma | 0 | 6 | 9 | 1 | 0.043 | - |
| CRC tissues | 0 | 2 | 13 | 1 | 0.001 | 0.164 |
Compared with the “Normal mucosa” group;
Compared with the “Adenoma” group.
Figure 2.
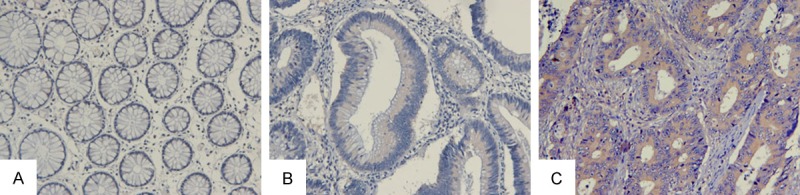
ITGB1 expression in normal mucosa, adenomas and tumor tissue from the same FAP patient. A. Normal mucosa; B. Adenoma tissue; C. Colorectal cancer. Magnification, ×200.
ITGB1 expression in patients with liver metastasis
In addition, ITGB1 expression was examined in the normal mucosa and primary or metastatic tumors of 21 patients with liver metastasis before surgery. Significant differences in ITGB1 expression in colorectal cancer or metastatic liver tumors compared to the normal mucosa were observed (P<0.05), while there was no difference in ITGB1 expression between colorectal and metastatic liver tumors (P>0.05) (Table 3; Figure 3).
Table 3.
Expression of ITGB1 in normal mucosa and tumors of patients with colorectal liver metastasis
| Characteristic | ITGB1 immunostaining (n) | P a | P b | |||
|---|---|---|---|---|---|---|
|
| ||||||
| - | + | ++ | +++ | |||
| Normal mucosa | 2 | 14 | 5 | 0 | - | - |
| Colorectal cancer | 2 | 3 | 13 | 3 | 0.002 | - |
| Liver metastasis | 1 | 3 | 16 | 1 | <0.001 | 0.865 |
Compared with the “normal mucosa” group;
Compared with the “colorectal cancer” group.
Figure 3.
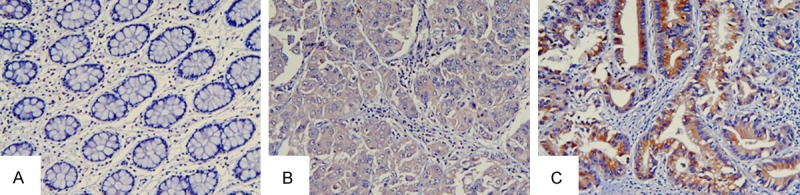
ITGB1 expression in normal mucosa, metastatic liver tissue and CRC from the same patient with primary metastasis. A. Normal tissue; B. Metastatic liver tissue; C. Colorectal cancer. Magnification, ×200.
ITGB1 expression and clinicopathological features of CRC patients
In order to evaluate the relationship between the ITGB1 expression and tumor biology, clinicopathologic features, including sex, age, bowel wall invasion, lymph node metastasis, distant metastasis, postoperative treatment, tumor differentiation, CEA and CA-199, and ITGB1 expression associations were investigated.
Patients with high ITGB1 expression were more likely to exhibit aggressive clinicopathological features, such as lymph node metastasis, distant metastasis, non-postoperative treatment and tumor differentiation (P<0.001, respectively). The details are shown in Table 4.
Table 4.
Relationship between the ITGB1 immunostaining and clinicopathological characteristics of patients with colorectal cancer
| Characteristic | ITGB1 immunostaining (n) | p | |||
|---|---|---|---|---|---|
|
| |||||
| - | + | ++ | +++ | ||
| Sex (n) | 0.479 | ||||
| Male | 23 | 122 | 164 | 21 | |
| Female | 23 | 96 | 114 | 19 | |
| Age (years) | 0.348 | ||||
| <60 | 29 | 106 | 139 | 18 | |
| ≥ 60 | 17 | 112 | 139 | 22 | |
| Bowel wall invasion | 0.534 | ||||
| T1 | 1 | 3 | 5 | 0 | |
| T2 | 5 | 35 | 29 | 4 | |
| T3 | 38 | 180 | 241 | 35 | |
| T4 | 2 | 0 | 3 | 1 | |
| Lymph node metastasis | <0.001 | ||||
| N0 | 29 | 154 | 100 | 21 | |
| N1 | 8 | 40 | 120 | 13 | |
| N2 | 9 | 24 | 58 | 6 | |
| Distant metastasis | 0.019 | ||||
| M0 | 38 | 208 | 253 | 28 | |
| M1 | 8 | 10 | 25 | 12 | |
| Postoperative treatment | <0.001 | ||||
| Yes | 32 | 136 | 235 | 33 | |
| No | 14 | 82 | 43 | 7 | |
| Tumor differentiation | <0.001 | ||||
| Well | 0 | 6 | 11 | 1 | |
| Moderate | 24 | 192 | 246 | 37 | |
| Poor | 9 | 13 | 15 | 2 | |
| Mucinous adenocarcinoma | 13 | 7 | 6 | 0 | |
| Serum CEA | 0.551 | ||||
| <5 ng/mL | 31 | 135 | 168 | 25 | |
| ≥5 ng/mL | 15 | 83 | 110 | 15 | |
| Serum CA-199 | 0.307 | ||||
| <37 U/ml | 41 | 182 | 229 | 32 | |
| ≥37 U/ml | 5 | 36 | 49 | 8 | |
ITGB1 expression and overall and disease-free survival of CRC patients
Kaplan-Meier analysis showed that patients with high ITGB1 expression (++ and +++) had a significantly lower DFS and OS compared to patients with low ITGB1 expression (+ and -) (Figure 4).
Figure 4.
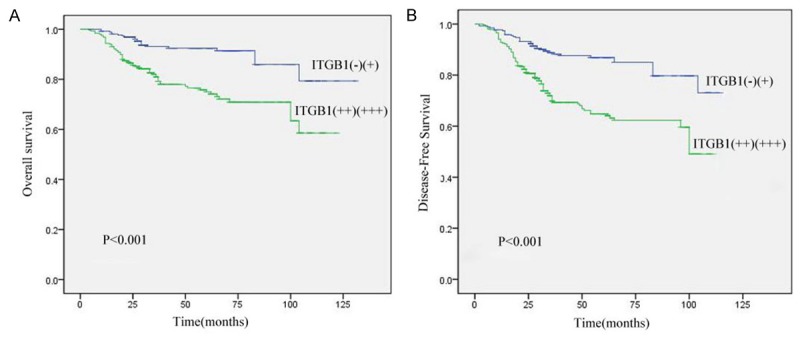
Prognostic significance assessed using Kaplan-Meier survival estimates and log-rank tests stratified by ITGB1. High ITGB1 expression was associated with decreased OS and DFS. A. Kaplan-Meier survival curves showed a significantly decreased OS among patients with high ITGB1 expression (++ and +++) compared with patients with low expression (+ and -). P<0.001, log-rank test; B. Kaplan-Meier survival curves showed a significantly reduced DFS among patients with high ITGB1 intensity scores compared with patients with low scores. P<0.001, log-rank test.
Univariate and multivariate analysis of predictive factors of OS in CRC patients
A total of six clinicopathological parameters, including patients’ bowel wall invasion, lymph node metastasis, distant metastasis, CEA, CA-199, postoperative treatment and ITGB1 expression, were included in the univariate analysis using the Cox proportional hazards model. All of these parameters were significant predictors for OS.
To further explore their independent predictive effect for OS, a multivariate Cox proportional-hazards model was performed within the same parameters. It was found that bowel wall invasion [hazard ratio (HR), 2.012; 95% confidence interval (CI), 1.065-3.800; P=0.031], lymph node metastasis (HR, 1.929; CI, 1.468-2.534; P<0.001), distant metastasis (HR, 3.648; CI, 2.287-5.820; P<0.001) and ITGB1 expression (HR, 1.537; CI, 1.147-2.059; P=0.004) were independent prognostic factors for OS. However, CEA, CA-199 and postoperative treatment were not found to be independent prognostic indicators for OS (Table 5).
Table 5.
Cox’s multivariate analysis for OS
| Characteristic | OS | ||
|---|---|---|---|
|
| |||
| HR | 95% CI | p | |
| Bowel wall invasion (T1-T2 vs T3-T4) | 2.012 | 1.065-3.800 | 0.031 |
| Lymph node metastasis (No vs Yes) | 1.929 | 1.468-2.534 | <0.001 |
| Distant metastasis (No vs Yes) | 3.648 | 2.287-5.820 | <0.001 |
| ITGB1 (Low vs High) | 1.537 | 1.147-2.059 | 0.004 |
| CEA (<5 vs ≥5 ng/mL) | 1.367 | 0.868-2.152 | 0.178 |
| CA-199 (<37 vs ≥37 U/mL) | 1.482 | 0.907-2.421 | 0.116 |
| Postoperative treatment (No vs Treatment) | 0.752 | 0.362-1.562 | 0.445 |
Abbreviation: OS: overall survival, HR: hazard ratio, CI: confidence interval.
From Table 5, we can find out that bowel wall invasion, lymph node metastasis, distant metastasis, ITGB1 expression, CEA and CA-199 are all risk factors for CRC patients. While, postoperative treatment (HR, 0.752; CI, 0.362-1.561; P=0.445) is a protective factor for CRC patients. Of all patients included, patients who expressed low ITGB1 expression and accepted postoperative treatment, including chemotherapy or/and radiotherapy showed better prognosis than that of patients who expressed high expression and without postoperative treatment (P<0.001).
Discussion
Integrins are heterodimeric cell-surface receptors consisting of α and β subunits, which integrate the extracellular matrix with the intracellular cytoskeleton to mediate cell adhesion, survival, differentiation and migration by a wide range of intracellular signaling pathways [13,18,19]. ITGB1 is the most important member of the integrin family because it facilitates cell-cell and cell-extracellular matrix interactions to mediate the survival, differentiation, angiogenesis, and invasion of cancer cells [10,20,21].
ITGB1 also acts as a signal transducer in signaling pathways involved in the regulation of survival and proliferation through the PI3K/Akt and p130Cas/paxillin/JNK signaling pathways [22,23]. In addition, ITGB1 has been reported to mediate the resistance to chemotherapy and radiation by enhancing cell survival and inhibition of apoptosis in several human cancers, and thus could be considered as an important therapeutic target for anti-cancer therapy [24-26]. Indeed, inhibition of ITGB1 has been shown to enhance radiotherapy efficacy and result in apoptosis in malignant breast cancer models [27,28]. All of these findings suggest that ITGB1 may be of great clinical significance in cancer patients.
At present, the potential effects of ITGB1 expression on clinical prognosis have been reported in breast cancer [14], ovarian cancer [29] and small-cell lung cancer [30,31]. However, correlation of ITGB1 expression and clinical prognosis in CRC patients on a large scale in China has been lacking. In the present study, we demonstrated that high ITGB1 expression was accompanied with aggressive clinicopathological features, including lymph node metastasis, liver metastasis and tumor differentiation, and advanced stages of CRC cancer (P<0.05). In addition, increased ITGB1 expression was closely associated with decreased OS and DFS of CRC patients. More importantly, in our study, ITGB1 remained an independent factor associated with OS (HR, 1.537; CI, 1.147-2.059; P=0.004) after multivariate regression analysis. Therefore, based on these findings we can conclude that ITGB1 is an attractive target for therapeutic strategies and a good predictor for clinical prognosis of CRC patients.
In several malignancies, such as melanoma, renal and breast cancer, integrin expression correlates with tumor progression and metastasis. Several experimental models have shown the efficacy of ITGB1 inhibitors in treatment of refractory tumors and advanced metastatic disease [32]. More so, a recent study has shown that stimulation of the TLR4/MD2 complex by lipopolysaccharide activates PI3K/AKT signaling and promotes downstream ITGB1 function, thereby increasing the adhesiveness and metastatic capacity of CRC cells [33]. The results of our study indicated that ITGB1 expression could be used as a potential biomarker to study the mechanism of tumor progression, based on the differences in ITGB1 expression in normal mucosa compared to primary liver metastasis tissue from the same patient. In addition, although ITGB1 is dispensable for the initiation of ErbB2 tumor induction, it plays an important role in the metastatic phase of tumor progression [34]. Therefore, ITGB1 expression may be useful in the evaluation of the potential for tumor metastasis.
FAP is an autosomal dominant inherited disease with relatively high mortality. It is caused by germline mutation of the adenomatous polyposis coli (APC) tumor-suppressor gene [35]. Thus far, congenital hypertrophy of the retinal pigment epithelium (CHRPE) is confirmed to be a clinical marker for FAP [36]. However, no definite molecular marker for clinical treatment has yet been established. Based on our results, which showed significant differences in ITGB1 expression in normal mucosa, adenomas and tumors of the same FAP patient, ITGB1 expression could be a potential marker from clinical progression from adenoma through early stage carcinoma to advanced stage carcinoma for FAP patients. In addition ITGB1 expression could be considered as a therapeutic target for FAP treatment.
There were some limitations in our study. Although this study was initially based on a large number of samples, many of them were excluded due to lack of information regarding post-operation adjuvant therapy and/or clinicopathological features.
In summary, using the tissue microarray method, we demonstrated that ITGB1 expression could mediate cancer progression and distinguish low- and high-risk patients after surgery in CRC. Nevertheless, the definite role of ITGB1, as well as its potential as a clinical marker of CRC, is still far from being unambiguously established. Thus, further studies are needed to fully understand its role in CRC development and progression.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008. Globocan 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Sugarbaker PH. Second-look surgery for colorectal cancer: revised selection factors and new treatment options for greater success. Int J Surg Oncol. 2011;2011:915078. doi: 10.1155/2011/915078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kodaira S. Treatment of post-operative recurrence of colorectal cancer. Nihon Geka Gakkai zasshi. 1999;100:206–10. [PubMed] [Google Scholar]
- 4.Storli KE, Sondenaa K, Bukholm IR, Nesvik I, Bru T, Furnes B, Hjelmeland B, Iversen KB, Eide GE. Overall survival after resection for colon cancer in a national cohort study was adversely affected by TNM stage, lymph node ratio, gender, and old age. Int J Colorectal Dis. 2011;26:1299–307. doi: 10.1007/s00384-011-1244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagtegaal ID, Gosens MJ, Marijnen CA, Rutten HJ, van de Velde CJ, van Krieken JH. Combinations of tumor and treatment parameters are more discriminative for prognosis than the present TNM system in rectal cancer. J. Clin. Oncol. 2007;25:1647–50. doi: 10.1200/JCO.2005.05.4825. [DOI] [PubMed] [Google Scholar]
- 6.Bates RC, Bellovin DI, Brown C, Maynard E, Wu B, Kawakatsu H, Sheppard D, Oettgen P, Mercurio AM. Transcriptional activation of integrin beta6 during the epithelial-mesenchymal transition defines a novel prognostic indicator of aggressive colon carcinoma. J Clin Invest. 2005;115:339–47. doi: 10.1172/JCI23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, Andre T, Bibeau F, Diebold MD, Rougier P, Ducreux M, Tomasic G, Emile JF, Penault-Llorca F, Laurent-Puig P. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J. Clin. Oncol. 2008;26:374–79. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 8.Deschoolmeester V, Boeckx C, Baay M, Weyler J, Wuyts W, Van Marck E, Peeters M, Lardon F, Vermorken JB. KRAS mutation detection and prognostic potential in sporadic colorectal cancer using high-resolution melting analysis. Br J Cancer. 2010;103:1627–36. doi: 10.1038/sj.bjc.6605959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–99. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 10.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 11.Collier ME, Ettelaie C. Induction of endothelial cell proliferation by recombinant and microparticle-tissue factor involves beta1-integrin and extracellular signal regulated kinase activation. Arterioscler Thromb Vasc Biol. 2010;30:1810–7. doi: 10.1161/ATVBAHA.110.211854. [DOI] [PubMed] [Google Scholar]
- 12.Koivisto L, Heino J, Häkkinen L, Larjava H. Integrins in wound healing. Adv Wound Care (New Rochelle) 2014;3:762–83. doi: 10.1089/wound.2013.0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Wang H. Integrin signalling and function in immune cells. Immunology. 2012;135:268–75. doi: 10.1111/j.1365-2567.2011.03549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao ES, Zhang H, Chen YY, Lee B, Chew K, Moore D, Park C. Increased β1 integrin is associated with decreased survival in invasive breast cancer. Cancer Res. 2007;67:659–64. doi: 10.1158/0008-5472.CAN-06-2768. [DOI] [PubMed] [Google Scholar]
- 15.Oshita F, Kameda Y, Ikehara M, Tanaka G, Yamada K, Nomura I, Noda K, Shotsu A, Fujita A, Arai H, Ito H, Nakayama H, Mitsuda A. Increased expression of integrin beta1 is a poor prognostic factor in small-cell lung cancer. Anticancer Res. 2002;22:1065–70. [PubMed] [Google Scholar]
- 16.Bottger TC, Maschek H, Lobo M, Gottwohl RG, Brenner W, Junginger T. Prognostic value of immunohistochemical expression of β-1 integrin in pancreatic carcinoma. Oncology. 1999;56:308–13. doi: 10.1159/000011984. [DOI] [PubMed] [Google Scholar]
- 17.Ding ZB, Shi YH, Zhou J, Shi GM, Ke AW, Qiu SJ, Wang XY, Dai Z, Xu Y, Fan J. Liver-intestine cadherin predicts microvascular invasion and poor prognosis of hepatitis B virus-positive hepatocellular carcinoma. Cancer. 2009;115:4753–65. doi: 10.1002/cncr.24513. [DOI] [PubMed] [Google Scholar]
- 18.Askari JA, Buckley PA, Mould AP, Humphries MJ. Linking integrin conformation to function. J Cell Sci. 2009;122:165–70. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malinin NL, Pluskota E, Byzova TV. Integrin signaling in vascular function. Curr Opin Hematol. 2012;19:206–11. doi: 10.1097/MOH.0b013e3283523df0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schooley AM, Andrews NM, Zhao H, Addison CL. beta1 integrin is required for anchorage-independent growth and invasion of tumor cells in a context dependent manner. Cancer Lett. 2012;316:157–67. doi: 10.1016/j.canlet.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 21.Nisticò P, Di Modugno F, Spada S, Bissell MJ. β1 and β4 integrins: from breast development to clinical practice. Breast Cancer Res. 2014;16:459. doi: 10.1186/s13058-014-0459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cordes N, Seidler J, Durzok R, Geinitz H, Brakebusch C. β1-integrin-mediated signaling essentially contributes to cell survival after radiation-induced genotoxic injury. Oncogene. 2006;25:1378–90. doi: 10.1038/sj.onc.1209164. [DOI] [PubMed] [Google Scholar]
- 23.Watt FM. Role of intrgrins in regulating epidermal adhesion, growth and differentiation. EMBO J. 2002;21:3919–26. doi: 10.1093/emboj/cdf399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurata M, Nakagawa Y, Yamamoto K, Suzuki K, Kitagawa M. Induction of integrin beta1 expression in bone marrow cells after chemotherapy correlates with the overexpression of lung resistance protein and poor outcome in patients with multiple myeloma. Am J Hematol. 2008;83:755–7. doi: 10.1002/ajh.21234. [DOI] [PubMed] [Google Scholar]
- 25.Cordes N, Seidler J, Durzok R, Geinitz H, Brakebusch C. beta1-integrin-mediated signaling essentially contributes to cell survival after radiation-induced genotoxic injury. Oncogene. 2006;25:1378–90. doi: 10.1038/sj.onc.1209164. [DOI] [PubMed] [Google Scholar]
- 26.Nam JM, Chung Y, Hsu HC, Park CC. beta1 integrin targeting to enhance radiation therapy. Int J Radiat Biol. 2009;85:923–8. doi: 10.3109/09553000903232876. [DOI] [PubMed] [Google Scholar]
- 27.Park CC, Zhang HJ, Yao ES, Park CJ, Bissell MJ. β1 integrin inhibition dramatically enhances radiotherapy efficacy in human breast cancer xenografts. Cancer Res. 2008;68:4398–405. doi: 10.1158/0008-5472.CAN-07-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park CC, Zhang H, Pallavicini M. β1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Res. 2006;66:1526–35. doi: 10.1158/0008-5472.CAN-05-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller-Klingspor V, Hefler L, Obermair A, Kaider A, Breitenecker G, Leodolte S, Kohlberger P. Prognostic value of beta1-integrin (=CD29) in serous adenocarcinomas of the ovary. Anticancer Res. 2001;21:2185–8. [PubMed] [Google Scholar]
- 30.Oshita F, Kameda Y, Ikehara M. Increased expression of integrin β1 is a poor prognostic factor in small-cell lung cancer. Anticancer Res. 2002;22:1065–70. [PubMed] [Google Scholar]
- 31.Lawson MH, Cummings NM, Rassl DM, Vowler SL, Wickens M, Howat WJ, Brenton JD, Murphy G, Rintoul RC. Bcl-2 and β1-integrin predict survival in a tissue microarray of small cell lung cancer. Br J Cancer. 2010;103:1710–15. doi: 10.1038/sj.bjc.6605950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barkan D, Chambers AF. β1-integrin: a potential therapeutic target in the battle against cancer recurrence. Clin Cancer Res. 2011;17:7219–23. doi: 10.1158/1078-0432.CCR-11-0642. [DOI] [PubMed] [Google Scholar]
- 33.Hsu RY, Chan CH, Spicer JD, Rousseau MC, Giannias B, Rousseau S, Ferri LE. LPS-induced TLR4 signaling in human colorectal cancer cells increases beta1 integrin-mediated cell adhesion and liver metastasis. Cancer Res. 2011;71:1989–98. doi: 10.1158/0008-5472.CAN-10-2833. [DOI] [PubMed] [Google Scholar]
- 34.Huck L, Pontier SM, Zuo DM, Muller WJ. beta1-integrin is dispensable for the induction of ErbB2 mammary tumors but plays a critical role in the metastatic phase of tumor progression. Proc Natl Acad Sci U S A. 2010;107:15559–64. doi: 10.1073/pnas.1003034107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su LK, Barnes CJ, Yao W, Qi Y, Lynch PM, Steinbach G. Inactivation of germline mutant APC alleles by attenuated somatic mutations: a molecular genetic mechanism for attenuated familial adenomatous polyposis. Am J Hum Genet. 2000;67:582–90. doi: 10.1086/303058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heyen F, Jagelman DG, Romania A, Zakov ZN, Lavery IC, Fazio VW, McGannon E. Predictive value of congenital hypertrophy of the retinal pigment epithelium as a clinical marker for familial adenomatous polyposis. Dis Colon Rectum. 1990;33:1003–08. doi: 10.1007/BF02139213. [DOI] [PubMed] [Google Scholar]


