Abstract
Spontaneous preterm labor is an important complication in perinatology characterized by early onset myometrium contractions leading to labor at preterm. However, the exact mechanism that maintain uterine quiescence and promote increased uterine contractility during labor were incompletely defined. MicroRNAs is a class of short non-coding RNAs that regulate gene expression at the post-transcriptional level by binding the 3’ untranslated region of target mRNAs and play an important role in biological process and cellular functions. We hypothesized we could find differentially expressed microRNAs in the myometrium of women in spontaneous preterm labor. Thus, a microarray analysis of miRNAs of preterm myometrium was performed. 18 out of the 2006 detected microRNAs were found to be significantly dysregulated in myometrium in labor verse not in labor at preterm. Biological validation by quantitative real-time polymerase chain reaction confirms us a consistence rate of 83.3% (5 out of 6) with microarray analysis. The target genes for validated microRNAs were predicted by three algorithms (PicTar, TargetScan, and miRanda). Most of the potential targets of the miRNAs were relevant to positive regulation of cardiac muscle hypertrophy, reduction of cytosolic calcium ion concentration and relaxation of cardiac muscle as well as prostate cancer, adherents junction, regulation of actin cytoskeleton and regulation and other factor-regulated calcium reabsorption. Our result illustrates a characteristic microRNA profile in myometrium tissues and provides a new understanding of the process involved in spontaneous preterm labor.
Keywords: Preterm labor, microRNA, myometrium
Introduction
Preterm labor, defined as birth before 37 weeks of gestation, is the leading cause of neonatal and maternal morbidity and mortality worldwide [1,2]. Approximately 70% of all preterm deliveries occur after spontaneous preterm labor [3]. Multiple factors may contribute to the etiology of spontaneous preterm labor including infections, multiple pregnancy and alcohol or narcotic addiction. However, the molecular mechanism that promotes uterine contractility before term labor remains unknown.
MiRNAs are a class of some 19-23 nucleotides, non-coding small RNA molecules that regulate gene expression at the post-transcriptional level by binding the 3’ untranslated region of target mRNAs and play an important role in biological process and cellular functions [4]. Dicer is an essential rib endonuclease coding gene required for microRNA biosynthesis and Dicer conditional mutant female mice is infertile with a reduction in the size of the oviducts and uterine horns as well as the structural alterations of myometrium, indicating us that microRNAs are important regulators in female reproductive tract [5]. The abnormal expression of miRNAs plays an important role in the preterm labor and the identification of miRNA profile is mainly investigate in the placentas, fetal membranes and myometrium of preterm labor, indicating us its extensively existed function in the pathophysiology of preterm [6].
Recently, a number of microRNAs (miRNAs) have become acknowledged as important regulators in myometrium contractility and preterm labor [7-11]. For example, Nora E. had shown us that miR-200 family could interact with ZEB1 and ZEB2 and act as unique P4- and PR-regulated modulators of uterine quiescence and contractility during pregnancy and labor at term or preterm [8]. Moreover, miR-200a serves a key role in the decline of progesterone receptor function by suppressing STAT5b and increasing the expression level of the P4-metabolizing enzyme 20-alpha-hydroxysteroid dehydrogenase (20-alpha-HSD) while overexpression of miR-199a/214 in cultured human myometrial cells inhibited cyclooxygenase-2 protein (COX-2) and blocked TNF-alpha-induced myometrial cell contractility [10,11].
To date, dysregulation of pregnancy myometrial miRNA expression was once evaluated in the mouse myometrium during late pregnancy [8]. MiRNA profiles in spontaneous preterm labor and the functional role for differentially expressed miRNA has never been determined. In this study, we aimed to find the unique expression profile of miRNAs in human myometrium at labor vs not in labor at preterm to provide valuable information for further investigation of parturition.
Material and method
Clinical sample collection
Lower uterine segment myometrial tissues were biopsied from pregnant women undergoing cesarean section because of spontaneous preterm labor (PL, n=6) and from these who underwent cesarean for other pregnancy associated complications and are not in labor (NL, n=6). In consideration of ethical demand of humanity, all of the samples were collected immediately after exogenous oxytocin injection into uterine during surgery to reduce postpartum hemorrhage. This Myometrial smooth muscle was dissected from each biopsy, snap frozen in liquid nitrogen, and stored at -80°C for subsequent analysis. This study was approved by the local ethics committees of Obstetrics and Gynecology Hospital of Fudan University (Shanghai, China), and written consent was obtained from each patients before surgery.
MicroRNA microarray analysis
Total RNA, including the miRNAs, was extracted from the myometrium tissue using the miRNeasy mini kit (QIAGEN, Germany) according to the manufacturer’s instructions. The RNA concentration was quantified by using a NanoDrop 2000 spectrophotometer (Thermo Fisher scientific, USA). The integrity and the quality of RNA were evaluated by Agilent 2100 Bioanalyzer (Agilent Technologies, USA). RNAs with a 2100RIN (RNA integrity number) ≥6.0 and 28S/18S ≥0.7 were used for the miRNA array analysis and reverse transcription (RT). An aliquot of 1.0 mg of total RNA was converted to cDNA by using the miScript II RT kit (QIAGEN, Germany).
The labeling and hybridization were performed at the Shanghai Biochip Company, according to the protocols of the Agilent miRNA microarray system. Agilent Scan Control software was used for scanning the microarray slides and Agilent Feature Extraction software (version 10.7.1.1, Agilent Technologies) was used for image analysis. Raw data were normalized by Quantile algorithm, Gene Spring Software 11.0 (Agilent technologies, US).
Quantitative real time polymerase chain reaction analysis (qRT-PCR)
The total RNA was isolated from myometrium tissues using TRIzol reagent (Invitrogen, USA) and then reversely transcribed using miScript II RT Kit (QIAGEN, Germany). Real time PCR was performed in triplicate by using miScript SYBR Green PCR kit (QIAGEN, Germany) on the ABI Prism 7900 HT detection system (Invitrogen, USA). U6 was used as a reference gene. The primers for RT-PCR are as shown in Table 1. A universal reverse primer was provided by the manufacturer (QIAGEN, Germany). The expression of miRNAs was expressed as a fold change based on the 2-ΔΔCt method and was then statistically analyzed.
Table 1.
qRT-PCR primers list
| Forward primers (5’-3’) | |
|---|---|
| has-mir-10b-3p | ACAGATTCGATTCTAGGGGAATAA |
| has-mir-936 | ACAGTAGAGGGAGGAATCGCAGAA |
| hsa-miR-5096 | GTTTCACCATGTTGGTCAGGCAA |
| hsa-miR-212-3p | TAACAGTCTCCAGTCACGGCCAA |
| hsa-miR-223-3p | TGTCAGTTTGTCAAATACCCCAA |
| has-miR-4743-5p | TTCCGGATGGGACAGGAGGCATAA |
| hsa-miR-U6 | GGATTAACGATACAGAGAAGATT |
Bioinformatics analysis and target prediction
Three online software programs, miRanda http://microrna.sanger.ac.uk, PicTar http://www.ncrna.org/KnowledgeBase/link-Database/mirna_target_database, and TargetScan http://www.targetscan.org, were used for bioinformatics analysis and target prediction for the qRT-PCR validated miRNAs. Go Ontology and KEGG pathway enrichment analysis were used to annotate the functions of miRNA targets.
Results
MiRNA expression profiles of preterm myometrium in labor verse not in labor
A total of 2006 miRNAs were detected by microarray in human myometrium tissues. Among them, 18 miRNAs were significantly dysregulated by greater than 2 times in the myometrium tissue in active labor. The hierarchical clustering heatmap shown 10 miRNAs were up-regulated while 8 miRNAs were down-regulated in myometrium in active labor compared with controls (Figure 1).
Figure 1.
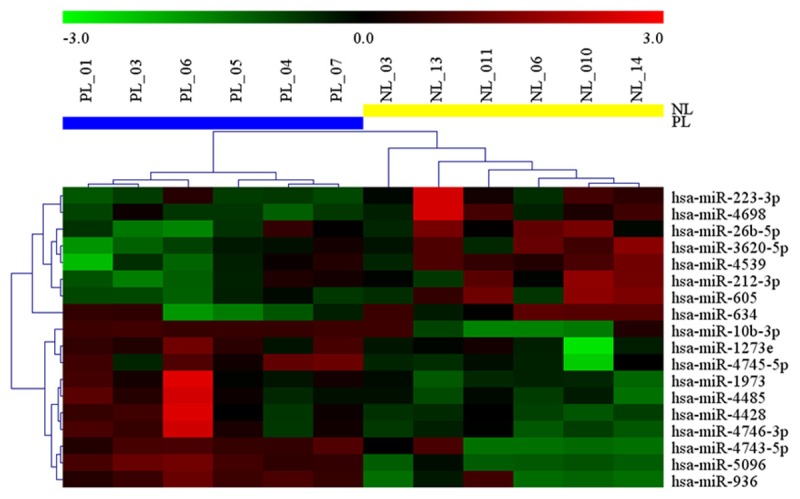
Heatmap generated by hierarchical clustering for differentially expressed miRNAs in the myometrium from spontaneous preterm labor versus these not in labor. Hierarchical clustering for differentially expressed miRNAs in spontaneous preterm labor (PL, n=6) vs. preterm not in labor (PL, n=6) (P-value < 0.05 and fold-change < 2 times). Red indicates high relative expression and Green indicates low relative expression.
Validation of differentially expressed miRNAs by qRT-PCR
We randomly chose six differentially expressed miRNAs to perform a biologic validation including miR-4743-5p, miR-223-3p, miR-10b-3p, miR-212-3p, miR-5096 and miR-936, According to the qRT-PCR results (Figure 2A-F), miR-4743-5p, miR-10b-3p, miR-212-3p, miR-5096 and miR-936 were overexpressed while miR-223-3p was depressed in myometrium at preterm labor (PL) compared with these not in labor (NL). Five of the tested 6 miRNAs have shown a consistent result in qRT-PCR with microarray data, with miR-212-3p being an exception, indicating a concordance rate of 83.3%.
Figure 2.
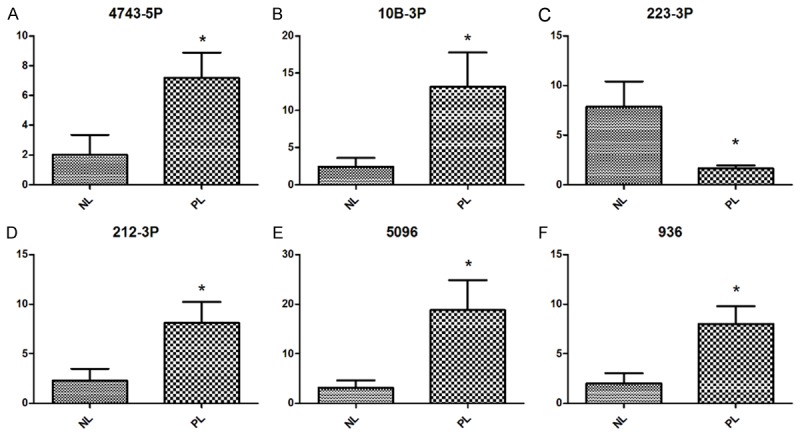
Validation of the microarray data using qRT-PCR. A-F. Six differentially expressed miRNAs were selected from microarray datasets and examined by qRT-PCR. Data was determined using the 2-ΔΔCT method and statistically analyzed by student t-test. *P < 0.05.
Bioinformatics analysis and functional prediction of the selected dysregulated miRNAs
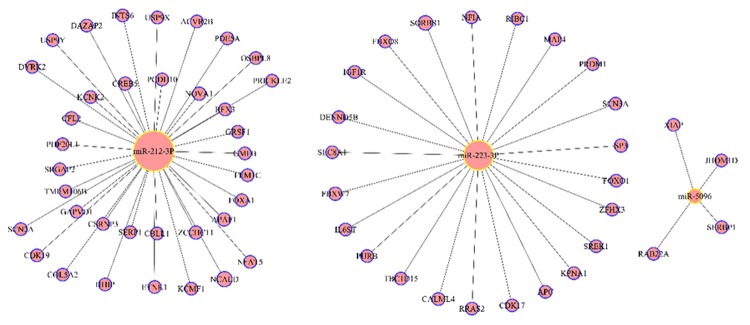
We used three online software programs including miRanda, PicTar and TargetScan to predict the targets of the validated miRNAs. To increase the specificity, we only acknowledged these predicted miRNA: mRNA target pairs if they were predicted by all of the three programs. Therefore, only three of them, miR-223-3p, miR-5096 and miR-212-3p, have predicted targets in our study. We ranked the miRNA-target gene interactions and generated a miRNA-mRNA interaction network using CytoScape (Figure 3). Because of our strict principles to select predicted miRNA: mRNA target pairs, only 63 protein coding gene were predicted to target the three miRNAs.
Figure 3.
A network of validated miRNAs (hsa-miR-212-3p, hsa-miR-223-3p and hsa-miR-5096) and their target genes.
According to our GO and KEGG pathway analysis, most of these target genes were enriched in the biological process of positive regulation of cardiac muscle hypertrophy, reduction of cytosolic calcium ion concentration and relaxation of cardiac muscle as well as in the KEGG pathway of prostate cancer, adherents junction, regulation of actin cytoskeleton and regulation and other factor-regulated calcium reabsorption (Figure 4A, 4B). Interestingly, these biological process and KEGG pathway are closely related with muscle contractions.
Figure 4.
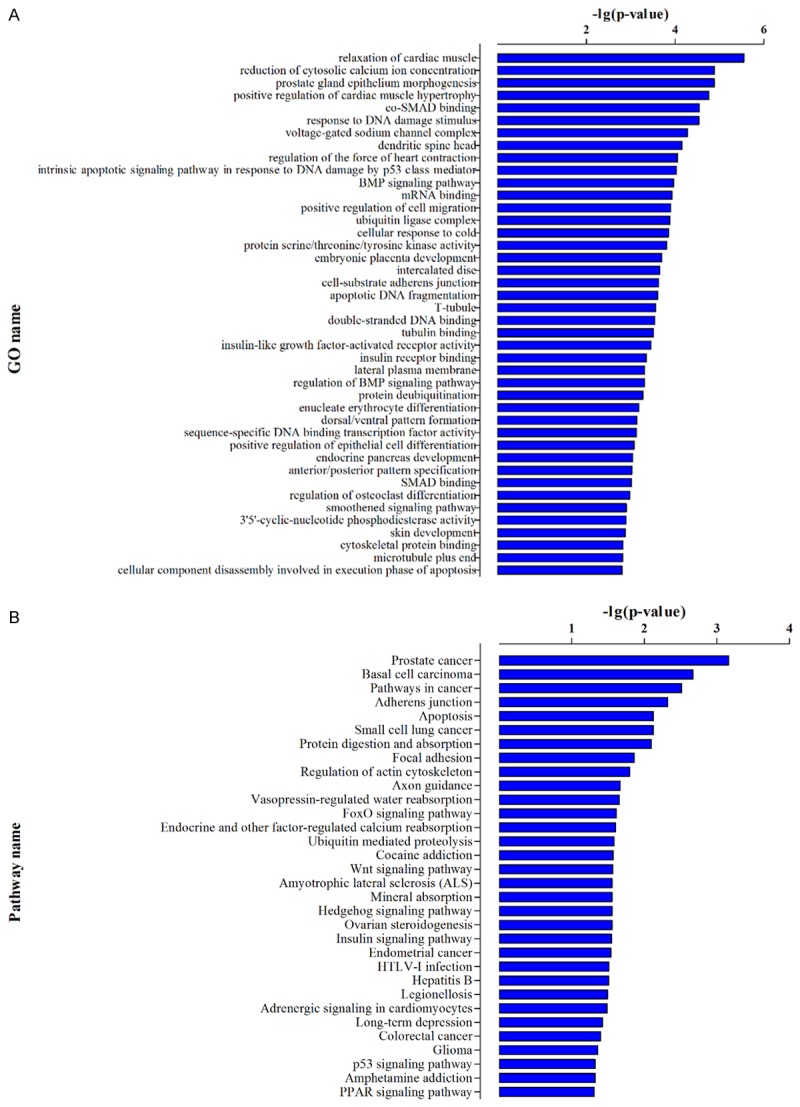
Functional prediction of validated miRNAs (hsa-miR-212-3p, hsa-miR-223-3p and hsa-miR-5096). A, B. GO and KEGG pathway enrichment analysis of the targets of validated miRNAs (hsa-miR-212-3p, hsa-miR-223-3p and hsa-miR-5096).
Discussion
To the best of our knowledge, this is the first study addressing miRNA expression profiling in human spontaneous preterm labor myometrium tissue. Our study shows a unique expression pattern of miRNA in the myometrium of spontaneous preterm labor with hsa-miR-4743-5p, hsa-miR-10b-3p, hsa-miR-212-3p, hsa-miR-5096, hsa-miR-936 being up-regulated and has-miR-223-3p being down-regulated in spontaneous preterm labor. Inspiringly, functional analysis of the selected miRNAs suggests us they might play a critical role by targeting several genes associated with important biological process and pathways during the transition of muscle functions.
The identification and functional role of aberrantly expressed miRNA and miRNA gene target in myometrium before and after the onset of labor is in a preliminary stage. To date, myometrial miRNA profiles have only been evaluated in a limited number of studies. For example, oxytocin alters the expression of a unique set of myometrial miRNA including hsa-miR-146b-3p, hsa-miR-196b-3p, hsa-miR-223-3p, hsa-miR-873-5p and hsa-miR-876-5p [12]. In the mouse myometrium, mmu-miR-200 family shows a dramatic increase during late pregnancy and labor [8]. Investigation of the uterine leiomyoma microRNAome also indicated us a number of miRNAs were implicated in the myometrial dysfunctions such as hsa-miR-363, hsa-miR-490, hsa-miR-137, hsa-miR-217, hsa-miR-4792 and hsa-miR-200c [13-15].
Hsa-miR-212 and has-miR-223 are dysregulated in our study and the biological function of two miRNAs has been reported to be associated with immune system. It is reported hsa-miR-212 modulates IL-10 production through the interaction with aryl hydrocarbon receptor in T cell and is positive correlated with IL-6 production in monocytes [16,17]. Moreover, recent study has shown has-miR-212 could be directly up-regulated by nuclear factor kappa B (NF-kB), which is an important inflammation-related transcription factor [18]. Similarly, hsa-miR-223-3p is believed to be a critical regulator in inflammasome activity and macrophage activation [19,20]. Through targeting NLRP3 and Pknox1, has-miR-223 regulates inflammatory cytokine IL-1β production and anti-inflammatory response [20,21]. Interestingly, inflammation and infection is one of the most important regulators in spontaneous preterm labor [22,23]. Additionally, according to Cook J, myometrial has-miR-223-3p expression is regulated by oxytocin which is a critical molecule in promoting myometrial contraction. However, more work needs to be done to illustrate the exact role of has-miR-212 and has-miR-223-3p in spontaneous preterm labor.
Hsa-miR-10b, has-miR-936, hsa-miR-4743-5p and hsa-miR-5096, another set of dysregulated miRNAs in our study, have been widely researched as well. Hsa-miR-10b is a metastatic related microRNA and was proved to be over expressed in some human cancers including breast tumor, gastric carcinoma, ovarian cancer and hepatocellular carcinoma [24-28]. Hsa-miR-10b directly interacts with transcription factor ZEB-1 and PIK3CA as well as affects pro-metastatic gene products such as MMP14 and RHOC to regulate epithelial cell and tumor cell invasiveness [24,27]. Has-miR-936 is over expressed in hyperplastic scar and may be closely correlated with the formation, development and evolution of hyperplastic scar [29]. There are few reports regarding hsa-miR-4743-5p and hsa-miR-5096 in human disease. However, hsa-miR-5096 is high conserved and is predicted to have some 725 target genes and binds to mRNA with high affinity, suggesting its important role in human evolution [30]. The mechanism how these miRNA involved in myometrial has yet to be determined.
We infer the functions of the miRNAs from its target mRNAs base on the fact that miRNAs are implicated in the regulation of protein coding gene expression by binding with MRE in the mRNA transcript. Because the current sequence-based available target prediction algorithms are known to have high false-positive rates and are not completely in agreement, we intersect the results of the three prediction algorithms to obtain more confident target genes for each miRNA [31].
In our study, we found the target genes of the validated miRNA to be significantly enriched in some biological process and pathways that closely related to muscle contractility, regulation of actin cytoskeleton and focal adhesion. For example, the target gene of has-miR-223-3p includes IL6ST and SLC8A1 which may be related to the reduction of cytosolic calcium ion concentration, and the target gene of has-miR-212-3p includes PDE5A which is involved the relaxation of cardiac muscle. As for has-miR-5096, the target is XIAP which is related to focal adhesion. Interestingly, our functional prediction of miRNAs indicates they are associated with inflammation and immune system as well. CREB5 and IL6ST, targets of hsa-miR-212-3p, are related with the TNF signaling pathway and negative regulation of interleukin-6-mediated signaling pathway. XIAP, a target of has-miR-5096, involves in the regulation of innate immune response and NF-kappa B signaling pathway. Not all validated miRNAs have predicted target genes in our study because they cannot be predicted by all three online programs. However, this does not mean they were not involved in preterm labor. More work needs to be done to elucidate the specific functions of each miRNA in myometrium.
There are several limitations in our study. Firstly, given ethical considerations, we only include limited numbers of samples in our study. More work can be done by using mouse preterm models. Moreover, limited by current clinical features, the patients we recruited in this study have other pregnancy complications, such as preelampsia and uterine scar, as well. However, we balance the confounders in the two groups equally to minimize their effects.
In summary, we describe a unique myometrial miRNA expression pattern in human spontaneous preterm labor and discuss their possible role by predicting the target genes of each miRNA. Our results suggest that miRNA play a critical role in the process of partuition in myometrium. The identification of myometrial miRNA profile throws a new light upon the molecule mechanism and provides an insight into the potential diagnosis and treatment of preterm labor.
Acknowledgements
This work was supported by Natural Science Foundation of China (Grant No. 81200440 to T.P, 81270712 to X-T. L and 81471470 to W-R. G) and the Science Foundation of Shanghai Municipal Health Bureau (Grant No. 20134031 to W-R. G).
Disclosure of conflict of interest
None.
References
- 1.Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, Lawn JE. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;379:2162–2172. doi: 10.1016/S0140-6736(12)60820-4. [DOI] [PubMed] [Google Scholar]
- 2.Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, Cousens S, Mathers C, Black RE. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. 2015;385:430–440. doi: 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 3.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez G, Behringer RR. Dicer is required for female reproductive tract development and fertility in the mouse. Mol Reprod Dev. 2009;76:678–688. doi: 10.1002/mrd.21010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eidem HR, Ackerman WEt, McGary KL, Abbot P, Rokas A. Gestational tissue transcriptomics in term and preterm human pregnancies: a systematic review and meta-analysis. BMC Med Genomics. 2015;8:27. doi: 10.1186/s12920-015-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montenegro D, Romero R, Kim SS, Tarca AL, Draghici S, Kusanovic JP, Kim JS, Lee DC, Erez O, Gotsch F, Hassan SS, Kim CJ. Expression patterns of microRNAs in the chorioamniotic membranes: a role for microRNAs in human pregnancy and parturition. J Pathol. 2009;217:113–121. doi: 10.1002/path.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc Natl Acad Sci U S A. 2010;107:20828–20833. doi: 10.1073/pnas.1008301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders AP, Burris HH, Just AC, Motta V, Svensson K, Mercado-Garcia A, Pantic I, Schwartz J, Tellez-Rojo MM, Wright RO, Baccarelli AA. microRNA expression in the cervix during pregnancy is associated with length of gestation. Epigenetics. 2015;10:221–228. doi: 10.1080/15592294.2015.1006498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams KC, Renthal NE, Condon JC, Gerard RD, Mendelson CR. MicroRNA-200a serves a key role in the decline of progesterone receptor function leading to term and preterm labor. Proc Natl Acad Sci U S A. 2012;109:7529–7534. doi: 10.1073/pnas.1200650109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams KC, Renthal NE, Gerard RD, Mendelson CR. The microRNA (miR)-199a/214 cluster mediates opposing effects of progesterone and estrogen on uterine contractility during pregnancy and labor. Mol Endocrinol. 2012;26:1857–1867. doi: 10.1210/me.2012-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook JR, MacIntyre DA, Samara E, Kim SH, Singh N, Johnson MR, Bennett PR, Terzidou V. Exogenous oxytocin modulates human myometrial microRNAs. Am J Obstet Gynecol. 2015;213:65, e61–69. doi: 10.1016/j.ajog.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Chuang TD, Khorram O. miR-200c regulates IL8 expression by targeting IKBKB: a potential mediator of inflammation in leiomyoma pathogenesis. PLoS One. 2014;9:e95370. doi: 10.1371/journal.pone.0095370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuang TD, Panda H, Luo X, Chegini N. miR-200c is aberrantly expressed in leiomyomas in an ethnic-dependent manner and targets ZEBs, VEGFA, TIMP2, and FBLN5. Endocr Relat Cancer. 2012;19:541–556. doi: 10.1530/ERC-12-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgieva B, Milev I, Minkov I, Dimitrova I, Bradford AP, Baev V. Characterization of the uterine leiomyoma microRNAome by deep sequencing. Genomics. 2012;99:275–281. doi: 10.1016/j.ygeno.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Chinen I, Nakahama T, Kimura A, Nguyen NT, Takemori H, Kumagai A, Kayama H, Takeda K, Lee S, Hanieh H, Ripley B, Millrine D, Dubey PK, Nyati KK, Fujii-Kuriyama Y, Chowdhury K, Kishimoto T. The aryl hydrocarbon receptor/microRNA-212/132 axis in T cells regulates IL-10 production to maintain intestinal homeostasis. Int Immunol. 2015;27:405–415. doi: 10.1093/intimm/dxv015. [DOI] [PubMed] [Google Scholar]
- 17.Weigelt K, Bergink V, Burgerhout KM, Pescatori M, Wijkhuijs A, Drexhage HA. Down-regulation of inflammation-protective microRNAs 146a and 212 in monocytes of patients with postpartum psychosis. Brain Behav Immun. 2013;29:147–155. doi: 10.1016/j.bbi.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 18.de la Rica L, Garcia-Gomez A, Comet NR, Rodriguez-Ubreva J, Ciudad L, Vento-Tormo R, Company C, Alvarez-Errico D, Garcia M, Gomez-Vaquero C, Ballestar E. NF-kappaB-direct activation of microRNAs with repressive effects on monocyte-specific genes is critical for osteoclast differentiation. Genome Biol. 2015;16:2. doi: 10.1186/s13059-014-0561-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol. 2012;189:4175–4181. doi: 10.4049/jimmunol.1201516. [DOI] [PubMed] [Google Scholar]
- 20.Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, Wu C, Zhou B. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125:2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 21.Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, Hammerschmidt W, O’Neill LA, Masters SL. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol. 2012;189:3795–3799. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 22.Gotsch F, Romero R, Kusanovic JP, Erez O, Espinoza J, Kim CJ, Vaisbuch E, Than NG, Mazaki-Tovi S, Chaiworapongsa T, Mazor M, Yoon BH, Edwin S, Gomez R, Mittal P, Hassan SS, Sharma S. The anti-inflammatory limb of the immune response in preterm labor, intra-amniotic infection/inflammation, and spontaneous parturition at term: a role for interleukin-10. J Matern Fetal Neonatal Med. 2008;21:529–547. doi: 10.1080/14767050802127349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazaki-Tovi S, Romero R, Vaisbuch E, Kusanovic JP, Erez O, Mittal P, Gotsch F, Chaiworapongsa T, Than NG, Kim SK, Pacora P, Yeo L, Dong Z, Hassan SS. Adiponectin in amniotic fluid in normal pregnancy, spontaneous labor at term, and preterm labor: a novel association with intra-amniotic infection/inflammation. J Matern Fetal Neonatal Med. 2010;23:120–130. doi: 10.3109/14767050903026481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Y, Lang X, Lu Z, Wang J, Li T, Liao Y, Jia C, Zhao W, Fang H. MiR-10b Directly Targets ZEB1 and PIK3CA to Curb Adenomyotic Epithelial Cell Invasiveness via Upregulation of E-Cadherin and Inhibition of Akt Phosphorylation. Cell Physiol Biochem. 2015;35:2169–2180. doi: 10.1159/000374022. [DOI] [PubMed] [Google Scholar]
- 25.Liao CG, Kong LM, Zhou P, Yang XL, Huang JG, Zhang HL, Lu N. miR-10b is overexpressed in hepatocellular carcinoma and promotes cell proliferation, migration and invasion through RhoC, uPAR and MMPs. J Transl Med. 2014;12:234. doi: 10.1186/s12967-014-0234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Z, Chen Y, Min L, Li L, Huang H, Li J, Yan Q, Song P, Dai L, Yao X. Augmented miR-10b expression associated with depressed expression of its target gene KLF4 involved in gastric carcinoma. Int J Clin Exp Pathol. 2015;8:5071–5079. [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama I, Shibazaki M, Yashima-Abo A, Miura F, Sugiyama T, Masuda T, Maesawa C. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int J Oncol. 2013;43:63–71. doi: 10.3892/ijo.2013.1935. [DOI] [PubMed] [Google Scholar]
- 28.Ma L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010;12:210. doi: 10.1186/bcr2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ning P, Liu DW, Mao YG, Peng Y, Lin ZW, Liu DM. [Differential expression profile of microRNA between hyperplastic scar and normal skin] . Zhonghua Yi Xue Za Zhi. 2012;92:692–694. [PubMed] [Google Scholar]
- 30.Ivashchenko A, Berillo O, Pyrkova A, Niyazova R, Atambayeva S. The properties of binding sites of miR-619-5p, miR-5095, miR-5096, and miR-5585-3p in the mRNAs of human genes. Biomed Res Int. 2014;2014:720715. doi: 10.1155/2014/720715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]