Abstract
Background: Leucine zipper/EF hand-containing transmembrane-1 (LETM1) is a mitochondrial inner membrane protein that was first identified in Wolf-Hirschhorn syndrome. However, high-level expression of LETM1 has been correlated with multiple human malignancies, suggesting roles in carcinogenesis and tumor progression. This study is aimed to explore the clinicopathological characteristics and prognostic value of LETM1 overexpression in breast cancer. Methods: Immunohistochemical (IHC) staining, and immunofluorescence (IF) were performed to examine LETM1 expression in breast cancer cell line/tissues compared with adjacent normal tissues. Statistical analysis was applied to evaluate the correlation between LETM1 overexpression and the clinicopathological features of breast cancer. Survival rates were calculated using the Kaplan-Meier method, and the relationship between prognostic factors and patient survival was analyzed using the Cox proportional hazard models. Results: LETM1 protein showed cytoplasmic staining pattern in breast cancer. The strongly positive rate of LETM1 protein was 61.6% (98/159) in breast cancer, which was significantly higher than in DCIS (29.7%, 11/37), hyperplasia (16.7%, 3/18) and adjacent normal breast tissues (15.9%, 7/44). High-level expression of LETM1 protein was correlated with lymph node metastasis, poor differentiation, late clinical stage, disease-free survival (DFS) and overall survival (OS) rates in breast cancer. Moreover, multivariate analysis suggested that LETM1 emerged as a significant independent prognostic factor along with clinical stage of patients with breast cancer. Conclusions: LETM1 plays an important role in the progression of breast cancer. High level expression of LETM1 is an independent poor prognostic factor of breast cancer.
Keywords: LETM1, immunohistochemistry, breast cancer
Introduction
For women, breast cancer has long been the most frequently diagnosed malignancy and one of the primary causes of cancer-related death all over the world. Every year, about 1.67 million new people were diagnosed with breast cancer, accounted for a quarter of all cancers [1,2]. According to the global statistics, the rate of breast cancer incidence especially in developing countries is increasing [3]. With the purpose of achieve better management of breast cancer, it is very important to find the identification of clinical, pathological and biological factors that have prognostic value, as those factors could be used to make contribution to the improvement of breast cancer prognosis [4].
Leucine zipper/EF hand-containing transmembrane-1 (LETM1) gene, which encodes a mitochondrial inner membrane protein, is located on the short arm of human chromosome 4 [5], and it was first identified in Wolf-Hirschhorn syndrome (WHS) [6,7]. LETM1 encodes for the human homologue of yeast Mdm38p, amitochondria-shaping protein which function is unknown. However, previous studies showed that LETM1 is acted as an anchor protein for complex formation between mitochondria and ribosome, which can also regulate the biosynthesis of mitochondria [8,9]. The increased expression of LETM1 in human cancer promotes that the disorder expression of LETM1 is a key feature of tumorigenesis [10]. Hwang et al. reported that LETM1 suppressed lung tumor growth through activation of AMPK activity and inhibition of Akt activity, suggesting that LETM1 may provide a useful target for designing lung tumor prevention and treatment [11]. Chen et al. demonstrated that LETM1 plays an important role in the progression of HNSCC, and high levels of LETM1 protein are significantly associated with the presence of lymph node metastasis, advanced stage, poor differentiation, and shortened survival of patients with HNSCC [12]. However, few studies have reported the clinicopathological significance of LETM1 protein overexpression in breast cancer.
In this study, we demonstrated the clinicopathological significance of LETM1 through prognostic evaluation of LETM1 overexpression in breast cancers. The results revealed that LETM1 protein is frequently up-regulated in breast cancers compared with DCIS, hyperplasia tissues and adjacent nontumor breast tissues. LETM1 may be a good independent predictor of prognosis for patients with breast cancer.
Materials and methods
Clinical samples
Total 258 tissue samples, including 159 of breast cancers, 37 of ductal carcinoma in situ (DCIS), 18 of hyperplasia, and 44 of normal tissues of breasts, were collected from the Department of Pathology, Yanbian Tumor Hospital. The normal breast tissues were obtained from the resection margins of modified radical mastectomy specimen of breast cancer. The tissues were routinely processed with 10% buffered formalin fixation and paraffin embedded. The pathological parameters were carefully reviewed in all of the cases and two experienced pathologists reviewed the H&E stained slides and one appropriate paraffin block was selected for this study.
Immunohistochemical (IHC) analysis
The immunostaining kits were purchased from DAKO Inc. (Glostrup, Denmark) and Nichirei Inc. (Tokyo, Japan). 4-μm tissue sections were prepared on silane-coated slides (Sigma, St Louis, MO, USA).
Tissue sections were deparaffinized, rehydrated and incubated with 3% H2O2 in methanol for 15 min at room temperature to deactivate endogenous peroxidase. The antigen was retrieved at 95°C for 20 min by placing the slices in 0.01 M sodium citrate buffer (pH 6.0). The slices were then incubated with primary antibody LETM1 (1:50, Abnova, Taipei, Taiwan) at 4°C overnight and then washed with PBS. After incubation at room temperature for 30 min with biotinylated secondary antibodies, the slides were incubated with streptavidin-peroxidase complex at room temperature for 30 min. Immunostaining was developed using 3,3’-diaminobenzidinechromogenand counterstained with Mayer’s hematoxylin. We used the Mouse IgG isotope controls which showed negative staining. Also, the positive tissue sections were processed omitting the primary antibody as the negative controls.
Two pathologists (Lin Z & Liu S) who did not possess knowledge of the clinical data examined and scored all tissue specimens. Briefly, the immunostaining for LETM1 was semi-quantitatively scored as ‘-’ (negative) (no or less than 5% positive cells), ‘+’ (5-25% positive cells), ‘++’ (26-50% positive cells), and ‘+++’ (more than 50% positive cells). Only cytoplasmic and membranous staining patterns were considered as positive staining, and the strong positive means ‘++’ and ‘+++’ positive cells. For survival analysis, the LETM1 expression level was denoted as high expression (‘++’ and ‘+++’) and low expression (‘-’ and ‘+’).
Immunofluorescence (IF) staining analysis
IF staining was used to detect the sub-cellular localization of LETM1 protein in MDA-MB-231 cells. All steps were performed at room temperature. MDA-MB-231 cells were grown on coverslips to 70-80% confluence, then fixed with 4% paraformaldehyde for 10 min, and after 24 h cells were permeabilized with 0.5% TritonX-100 for 10 min. Blocking was performed with 3% Albumin Bovine V (A8020, Solarbio, Beijing, China) for 1 h. After washing with PBS, cells were incubated with the LETM1 antibody (1:500) at 4°C overnight, followed by incubation with Alexa Fluor® 568 goat anti-mouse IgG (H+L) (A11004, 1:1000, Invitrogen, USA) for 1 h. After washing with PBS, cells were counterstained with DAPI (C1006, Beyotime, Shanghai, China) and the coverslips were mounted with Antifade Mounting Medium (P0126, Beyotime, Shanghai, China). Finally, the IF signals were visualized and recorded with a Leica SP5II confocal microscope (Heidelberg, Germany).
Statistical analysis
Statistical analyses of the data were performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Correlations between LETM1 expression and clinicopathological characteristics were analyzed by Chi-square tests (χ2) and Fisher’s exact tests. Disease-free survival (DFS) and overall survival (OS) rates after tumor removal were calculated using the Kaplan-Meier method, and differences in survival curves were analyzed using the Log-rank tests. The Cox proportional hazards regression model was used for univariate and multivariate survival analysis. P<0.05 was considered statistically significant.
Results
LETM1 expression in breast cancer and normal breast
IF staining showed that LETM1 mainly located in the cytoplasm of MDA-MB-231 breast cancer cells (Figure 1). IHC staining also showed that LETM1 is mainly located in the cytoplasm of breast cancers (Figure 2). The positive rate of LETM1 protein was significantly higher in breast cancer 79.2% (126/159) than that in DCIS (45.9%, 17/37), hyperplasia (33.3%, 6/18) and adjacent normal breast tissues (29.5%, 13/44) (P<0.01) (Table 1). Similarly, the strongly positive rate of LETM1 protein was 61.6% (98/159) in breast cancers, which was also significantly higher than that in DCIS (29.7%, 11/37), hyperplasia (16.7%, 3/18) and adjacent normal breast tissues (15.9%, 7/44) (P<0.01) (Table 1).
Figure 1.
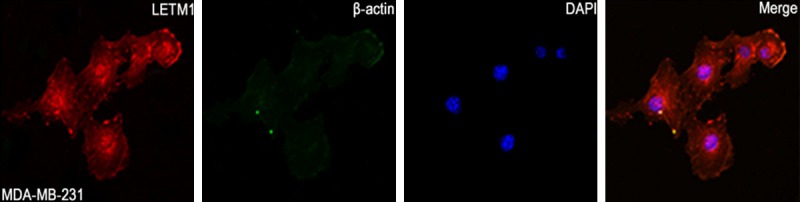
Immunofluorescent staining of LETM1 in MDA-MB-231 human breast cancer cells. LETM1 protein is mainly located in the cytoplasm of MDA-MB-231 cancer cells. (Red for LETM1, Green for β-actin, and Blue for DAPI).
Figure 2.
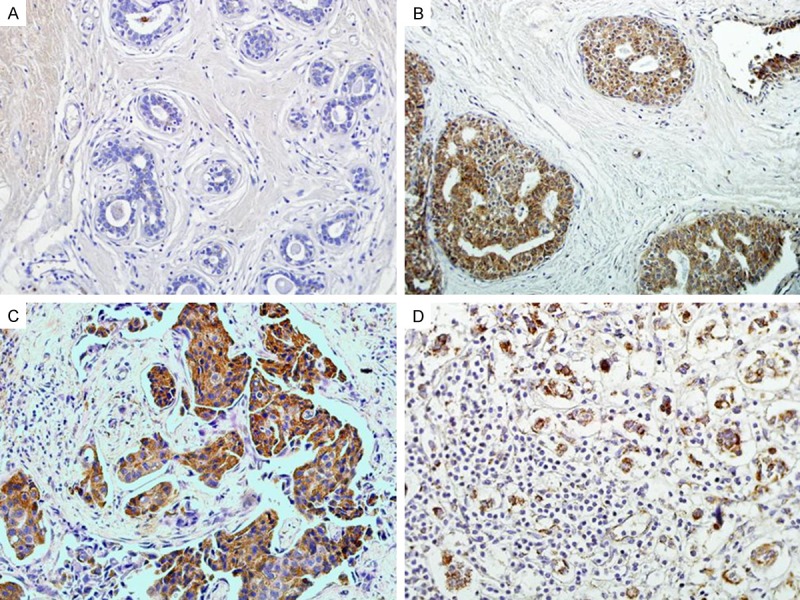
Immunohistochemical staining for LETM-1 protein expression. A. LETM-1 protein is absolutely negative in normal breast tissue. B. LETM-1 protein showed positive signals in DCIS. C. LETM1 protein showed strongly cytoplasmic positive signals in invasive breast cancer tissues. D. LETM-1 protein is strongly positive in lymph node with metastasis breast cancer. (Original magnification, 200× in A-D).
Table 1.
LETM-1 proteinexpression in breast cancers
| Diagnosis | No. of cases | LETM1 expression | Positive rate | Strongly positive rate | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| - | + | ++ | +++ | ||||
| Breast cancers | 159 | 33 | 28 | 65 | 33 | 79.2%** | 61.6%** |
| DCIS | 37 | 20 | 6 | 9 | 2 | 45.9% | 29.7% |
| Hyperplasia | 18 | 12 | 3 | 3 | 0 | 33.3% | 16.7% |
| Adjacent normal breast | 44 | 31 | 6 | 7 | 0 | 29.5% | 15.9% |
Positive rate: percentage of positive cases with +, ++, and +++ staining score. Strongly positive rate: (high-level expression): percentage of positive cases with ++ and +++ staining score.
P<0.01 compared with adjacent normal breast.
Clinicopathological significance of LETM1 overexpression in breast cancers
LETM1 overexpression was significantly correlated with tumor differentiation, clinical stage and LN metastasis of breast cancer. However, LETM1 expression was not related with patients’ age, menopausal status, and tumor size in breast cancer (Table 2).
Table 2.
Relationship between LETM-1 protein overexpression and the clinicopathological features of breast cancer
| Clinical features | No. of cases | Strongly positive cases (%) | χ2 | P value |
|---|---|---|---|---|
| Age | 0.207 | 0.650 | ||
| ≥50 | 85 | 51 (60.0%) | ||
| <50 | 74 | 47 (63.5%) | ||
| Menopausal status | 0.468 | 0.494 | ||
| Premenopausal | 65 | 38 (58.5%) | ||
| Postmenopausal | 94 | 60 (63.8%) | ||
| Tumor size | 1.450 | 0.229 | ||
| T1 | 80 | 53 (66.3%) | ||
| T2 | 79 | 45 (60.0%) | ||
| Tumor differentiation | 9.854 | 0.007** | ||
| Well | 60 | 28 (46.7%) | ||
| Moderate | 76 | 52 (68.4%) | ||
| Poor | 23 | 18 (78.3%) | ||
| Clinical stage | 4.556 | 0.033** | ||
| 0-II | 82 | 44 (53.7%) | ||
| III-IV | 77 | 54 (70.1%) | ||
| LN metastasis | 8.975 | 0.003** | ||
| T1 | 91 | 47 (51.6%) | ||
| T2 | 68 | 51 (75.0%) |
P<0.01.
For tumor differentiation, the strongly positive rate of LETM1 was significantly higher in poorly differentiated breast cancer (78.3%, 18/23) than in well (46.7%, 28/60) and moderately (68.4%, 52/76) differentiated cases (P=0.007). Similarly, the strongly positive rate of LETM1-was higher in advanced-stage breast cancer (stage III-IV) (70.1%, 54/77) than in early-stage cases (stage 0-II) (53.7%, 44/82) (P=0.033). In addition, breast cancer with lymph node metastasis had higher LETM1 expression (75.0%, 51/68) compared with the cases with nonmetastasis (51.6%, 47/91) (P=0.003).
Association between LETM1 expression and patient survival
Survival analysis showed that breast cancer patients with high LETM1 expression had lower disease-free survival (DFS) and overall survival (OS) rates than those with low LETM1 expression as determined by the Kaplan-Meier method (both P<0.01) (Figure 3). By combination analysis, we found that for breast cancer patients with early stage (I-II), the LETM1 level was associated with lower DFS and OS rates (P=0.030, P=0.025, respectively) (Figure 4A, 4B). However, the LETM1 expression were not correlate with the survival rates of breast cancer patients with advanced stage (III-IV) (P=0.143 and P=0.065, respectively) (Figure 4C, 4D).
Figure 3.
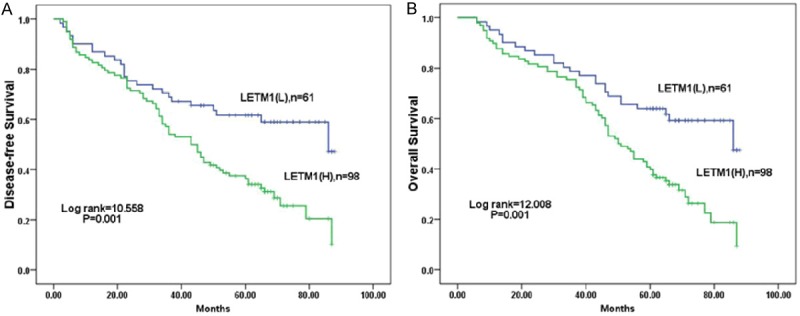
Kaplan-Meier analysis of DFS and OS rates in 159 breast cancer patients in relation to LETM1 protein overexpression. Breast cancer patients with high LETM1 expression had lower DFS (A) and OS (B) rates compared with breast cancer patients with low LETM1 expression (both P=0.001).
Figure 4.
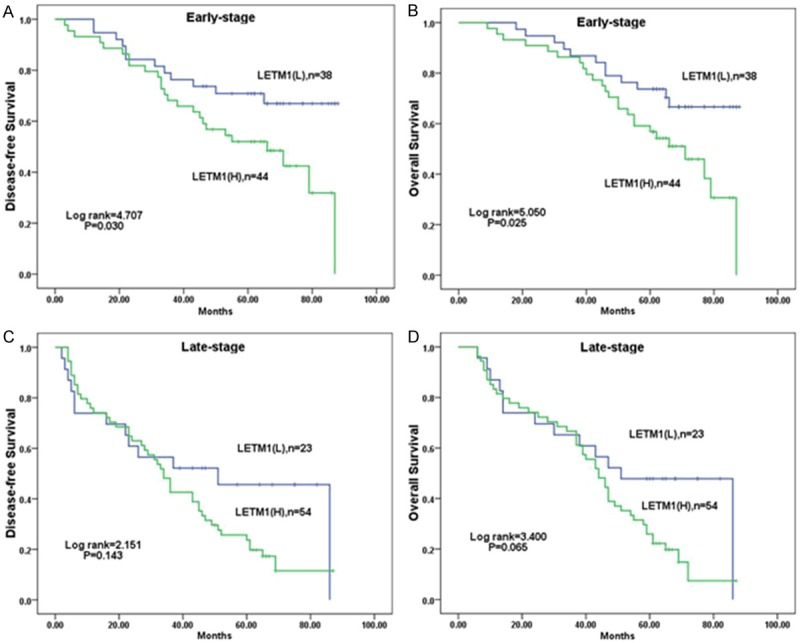
Kaplan-Meier analysis of DFS and OS rates in 159 breast cancer patients with or without LETM1 highly expressed in relation to clinical stage. In early stage (stages 0-II, n=82), patients with high LETM1 expression had significantly reduced DFS (A) and OS (B) rates compared with those with low LETM1 expression (P=0.030 and P= 0.025, respectively). The DFS (C) and OS (D) rates in patients were not related with LETM1 expression in late stage (stages III-IV, n=77) (P=0.143, P=0.065, respectively).
Moreover, univariate analysis demonstrated that clinical stage (P=0.000) and LETM1 expression status (P=0.008) were significantly associated with DFS and OS in patients with breast cancer (Table 3). These data suggest that LETM1 could be a valuable prognostic factor in breast cancer. Multivariate analysis was performed using the Cox proportional hazards model for all significant variables examined in the univariate analysis. We found that clinical stage (HR: 1.899, 95% CI: 1.378-2.617, P=0.000) proved to be an independent prognostic factor for survival in breast cancer. Importantly, LETM1 high expression also emerged as a significant independent poor prognostic factor in breast cancer (HR: 1.448, 95% CI: 1.042-2.011, P=0.027) along with clinical stage (Table 3).
Table 3.
Cox regression model analysis of the clinicopathological features in 159 patients with breast cancer
| Characteristics | B | SE | Wald | HR | 95% CI | P value | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Lower | Upper | ||||||
| Univariate | |||||||
| Age | 0.037 | 0.160 | 0.054 | 1.038 | 0.759 | 1.419 | 0.816 |
| Menopausal status | 0.264 | 0.163 | 2.628 | 1.302 | 0.946 | 1.792 | 0.105 |
| Tumor size | 0.204 | 0.161 | 1.611 | 1.226 | 0.895 | 1.680 | 0.204 |
| Tumor differentiation | 0.150 | 0.114 | 1.724 | 1.162 | 0.929 | 1.454 | 0.189 |
| Clinical stage | 0.684 | 0.162 | 17.722 | 1.981 | 1.441 | 2.724 | 0.000** |
| LN metastasis | 0.075 | 0.161 | 0.218 | 1.078 | 0.786 | 1.477 | 0.641 |
| LETM-1 | 0.442 | 0.166 | 7.076 | 1.555 | 1.123 | 2.154 | 0.008** |
| Multivariate | |||||||
| Clinical stage | 0.641 | 0.164 | 15.340 | 1.899 | 1.378 | 2.617 | 0.000** |
| LETM-1 | 0.370 | 0.168 | 4.870 | 1.448 | 1.042 | 2.011 | 0.027** |
B: coefficient; SE: standard error; Wald: Wald statistic; HR: hazard ratio; CI: confidence interval.
P<0.01.
Discussion
The number of younger patients (aged ≤30 years) with breast cancer is increasing, and have a greater chance of having an endocrine-unresponsive tumor and a significantly poor prognosis compared with patients aged between 31 and 50 years [13]. In order to reduce the number of deaths and improve quality of life for patients with breast cancer, it is necessary to find new biomarkers.
As an organelle, mitochondria participate in a variety of cellular functions, such as growth, division, apoptosis, and energy metabolism [14], which is crucial for life and death of the cell. Mitochondria can produce most of the ATP, participate in Ca2+ signaling and integrate diverse apoptotic stimuli by releasing protein cofactors needed in the cytosol for activation of effector caspases [15,16]. The role of mitochondria in the development and progression of cancer is a rapidly expanding topic [17-19].
In addition, current study has provided clear evidence that LETM1 regulates mitochondrial biogenesis and translation system which can be implicated in tumorigenesis and cancer. Piao et al. [10] found that LETM1 is overexpressed in various human cancers compared with normal tissue from breast, colon, esophagus, lung, ovary, rectum, stomach, and uterine cervix by immunohistochemical analysis. They also found that the overexpression of LETM1 can induce necrotic cell death in HeLa cells, and the inhibition of mitochondrial biogenesis and ATP production seems to be the main cause. Hwang et al. [11] found that the overexpression of LETM1 can induce destruction of mitochondria in lung cancer cells through depleting ATP and AMPK activation and can also facilitating apoptosis via alter Akt signaling and inhibit the cell cycle. Shin et al. [20] showed that the overexpression of the two mitochondria-targeting genes, LETM1 and carboxyl-terminal modulator protein (CTMP), significantly reduced the incidence of tumorigenesis in H-ras12V liver cancer model mice. These results suggested its potential as a tool for gene therapy. Taken together, these findings indicated that LETM1 may play an important role in carcinogenesis and tumor progression. Interestingly, our team found that the overexpression of LETM1 was related with clinical features of triple-negative breast cancer (TNBC) patients [21]. In this study, we want to investigate the clinicopathological significance of LETM1 expression in other more types of breast cancer.
We first performed IF staining of LETM1 in MDA-MB-231 breast cancer cells. In agreement with previous studies, we found that staining of LETM1 is mainly localized in the cytoplasm, which was also reinforced by IHC staining in breast cancer tissue. Through IHC staining of LETM1 and survival data analysis, we found that the positive and strongly positive rates of LETM1 were significantly higher in breast cancer tissues than in either DCIS or nontumor tissues. These findings suggest that LETM1 potentially plays an important role in breast cancer progression and aggressiveness.
In our previous studies, LETM1 expression was found to be elevated and correlate closely with aggressive features, and/or poor prognosis in many human cancers [12,21]. A clinical study involving 107 TNBC patients showed that the LETM1 could serve as an independent predictor for disease-free survival and overall survival in patients with TNBC [21]. In 176 cases of head and neck squamous cell carcinoma (HNSCC), the strongly positive expression of LETM1 was observed in 65.3% of the cases, and it was significantly correlated with poor differentiation, presence of lymph node metastasis and advanced stage, and it was a poor prognostic predictor of the patients [12].
Similarly, we found that the strongly positive rate of LETM1 was correlated closely with tumors poorer differentiation, lymph node metastasis and later clinical stages. These results suggested that LETM1 up-regulation promotes the invasion and/or metastasis of breast cancer cells. However, LETM1 expression levels were not correlated with age, menopausal status and tumor size of patients with breast cancer. Importantly, positive expression of LETM1 was a strong and independent predictor of short overall survival of breast cancer patients, as evidenced by the Kaplan-Meier curves and multivariate Cox proportional hazards regression analysis. Furthermore, stratified survival analysis of breast cancer clinical stage showed that LETM1 expression was helpful for predicting the poor survival of patients with early stages of breast cancer.
In conclusion, LETM1 overexpression appears to be associated with breast cancer progression, and may potentially be used as a biomarker for prognostic evaluation in breast cancer. Therefore, understanding the role of LETM1 may be important in developing effective therapeutics for breast cancer and it may be taken as a candidate for breast cancer therapy. A comprehensive analysis of the molecular mechanism of LETM1 involved in the development and progression of breast cancer is eagerly required.
Conclusions
In summary, LETM1 plays a key role in the progression of breast cancer, and high level of LETM1 protein are strongly associated with the tumor differentiation, advanced stage and lymph node metastasis of patients with breast cancer. The high proportion and prognostic value of LETM1 expression in breast cancer suggests that LETM1 may be a significant biomarker and a potential therapeutic target for breast cancer.
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 31301065), and Grant from Key Laboratory Nature Resources of Changbai Mountain & Functional Molecules, Jilin P. R. of China.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Stopeck AT, Brown-Glaberman U, Wong HY, Park BH, Barnato SE, Gradishar WJ, Hudis CA, Rugo HS. The role of targeted therapy and biomarkers in breast cancer treatment. Clin Exp Metastasis. 2012;29:807–819. doi: 10.1007/s10585-012-9496-y. [DOI] [PubMed] [Google Scholar]
- 3.Okobia M, Bunker C, Zmuda J, Kammerer C, Vogel V, Uche E, Anyanwu S, Ezeome E, Ferrell R, Kuller L. Case-control study of risk factors for breast cancer in Nigerian women. Int J Cancer. 2006;119:2179–2185. doi: 10.1002/ijc.22102. [DOI] [PubMed] [Google Scholar]
- 4.Patani N, Martin LA, Dowsett M. Biomarkers for the clinical management of breast cancer: international perspective. Int J Cancer. 2013;133:1–13. doi: 10.1002/ijc.27997. [DOI] [PubMed] [Google Scholar]
- 5.Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Hart L, Rauch A, Carr AM, Vermeesch JR, O’Driscoll M. LETM1 haploinsufficiency causes mitochondrial defects in cells from humans with Wolf-Hirschhorn syndrome: implications for dissecting the underlying pathomechanisms in this condition. Dis Model Mech. 2014;7:535–545. doi: 10.1242/dmm.014464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endele S, Fuhry M, Pak SJ, Zabel BU, Winterpacht A. LETM1, a novel gene encoding a putative EF-hand Ca(2+)-binding protein, flanks the Wolf-Hirschhorn syndrome (WHS) critical region and is deleted in most WHS patients. Genomics. 1999;60:218–225. doi: 10.1006/geno.1999.5881. [DOI] [PubMed] [Google Scholar]
- 8.Frazier AE, Taylor RD, Mick DU, Warscheid B, Stoepel N, Meyer HE, Ryan MT, Guiard B, Rehling P. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J Cell Biol. 2006;172:553–564. doi: 10.1083/jcb.200505060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlickum S, Moghekar A, Simpson JC, Steglich C, O’Brien RJ, Winterpacht A, Endele SU. LETM1, a gene deleted in Wolf-Hirschhorn syndrome, encodes an evolutionarily conserved mitochondrial protein. Genomics. 2004;83:254–261. doi: 10.1016/j.ygeno.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Piao L, Li Y, Kim SJ, Byun HS, Huang SM, Hwang SK, Yang KJ, Park KA, Won M, Hong J, Hur GM, Seok JH, Shong M, Cho MH, Brazil DP, Hemmings BA, Park J. Association of LETM1 and MRPL36 contributes to the regulation of mitochondrial ATP production and necrotic cell death. Cancer Res. 2009;69:3397–3404. doi: 10.1158/0008-5472.CAN-08-3235. [DOI] [PubMed] [Google Scholar]
- 11.Hwang SK, Piao L, Lim HT, Minai-Tehrani A, Yu KN, Ha YC, Chae CH, Lee KH, Beck GR, Park J, Cho MH. Suppression of lung tumorigenesis by leucine zipper/EF handcontaining transmembrane-1. PLoS One. 2010;5:e12535. doi: 10.1371/journal.pone.0012535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Yang Y, Liu S, Piao L, Zhang Y, Lin Z, Li Z. High expression of leucine zipper-EF-hand containing transmembrane protein 1 predicts poor prognosis in head and neck squamous cell carcinoma. Biomed Res Int. 2014:850316. doi: 10.1155/2014/850316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao Y, Cao M, Fang H, Xie J. Breast cancer in 30-year-old or younger patients: clinicopathologic characteristics and prognosis. World J Surg Oncol. 2015;13:38. doi: 10.1186/s12957-015-0462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 16.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 17.Hall DD, Wu Y, Domann FE, Spitz DR, Anderson ME. Mitochondrial calcium uniporter activity is dispensable for MDA-MB-231 breast carcinoma cell survival. PLoS One. 2014;9:e96866. doi: 10.1371/journal.pone.0096866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makowska K, Estan MC, Ganan-Gomez I, Boyano-Adanez MC, Garcia-Perez AI, Sancho P. Changes in mitochondrial function induced by dequalinium precede oxidative stress and apoptosis in the human prostate cancer cell line PC-3. Mol Biol (Mosk) 2014;48:416–428. [PubMed] [Google Scholar]
- 19.Hou Y, Xu J, Liu X, Xia X, Li N, Bi X. Shikonin induces apoptosis in the human gastric cancer cells HGC-27 through mitochondria-mediated pathway. Pharmacogn Mag. 2015;11:250–256. doi: 10.4103/0973-1296.153074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin JY, Chung YS, Kang B, Jiang HL, Yu DY, Han K, Chae C, Moon JH, Jang G, Cho MH. Co-delivery of LETM1 and CTMP synergistically inhibits tumor growth in H-ras12V liver cancer model mice. Cancer Gene Ther. 2013;20:186–194. doi: 10.1038/cgt.2013.6. [DOI] [PubMed] [Google Scholar]
- 21.Wang CA, Liu Q, Chen Y, Liu S, Xu J, Cui X, Zhang Y, Piao L. Clinical implication of leucine zipper/EF hand-containing transmembrane-1 overexpression in the prognosis of triple-negative breast cancer. Exp Mol Pathol. 2015;98:254–259. doi: 10.1016/j.yexmp.2014.12.012. [DOI] [PubMed] [Google Scholar]


