Abstract
Mixed linage leukemia gene 2 (MLL2) is identified as a novel mutation gene in diffuse large B cell lymphoma (DLBCL). However, the significance of MLL2 protein expression for the prognosis of DLBCL is unclear. In this study, we detected MLL2 protein expression in primary gastrointestinal diffuse large B cell lymphoma (PGI-DLBCL) samples by using tissue microarray immunohistochemistry, and analyzed the correlation between MLL2 protein expression and tumor proliferation activity. In addition, we investigated clinical significance of MLL2 protein expression for PGI-DLBCL prognosis. We found that there was significant difference in MLL2 protein expression between PGI-DLBCL and reactive hyperplasia of lymph node. High expression of MLL2 protein indicated higher clinical stage. In older patients (>60 years) with PGI-DLBCL, MLL2 protein expression was positively correlated with Ki-67 expression and negatively correlated with patient survival. Our data suggest that MLL2 protein is overexpressed in PGI-DLBCL and appears as a prognostic factor for patients of PGI-DLBCL, especially for those older than 60 years old.
Keywords: Primary gastrointestinal lymphoma, MLL2 protein, tissue microarray, immunohistochemistry, prognosis
Introduction
Mixed lineage leukemia (MLL) indicates an evolutionarily conserved trithorax family of human genes that play critical roles in gene regulation and embryonic development. MLL2 is a member of MLL that possesses histone H3 lysine 4 (H3K4)-specific methyltransferase activity and plays important role in epigenetic regulation of transcription [1,2]. Natarajan et al. reported that the level of MLL2 protein was increased in the nucleus and cytoplasm of breast and colorectal cancer cells [1]. Furthermore, abnormal MLL2 expression could be found in prostate cancer and gastric carcinoma compared with normal tissues [2,3]. On the other hand, MLL2 mutations have been detected in small cell lung cancer and renal carcinoma [4,5]. Recurrent mutation of MLL2 gene could be found in 16% of medulloblastoma by exon sequencing [6]. In addition, MLL2 mutations and abnormal expression have been detected in diffuse large B cell lymphoma (DLBCL) [7,8].
The gastrointestinal (GI) tract is the most frequent site of primary extranodal non-Hodgkin’s lymphoma. The main pathology type of GI lymphoma is DLBCL [9]. Primary gastrointestinal diffuse large B cell lymphoma (PGI-DLBCL) is a highly heterogeneous disease with different clinical characteristics and prognosis. PGI-DLBCL is highly aggressive and not sensitive to chemotherapy agents in some patients, leading to poor prognosis. However, some patients with PGI-DLBCL respond well to chemotherapy, such as CHOP-like regimen. Although molecular typing that divides DLBCL into germinal center (GC) and non-germinal center (non-GC) can predict the response to therapy and prognosis, the prognosis of PGI-DLBCL remains difficult.
Considering MLL2 mutations and abnormal expression in DLBCL, we wondered whether MLL2 protein could be a new prognostic factor for PGI-DLBCL. In this study, we detected MLL2 protein expression in PGI-DLBCL samples by using tissue microarray immunohistochemistry, and analyzed the correlation between MLL2 protein expression and tumor proliferation activity. In addition, we investigated the clinical significance of MLL2 protein expression for PGI-DLBCL prognosis.
Material and methods
Subjects
Total 52 cases of PGI-DLBCL and 12 control cases of reactive hyperplasia of lymph node were randomly selected from the patients who visited the First Affiliated Hospital of Wenzhou Medical University. The study protocols were approved by Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University, and signed consent form was obtained from each patient. Follow-up data were obtained from the outpatient reexamination and telephone. The survival of dead patients was defined as the time from pathological diagnosis to the date of death. The survival of alive patients was defined as the time from pathological diagnosis to the last follow-up day. Clinical data were collected from the patients’ medical records, including the age, lactate dehydrogenase (LDH) level, B symptoms, gastric or intestinal involvement, bulky mass size, infiltration depth, lymph node metastasis, Ann Arbor stage, international prognostic index (IPI) and Eastern Cooperative Oncology Group performance status (ECOG). All cases were de novo DLBCL, without histological evidence of mucosa-associated lymphoid tissue (MALT) lymphoma. No one had hepatitis C virus (HCV) and helicobacter pylori (H pylori) infection.
Treatment
Total 46 cases had radical or palliative operation, while the other 6 cases were treated with rituximab plus CHOP chemotherapy (Rituximab 375 mg/m2 d0, Cyclophosphamide 750 mg/m2 d1, Epirubicin 50 mg/m2 d1, Vincristine 1.4 mg/m2 d1, Prednison 100 mg d1-5) after biopsy. 46 cases received chemotherapy after operation, including CHOP or CHOP-like regimen for 27 cases, R-CHOP for 19 cases.
Construction of tissue microarrays (TMAs)
Two 12×8 TMAs were constructed from 52 cases of PGI-DLBCL and 12 control cases of reactive hyperplasia of lymph node. In each case, 3 cases with a diameter of 1.2 mm were obtained from 3 different areas of the tissue blocks and embedded into a single microarray paraffin block. TMAs were kept at 4°C until they were ready for analysis.
Immunohistochemistry
TMAs were cut into 4 μm sections, deparaffinized and rehydrated through graded alcohol series. Slides were pretreated in the microwave with citrate buffer (pH 6.0) at 90°C for 10 min to retrieve antigen, immersed in 3% hydrogen peroxide for 10 min to block endogenous peroxidase activity, and incubated overnight at 4°C with normal bovine serum to reduce non-specific binding. Subsequently, each TMA block was incubated with antibodies against MLL2 (1:30, clone I-18, Santa Cruz Biotechology, CA, USA) or Ki-67 (1:100, clone SP6, Maixin Biotechology, Fuzhou, China), followed by incubation with secondary antibody. The stained sections of the TMA blocks were independently evaluated by 3 hematopathologists. The cases were divided into germinal center B-cell-like (GCB) or non-GCB DLBC as described previously [10]. Dark brown stain granules in nucleus were defined as positive. Stains were scored as low expression for the <80% positive tumor cells, high expression for ≥80% positive tumor cells [11]. Ki-67 located in nucleus. Claybank or yellow brown stain granules in nucleus were defined as positive. Stains were scored as low expression for the <60% positive tumor cells, high expression for ≥60% positive tumor cells [12].
Statistical analysis
Statistical analyses were performed using SPSS19.0 statistical software. χ2 test was used to compare different clinical features between high and low MLL2 protein expression groups. The relationship between binary variable of high and low expression of MLL2 and Ki-67 were analyzed by using binary Logistic regression. Univariate analyses of prognostic factors by using Kaplan-Meier and Log-rank test. A multivariate survival analysis was performed by Cox’s proportional hazards regression model. A P value of less than 0.05 was considered statistically significant.
Results
Survival analysis
Of 52 cases of PGI-DLBCL, 8 cases were positive for CD10 expression, 33 cases were positive for B cell lymphoma-6 (Bcl-6) expression and 43 cases were positive for multiple myeloma oncogene 1 (MUM1) expression (Figure 1A-C). According to immunohistochemistry analysis, PGI-DLBCL was divided into 11 cases of GCB and 41 cases of non-GCB.
Figure 1.
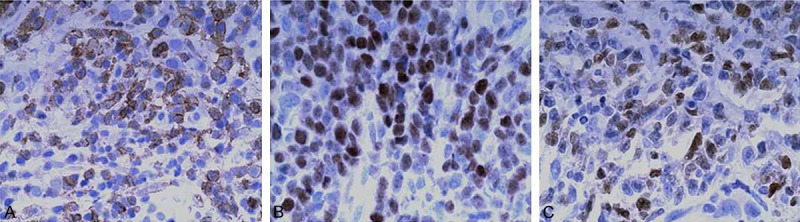
Immunohistochemical staining of PGI-DLBCL tissues. A. Positive staining of CD10. B. Positive staining of Bcl-6. C. Positive staining of MUM1. Magnification: 400 ×.
The median length of follow-up was 28 months (range 2-102 months). Median survival was (67±5) months (95% confidence interval: 58-77), including 20 cases of death and 32 cases of survival. In patients >60 years old, 25 cases had high expression of MLL2 protein, including 8 cases of survival and 11 cases of death, whereas 7 cases had low expression of MLL2 protein, including 5 cases of survival and 2 cases of death. Between the two groups of high and low expression of MLL2 protein in older patients, there are no meaningful difference in ECOG, Ann-Arbor stage, B symptoms, Site involvement, LDH level, IPI score, GCB type and treatment.
Immunohistochemical analysis of MLL2
MLL2 protein was stained both in the cytoplasm and in the nucleus of PGI-DLBCL tissues (Figure 2A). Twelve cases of reactive hyperplasia lymph node tissues had low expression of MLL2 protein (Figure 2B), without differences within the reactive follicles/germinal centers versus interfollicular areas. In 52 cases of PGI-DLBCL, 12 cases had low expression of MLL2 protein, while 40 cases had high expression of MLL2 protein. MLL2 protein expression in PGI-DLBCL and reactive hyperplasia lymph node tissues was statistically significant.
Figure 2.
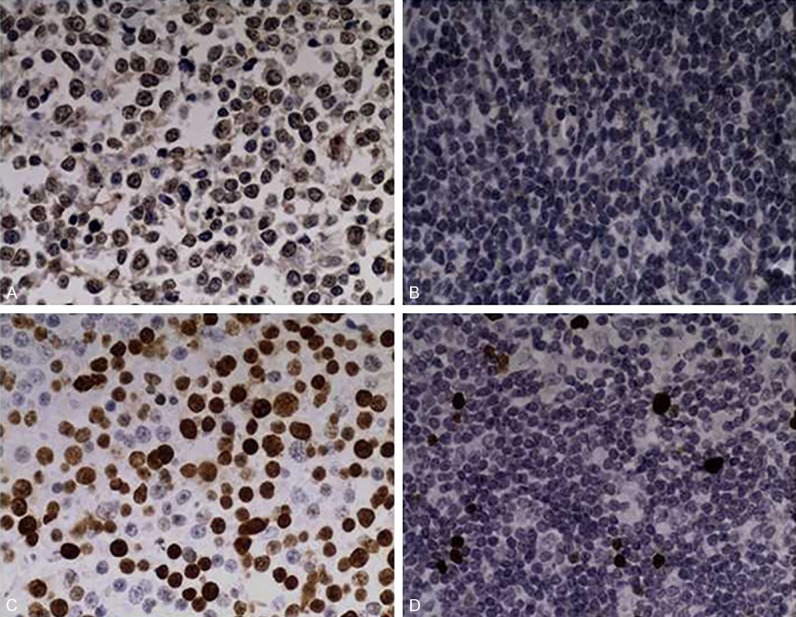
Immunohistochemical staining of MLL2 in PGI-DLBCL and reactive hyperplasia of lymph node tissues. A. Positive staining of MLL2 in PGI-DLBCL. B. Positive staining of MLL2 expression in reactive hyperplasia of lymph node. C. Positive staining of Ki-67 in PGI-DLBCL. D. Positive staining of Ki-67 expression in reactive hyperplasia of lymph node. Positive staining of MUM1. Magnification: 400 ×.
To characterize the proliferation ability of PGI-DLBCL tissues and reactive hyperplasia lymph node tissues, we performed Ki-67 staining. Positive expression of Ki-67 was located in the nucleus of PGI-DLBCL tissues, stained as dark brown or yellow brown granules (Figure 2C). Among 52 cases of PGI-DLBCL, 30 cases had high expression of Ki-67, which accounted for 58% (30/52), while 22 cases had low expression of Ki-67, which accounted for 42% (22/52). In reactive hyperplasia lymph node tissues, Ki-67 expression was located in lymphoid follicle germinal center of mature lymphocyte nucleus (Figure 2D).
Relationship between MLL2 protein expression and clinical features of PGI-DLBCL
MLL2 protein expression was correlated with clinical stages of PGI-DLBCL. The expression rate of MLL2 protein was 57.9% in 11 cases with I/II stage but was significantly higher in 29 cases with III/IV stages (87.8%) (P<0.05). MLL2 protein expression had no correlation with the age, B symptom, site involvement, the size of mass, infiltration depth, lymph node involvement, LDH, IPI and immunophenotyping of PGI-DLBCL (Table 1).
Table 1.
Comparison of clinical features between patients with high and low MLL2 protein expression in all patients
| MLL2 expression | |||
|---|---|---|---|
|
|
|||
| Characteristics | <80% (n=12) | ≥80% (n=40) | P-value |
| Age >60 years | 7 (58.3%) | 25 (62.5%) | 0.795 |
| Performance, ECOG >2 | 3 (25.0%) | 12 (30.0%) | 0.650 |
| Ann-Arbor stage III/IV | 4 (33.3%) | 29 (72.5%) | 0.013* |
| B symptoms | 4 (33.3%) | 15 (37.5%) | 0.793 |
| Site involvement | 0.838 | ||
| Gastric | 7 (58.3%) | 22 (55.0%) | |
| Intestinal | 5 (41.7%) | 18 (45.0%) | |
| Bulky tumor (>5 cm) | 8 (66.7%) | 30 (75.0%) | 0.568 |
| Lymph node involvement | 7 (58.3%) | 31 (77.5%) | 0.189 |
| LDH elevation | 2 (16.7%) | 11 (27.5%) | 0.447 |
| IPI | 0.128 | ||
| 0-2 | 11 (91.7%) | 28 (70.0%) | |
| 3-5 | 1 (8.33%) | 12 (30.0%) | |
| GCB type | 4 (33.3%) | 7 (17.5%) | 0.838 |
ECOG, eastern cooperative oncology group; LDH, lactate dehydrogenase; IPI, international prognostic index; GCB, germinal center B-cell.
P<0.05.
In 52 cases of PGI-DLBCL, the correlation between MLL2 and Ki-67 protein expression was not significant (P>0.05). Meanwhile, low expression of MLL2 protein in reactive tissue also had no significant correlation with Ki-67 expression (P>0.05). Additionally, the survival of the patients between those with high expression of MLL2 protein (>80%) and those with low expression of MLL2 protein (<80%) was not significantly different (P>0.05, Figure 3A). Cox multivariate analysis revealed that IPI, immunophenotyping and LDH were significantly related to the prognosis (P<0.05). However, in older patients (age >60 years), 25 cases had high expression of MLL2 protein and 7 cases had low expression of MLL2 protein, while 19 cases had high expression of Ki-67 (>60%) and 13 cases had low expression of MLL2 protein (<60%). In the older patient group, MLL2 protein expression was positively correlated with Ki-67 protein expression (P=0.03). In addition, patients with high expression of MLL2 protein had shorter survival (Figure 3B).
Figure 3.
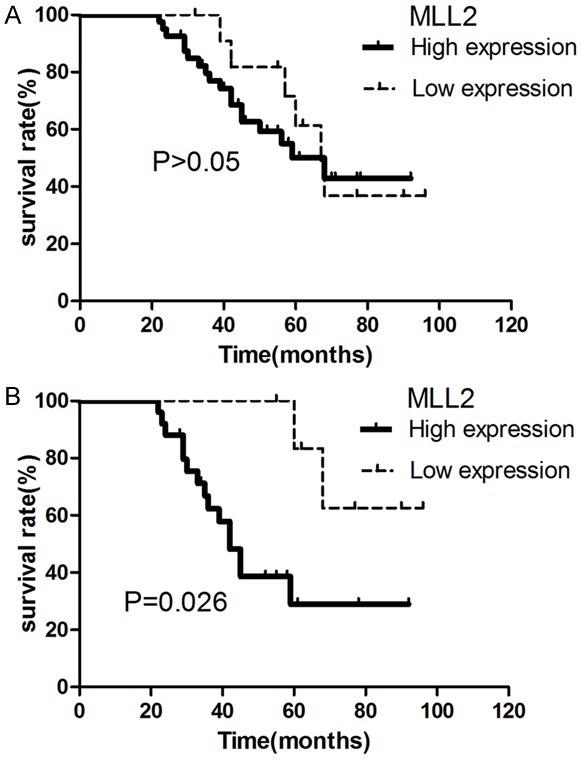
Survival curve analysis of PGI-DLBCL patients. A. The curve of all 52 PGI-DLBCL patients with different expression level of MLL2. B. The curve of older PGI-DLBCL patients (>60 years, n=32) with different expression level of MLL2.
Discussion
Histone methyltransferases regulate a variety of cellular processes such as the formation of heterochromatin, transcription, cell proliferation and adhesion, apoptosis and DNA damage repair, and are crucially involved in cancer progression [13-15]. MLL2 as a histone methyltransferase has been shown to be mutated or overexpressed in tumors such as breast cancer and colorectal cancer [16]. Previous data showed that MLL2 is a common target for chromosomal translocations associated with human acute leukemias [16].
In this study we examined MLL2 protein expression in PGI-DLBCL by using tissue microarray immunohistochemistry. Tissue microarray combines the protein expression level of gene and morphology of tissue, which different from gene chip and protein array. In addition, tissue microarray can save tissue specimen, reagent, time, and reduce systematic error.
Our data demonstrated that MLL2 proteins expression in reactive hyperplasia of lymph node tissue is significantly different from that in PGI-DLBCL. MLL2 mutation in lymphoma indicate that chromatin dysfunction play an important role in lymphoma [8]. Recently, a study showed that t(14; 18)(q32; q21) gene rearrangement occurred from the early benign B cell clone to tumor precursor B cells clone. However, MLL2 gene mutation occurred in the late stage of malignant B cell clone. Therefore, MLL2 gene mutation was considered as a secondary event in follicular lymphoma, and MLL2 gene mutation work as a helper or a passerby in the pathogenesis of lymphoma, but not as a caustic pathogenic factor [17].
Our study showed that MLL2 protein expression was positively correlated with Ki-67 expression in old patients with PGI-DLBCL (age >60 years). Natarajan et al. found that MLL2 expression level was positively correlated with malignancy of breast cancer and colorectal cancer [1]. We also investigated the clinical outcome of the patients with high MLL2 protein expressions. All cases had no underlying conditions, such as H pylori or HCV infections. For the entity of PGI-DLBCL with H. pylori positive is biologically distinct from H. pylori negative cases and has a better clinical outcome [18]. HCV infection may influence the outcome of B cell lymphoma, notwithstanding there is no statistical evidence that the prognosis of HCV-positive patients is inferior to that of HCV-negative subjects [19]. We found that MLL2 protein expressions were correlated to prognosis in elderly patients (age >60 years), which indicated that abnormal methylation regulated by MLL2 may be involved in elderly patients with PGI-DLBCL.
Although genome abnormalities such as chromosome translocation are the prominent presentation in lymphoma, abnormal methylation in lymphoma has been reported. Choi et al. found that abnormal methylation was frequently detected in promoter region in lymphoma [20]. Methylation is involved in lymphoma through influence on gene transcription by regulation of chromosome structure [21]. Moreover, the degree of methylation is related to clinical stage of DLBCL [22]. Amara et al. found that p16 methylation in DLBCL was related to clinical stage and B symptom, and DLBCL patients with p16 methylation had shorter overall survival (OS) and disease free survival (DFS) [23]. However, DLBCL patients with GSTPl gene methylation had low clinical stage and high complete remission [24]. Therefore, the role of methylation in lymphoma needs further investigation, which will help develop novel demethylation therapy for elderly lymphoma patients.
Currently, immunohistochemistry test for CD10, BCL-6 and MUM1 is used to distinguish between GCB and activated B-cell (ABC) type of DLBCL. Our data showed that there was no significantly difference in MLL2 protein expression between GCB and non-GCB groups. However, Shaknovich et al. sorted GCB and non-GCB groups based on the methylation of 263 genes and the accurate rate was up to 91% [25]. Therefore, it is interesting to investigate whether MLL2 combined with methylation of other genes could be used to distinguish between GCB and non-GCB.
Several limitations of this study need to be pointed out. First, we still do not understand why MLL2 protein expression was positively correlated with Ki-67 expression and negatively correlated with patient survival only in older patients (>60 years) with PGI-DLBCL, but not in all PGI-DLBCL patients. Further mechanistic studies are necessary. Second, the sample size of this study is small. Studies that employ larger samples are needed to confirm our conclusion.
In conclusion, our data suggest that MLL2 protein is overexpressed in PGI-DLBCL and appears as a prognostic factor for patients of PGI-DLBCL, especially for those older than 60 years old.
Acknowledgements
This study was supported by grants from the Wenzhou Municipal Sci-Tech Bureau’s program (No. Y20120045) , Zhejiang Provincial Natural Science Foundation of China (No. Y13H080014 to Hai-ge Ye; No. LY12H08003 to Shen-meng Gao). We acknowledge the pathology department of the Second Affiliated Hospital of Wenzhou Medical University, for assistance in the collection of some of the paraffin block and HE (hematoxylin-eosin) pathological section samples.
Disclosure of conflict of interest
None.
References
- 1.Natarajan TG, Kallakury BV, Sheehan CE, Bartlett MB, Ganesan N, Preet A, Ross JS, Fitzgerald KT. Epigenetic regulator MLL2 shows altered expression in cancer cell lines and tumors from human breast and colon. Cancer Cell Int. 2010;10:13. doi: 10.1186/1475-2867-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Je EM, Lee SH, Yoo NJ, Lee SH. Mutational and expressional analysis of MLL genes in gastric and colorectal cancers with microsatellite instability. Neoplasma. 2013;60:188–195. doi: 10.4149/neo_2013_025. [DOI] [PubMed] [Google Scholar]
- 3.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, Varela I, Nik-Zainal S, Davies HR, Ordonez GR, Mudie LJ, Latimer C, Edkins S, Stebbings L, Chen L, Jia M, Leroy C, Marshall J, Menzies A, Butler A, Teague JW, Mangion J, Sun YA, McLaughlin SF, Peckham HE, Tsung EF, Costa GL, Lee CC, Minna JD, Gazdar A, Birney E, Rhodes MD, McKernan KJ, Stratton MR, Futreal PA, Campbell PJ. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O’Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, Novak AJ, Dogan A, Ansell SM, Link BK, Zou L, Gould J, Saksena G, Stransky N, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Hernandez-Lemus E, Schwarz-Cruz y Celis A, Imaz-Rosshandler I, Ojesina AI, Jung J, Pedamallu CS, Lander ES, Habermann TM, Cerhan JR, Shipp MA, Getz G, Golub TR. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, Jackman S, Krzywinski M, Scott DW, Trinh DL, Tamura-Wells J, Li S, Firme MR, Rogic S, Griffith M, Chan S, Yakovenko O, Meyer IM, Zhao EY, Smailus D, Moksa M, Chittaranjan S, Rimsza L, Brooks-Wilson A, Spinelli JJ, Ben-Neriah S, Meissner B, Woolcock B, Boyle M, McDonald H, Tam A, Zhao Y, Delaney A, Zeng T, Tse K, Butterfield Y, Birol I, Holt R, Schein J, Horsman DE, Moore R, Jones SJ, Connors JM, Hirst M, Gascoyne RD, Marra MA. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC, et al. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lym- phoma Study Group. Blood. 1994;84:1361–1392. [PubMed] [Google Scholar]
- 10.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 11.Yoon DH, Choi DR, Ahn HJ, Kim S, Lee DH, Kim SW, Park BH, Yoon SO, Huh J, Lee SW, Suh C. Ki-67 expression as a prognostic factor in diffuse large B-cell lymphoma patients treated with rituximab plus CHOP. Eur J Haematol. 2010;85:149–157. doi: 10.1111/j.1600-0609.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 12.Jovanovic MP, Jakovic L, Bogdanovic A, Markovic O, Martinovic VC, Mihaljevic B. Poor outcome in patients with diffuse large B-cell lymphoma is associated with high percentage of bcl-2 and Ki 67-positive tumor cells. Vojnosanit Pregl. 2009;66:738–743. doi: 10.2298/vsp0909738p. [DOI] [PubMed] [Google Scholar]
- 13.Jun HJ, Kim J, Hoang MH, Lee SJ. Hepatic lipid accumulation alters global histone h3 lysine 9 and 4 trimethylation in the peroxisome proliferator-activated receptor alpha network. PLoS One. 2012;7:e44345. doi: 10.1371/journal.pone.0044345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Champagne KS, Kutateladze TG. Structural insight into histone recognition by the ING PHD fingers. Curr Drug Targets. 2009;10:432–441. doi: 10.2174/138945009788185040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, Imbert J, Andrau JC, Ferrier P, Spicuglia S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 17.Green MR, Gentles AJ, Nair RV, Irish JM, Kihira S, Liu CL, Kela I, Hopmans ES, Myklebust JH, Ji H, Plevritis SK, Levy R, Alizadeh AA. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paydas S. Helicobacter pylori eradication in gastric diffuse large B cell lymphoma. World J Gastroenterol. 2015;21:3773–3776. doi: 10.3748/wjg.v21.i13.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox MC, Aloe-Spiriti MA, Cavalieri E, Alma E, Gigante E, Begini P, Rebecchini C, Delle Fave G, Marignani M. HCV infection, B-cell non-Hodgkin’s lymphoma and immunochemotherapy: Evidence and open questions. World J Gastrointest Oncol. 2012;4:46–53. doi: 10.4251/wjgo.v4.i3.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi JH, Li Y, Guo J, Pei L, Rauch TA, Kramer RS, Macmil SL, Wiley GB, Bennett LB, Schnabel JL, Taylor KH, Kim S, Xu D, Sreekumar A, Pfeifer GP, Roe BA, Caldwell CW, Bhalla KN, Shi H. Genome-wide DNA methylation maps in follicular lymphoma cells determined by methylation-enriched bisulfite sequencing. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaknovich R, Melnick A. Epigenetics and B-cell lymphoma. Curr Opin Hematol. 2011;18:293–299. doi: 10.1097/MOH.0b013e32834788cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zainuddin N, Kanduri M, Berglund M, Lindell M, Amini RM, Roos G, Sundstrom C, Enblad G, Rosenquist R. Quantitative evaluation of p16(INK4a) promoter methylation using pyrosequencing in de novo diffuse large B-cell lymphoma. Leuk Res. 2011;35:438–443. doi: 10.1016/j.leukres.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Amara K, Trimeche M, Ziadi S, Laatiri A, Hachana M, Korbi S. Prognostic significance of aberrant promoter hypermethylation of CpG islands in patients with diffuse large B-cell lymphomas. Ann Oncol. 2008;19:1774–1786. doi: 10.1093/annonc/mdn374. [DOI] [PubMed] [Google Scholar]
- 24.Nakamichi I, Tomita Y, Zhang B, Sugiyama H, Kanakura Y, Fukuhara S, Hino M, Kanamaru A, Ogawa H, Aozasa K. Correlation between promoter hypermethylation of GSTP1 and response to chemotherapy in diffuse large B cell lymphoma. Ann Hematol. 2007;86:557–564. doi: 10.1007/s00277-007-0299-1. [DOI] [PubMed] [Google Scholar]
- 25.Shaknovich R, Geng H, Johnson NA, Tsikitas L, Cerchietti L, Greally JM, Gascoyne RD, Elemento O, Melnick A. DNA methylation signatures define molecular subtypes of diffuse large B-cell lymphoma. Blood. 2010;116:e81–89. doi: 10.1182/blood-2010-05-285320. [DOI] [PMC free article] [PubMed] [Google Scholar]


