Abstract
As one of the most common malignant tumors, gastric cancer still lacks tumor markers with enough specificity and sensitivity. Therefore the development of novel tumor markers is necessary for early diagnosis in clinics. MicroRNA (miR) has been known to be of unique expressional patterns in various tumors and may work as potential tumor markers for clinical use. This study thus explored the significance of plasma miR-106a in clinical diagnosis of gastric cancer and its effects on proliferation of cancer cells. Plasma miR-106a levels were quantified by real-time quantitative fluorescent PCR methods in 80 cases of gastric cancer patients and healthy individuals to analyze the correlation between miR-106a and clinical features. MiR-106 inhibitor was further transfected into human gastric carcinoma cells for further cell proliferation using CCK-8 approach. MiR-106a was significantly up-regulated in gastric cancer patient plasma samples compared to healthy individuals (P<0.01). The area under ROC curve was 0.895 (95% CI: 0.846~0.943). It has a specificity of 93.8% and a sensitivity of 77.5% in diagnosing gastric cancer. MiR-106a level was also correlated with cancer differentiation stage, lymph node metastasis, TNM stage and tumor size (P<0.05). The down-regulation of miR-106 in gastric carcinoma cells inhibited cell proliferation (P<0.05). MiR-106a was significantly up-regulated in gastric cancer patients and can facilitate the in vitro proliferation of tumor cells. It may work as a biological marker for gastric cancer.
Keywords: MicroRNA-106a, gastric carcinoma, plasma, cell proliferation
Introduction
As one of most common malignant tumors, gastric carcinoma is the four popular and second deadly cancers worldwide, although displaying an decreasing trend of incidence [1,2]. Patients were often at the late stage at the time of first diagnosis, causing high treatment cost and low survival rate. The 5-year overall survival rate of gastric carcinoma is closely correlated with the tumor stage, as stage I patients had 5-year survival rate over 90%, whilst stage IV patients had less than 5% survival after 5 years [3,4]. Therefore, the early diagnosis and treatment are crucial for improving the prognosis of gastric cancer. Classical serum tumor markers including CEA, CA19-9 had certain implication for gastric cancer diagnosis but lack enough specificity and sensitivity [5]. Novel tumor markers are thus required for improving the diagnostic rate of gastric carcinoma.
MicroRNA (miR) is a type of endogenous non-coding small RNA molecule with 19~24 nt length. It can bind onto the 3’-untranslated region (UTR) of mRNA of target genes, and modulate gene expression via the degradation of target mRNAs. MiRNA plays a critical role in cellular function, including cell apoptosis, proliferation, differentiation and tumor cell infiltration [6-8]. The expression of miRNA has unique patterns across different tumor tissues, suggesting the correlation between miRNA expression and tumor progression. The findings that miRNA expressional profile was correlated with tumor tissue type [9] indicated the potency of miRNA as clinical markers for tumor. The expression of miRNA in peripheral blood is relatively stable, as those in the plasma samples from tumor patients. Unique expressional spectrum of miRNA has been confirmed in plasma of tumor patients [10,11], indicating its potency in diagnosis, treatment and prognosis prediction of cancer.
Previous study has reported the altered expression of miR-106a in plasma from gastric cancer patients [12], indicating its potency as a gastric cancer diagnostic marker. To further illustrate the applicable value of miR-106a in gastric cancer diagnosis, this study utilized quantitative real-time PCR approach to detect the expressional profile of miR-106a in plasma from both gastric cancer and healthy people, in order to analyze the clinical implication of plasma miR-106a. Further studies were performed to analyze the correlation between miR-106a and gastric carcinoma pathogenesis, in addition to in vitro effect of miR-106a on human gastric carcinoma cells, in an attempt to elucidate the role of miR-106a on gastric cancer occurrence and its potency as a novel tumor marker.
Materials and methods
General information of patients
A total of 80 gastric cancer patients in the first affiliated hospital of Harbin medical university between May, 2014 and May, 2015 were recruited in this study as the disease group. All patients had confirmed diagnosis of gastric carcinoma by pathological examination. There were 46 males and 34 females in the disease group, with a medium age at 68 years old. According to the TNM staging system (7th version, 2010) stipulated by American Joint Committee on Cancer (AJCC), there were 45 cases of stage I~II, and 35 patients at stage III~IV. No treatment has been performed on patients before collecting plasma samples. Another cohort of 80 healthy individuals (44 males, 36 females, medium age =67 years) were recruited from routine health examination of the first affiliated hospital of Harbin medical university. This study has been approved by the ethical committee of the first affiliated hospital of Harbin medical university and has obtained written consents from all participants.
Plasma collection and RNA extraction
Peripheral blood samples were collected in EDTA-containing tubes. After room temperature incubation for 30 min, blood samples were centrifuged at 2,000 g for 10 min. Plasma at the upper phase was transferred to a new tube for another centrifugation at 12,000 g for 10 min and stored at -80°C for further use.
Trizol kit (Invitrogen, US) was used to extract total RNA from plasma according to the manual instruction. After dissolving in RNAse-free water, RNA contents were quantified by ultraviolet spectrophotometer and stored at -80°C for further use.
MiR-106a quantification
Total RNA extracted from plasma was used in in vitro reverse transcription using TaqMan microRNA Reverse Transcription Kit (ABI, US) following the manual instruction. The reaction condition was: 16°C for 30 min, 42°C for 30 min, followed by 85°C for 5 min and 4°C storage. Real-time fluorescent quantitative PCR was performed using cDNA as the template and TaqMan Universal Master Mix (ABI, US) in an ABI7500 RT-PCR cycler. The amplification parameters were: 95°C denature for 10 min, followed by 40 cycles each containing 95°C denature for 10 sec, 60°C annealing for 1 min and 72°C elongation for 1 min. Using U6 gene as the internal reference, miR-106a level was quantified using 2-ΔCt method (ΔCt=CtmiR-106a-CtU6).
Cell culture and transfection
Human gastric carcinoma cell line MGC-803 (Cell bank of Chinese Academy of Science, Shanghai, China) was incubated in PRIM 1640 medium (Gibco, US) with 10% fetal bovine serum (FBS, Gibco, US), 100 U/mL penicillin and 100 μg/mL streptomycin under a 37°C humidified chamber with 5% CO2. Cells at log phase were seeded into 6-well plate until reaching confluence of 70%~80%. Transfection was performed using Lipofectamine 2000 reagent (Invitrogen, US) with miR-106a inhibitor (Ruibo Biotech, China) or controlled vector (50 nM). Six hours after transfection, cells were continuously incubated with complete medium.
Cell proliferation assay
48 hours after transfection, all cells were seeded into 96-well plate (0.1 mL each well, 10,000 per mL) for continuous incubation at 37°C humidified chamber. 10 μL CCK-8 working solutions (Dojindo, Japan) were added into each well at 1 day, 2 days, 3 days, 4 days and 5 days after transfection. Optical density (OD) values at 450 nm were measured in a microplate reader. Cell growth curve was plotted using OD values against incubation time.
Statistical analysis
SPSS 19.0 software package was used to analyze all collected data, of which measurement data were presented as mean ± standard deviation (SD). Independent 2-sample student t-test was used to compare means between two groups. Receiver operator characteristic (ROC) curve was used to analyze the clinical significance of miR-106a in gastric cancer diagnosis. Analysis of variance (ANOVA) was used to compare means across multiple groups. A statistical significance was defined when P<0.05.
Results
Plasma miR-106a expression in gastric cancer
After extracting total RNA from 80 gastric cancer patients and 60 healthy individuals, real-time fluorescent quantitative PCR was used to detect plasma miR-106 levels. Results showed significantly elevated plasma miR-106a in gastric carcinoma patients compared to controlled individuals (P<0.01, Figure 1).
Figure 1.
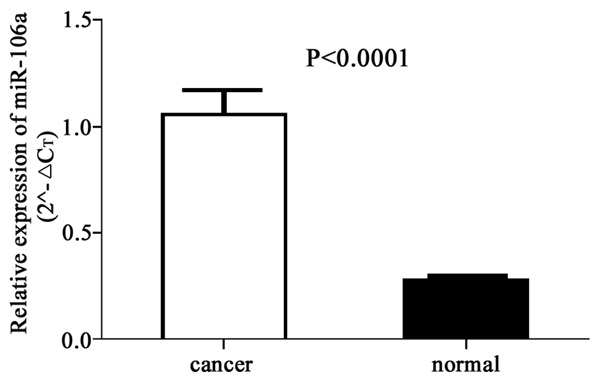
Plasma miR-106a levels in gastric cancer patients.
Correlation between plasma miR-106a level and clinical features of gastric cancer
We performed a statistical analysis regarding the correlation between plasma miR-106a level in gastric cancer patients and their clinical parameters. Results showed elevated miR-106 level in moderate to low differentiated cancer patients compared to those in high differentiated cancers (P<0.01, Table 1). Moreover, patients with tumors larger than 5 cm had higher plasma miR-106a levels compared to those with smaller tumors (P<0.05). MiR-106a level was also elevated in patients with lymph node metastasis (P<0.01). When examining TNM stage, it is found that stage I and stage II gastric cancers had lower miR-106a levels compared to late stage (stage III and IV) patients (P<0.01). Other parameters such as age or sex had no significant correlation with miR-106a levels (P>0.05, Table 1).
Table 1.
Correlation of plasma miR-106a level with clinical features of gastric carcinoma
| Parameter | N | miR-106a relative level | P value |
|---|---|---|---|
| Sex | |||
| Male | 42 | 0.628 (0.544~0.932) | 0.832 |
| Female | 38 | 0.782 (0.331~1.090) | |
| Age (years) | |||
| ≥68 | 43 | 0.709 (0.525~1.086) | 0.915 |
| <68 | 37 | 0.648 (0.518~1.042) | |
| Differentiation | |||
| Low to moderate | 46 | 0.939 (0.706~1.806) | 0.0005** |
| High | 34 | 0.633 (0.423~0.775) | |
| Tumor size | |||
| ≥5 cm | 30 | 0.764 (0.608~1.806) | 0.017* |
| <5 cm | 50 | 0.625 (0.328~0.919) | |
| Lymph node metastasis | |||
| Yes | 46 | 0.765 (0.528~1.209) | 0.001** |
| No | 34 | 1.034 (0.714~2.462) | |
| TNM stage | |||
| I + II | 45 | 0.759 (0.528~0.904) | 0.0001** |
| III + IV | 35 | 1.086 (0.744~2.462) |
P<0.05 compared to control group;
P<0.01 compared to control group.
Diagnostic implication of miR-106a
ROC curve showed certain clinical values of plasma miR-106a in diagnosis of gastric cancer. The area under ROC curve was 0.895 (95% CI: 0.846~0.943). The specificity and sensitivity of diagnosing gastric carcinoma were 93.8% and 77.5%, respectively (Figure 2).
Figure 2.
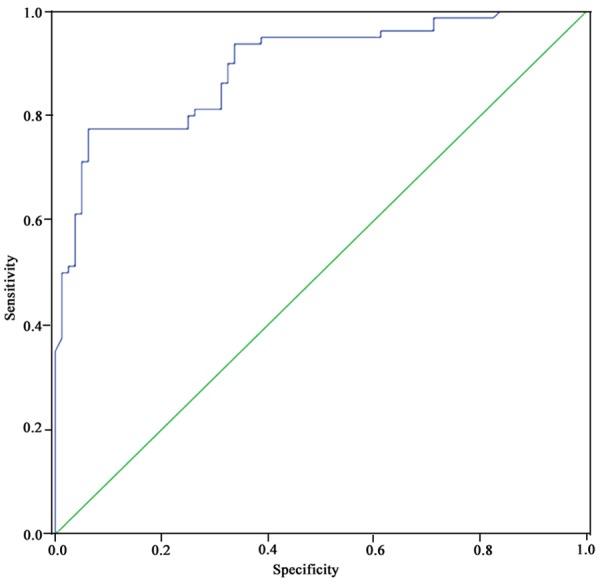
Receiver operator characteristic (ROC) curve.
Gastric carcinoma cell proliferation under miR-106a inhibition
Using CCK-8 assay, we found significantly depressed OD values in gastric cancer cells transfected with miR-106 inhibitor compared to negative control cells (P=0.017, Figure 3), suggesting the inhibitor function on MGC-803 cell proliferation by silencing miR-106a expression.
Figure 3.
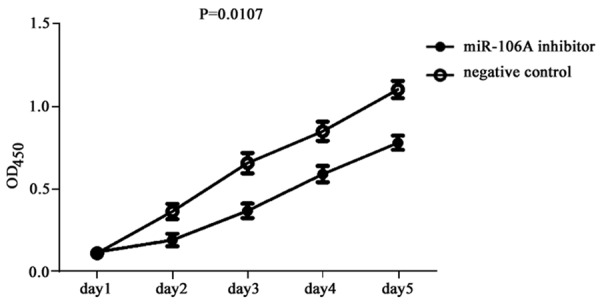
MGC-803 cell proliferation after miR-106a inhibition.
Discussion
As one of the most common malignant tumors, gastric cancer severely affects the public health. About 70% of mortality cases occur in developing countries, and almost half of tumor patients reside in east Asian region [13]. Recently, the occurrence mortality rate of gastric cancer has been decreased, but still resides within most popular cancers. Due to the lack of significant clinical features, most gastric carcinoma patients are already at the terminal stage at the time of first prognosis, causing unfavorable prognosis. Therefore the early diagnosis of gastric cancer is of critical importance for improving patient prognosis [14,15].
Currently available tumor markers are better fitted for monitoring recurrence of tumors but lack enough specificity and sensitivity for early diagnosis of tumors. The searching for tumor markers with high sensitivity and specificity thus can help to improve the early diagnostic rate of gastric carcinoma, in addition to evaluating treatment efficacy. Currently widely used tumor markers such as CEA and CA19-9 had insufficient sensitivity and specificity, thus limiting its application in early diagnosis of gastric cancer. The establishment of gastric cancer markers with enough sensitivity and specificity is thus of critical importance. Although a wide array of studies have been performed regarding the molecular mechanism of gastric cancer pathogenesis, it is still unclear for the exact pathogenesis mechanism.
Current studies have found the involvement of multiple miRNA molecules in occurrence and progression of gastric cancer [4,16,17]. As the most widely studied small non-coding RNA, miRNA can bind with target mRNA sequence to form RNA-induced silencing complex, which is further degraded for mediating gene expression at translational level. Studies have confirmed the participation of miRNA in various gene expression regulation and its crucial roles in various physiological or pathological processes. MiRNA has been found to be abnormally expressed in multiple tumor tissues, supporting its potency as tumor markers [18-20]. Previous study has found multiple miRNAs as independent predictive index for prognosis of gastric cancer [21], further supporting the potency of miRNA as novel markers for early diagnosis of gastric carcinoma.
Current studies regarding the correlation between miRNAs and gastric cancer mainly focused on tissue samples, which is difficult to be directly examined in clinical practice. As miRNA has a stable expressional prolife in plasma and correspondingly altered expressional spectrum after tumor occurrence [22,23], plasma miRNA level may be used for diagnosis and prognosis prediction of malignant tumors in clinics.
MiR-106a is one member of miR-17 family, which has been observed in multiple tumors with over-expression and may facilitate the occurrence and progression of tumors via facilitating tumor cell proliferation, inhibiting apoptosis, inducing tumor angiogenesis and resisting tumor suppressor genes [24]. This study found elevated plasma miR-106a in gastric cancer patients compared to healthy people. Further clinical-based analysis found the correlation between miR-106a and cancer differentiation stage, lymph node metastasis, TNM staging and tumor size. Those results suggested the potential relationship between high plasma miR-106a and high malignancy or unfavorable prognosis of tumors. ROC curve analysis identified the area under the curve of 0.895 (95% CI: 0.846~0.943), with 93.8% specificity and 77.5% sensitivity. These results indicated satisfactory diagnostic efficacy of plasma miR-106a, thus suggesting the potency of plasma miR-106 as one novel marker for gastric cancer with high sensitivity and specificity.
To further explore the possible mechanism of miR-106a during the occurrence and progression of gastric carcinoma, we artificially silenced the miR-106a expression in human gastric carcinoma cell line MGC-80, whose proliferation status was observed using CCK-8 assay. Results found the significantly proliferation inhibition by miR-106a inhibitor. These results supported the ideas that miR-106a might play a role in the occurrence and progression of gastric cancer.
Our results showed the correlation between plasma miR-106a expression level and various clinical features of gastric cancer, suggesting its potency as a novel tumor marker for clinical diagnosis of gastric carcinoma. Further in vitro cell study found inhibition of gastric carcinoma cell proliferation by miR-106 inhibitor transfection, revealing its possible function in tumor progression. These results collectively supported the potency of plasma miR-106a as a novel biological marker for gastric cancer, thus providing new insights for both early diagnosis and cellular therapy of gastric cancer.
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Hamashima C. Current issues and future perspectives of gastric cancer screening. World J Gastroenterol. 2014;20:13767–74. doi: 10.3748/wjg.v20.i38.13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuda T, Ajiki W, Marugame T, Ioka A, Tsukuma H, Sobue T Research Group of Population-Based Cancer Registries of Japan. Population-based survival of cancer patients diagnosed between 1993 and 1999 in Japan: a chronological and international comparative study. Jpn J Clin Oncol. 2011;41:40–51. doi: 10.1093/jjco/hyq167. [DOI] [PubMed] [Google Scholar]
- 4.Yin Y, Li J, Chen S, Zhou T, Si J. MicroRNAs as diagnostic biomarkers in gastric cancer. Int J Mol Sci. 2012;13:12544–55. doi: 10.3390/ijms131012544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He CZ, Zhang KH, Li Q, Liu XH, Hong Y, Lv NH. Combined use of AFP, CEA, CA125 and CAl9-9 improves the sensitivity for the diagnosis of gastric cancer. BMC Gastroenterol. 2013;13:87. doi: 10.1186/1471-230X-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song B, Ju J. Impact of miRNAs in gastrointestinal cancer diagnosis and prognosis. Expert Rev Mol Med. 2010;12:e33. doi: 10.1017/S1462399410001663. [DOI] [PubMed] [Google Scholar]
- 7.Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29:5761–71. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 8.Cadamuro AC, Rossi AF, Maniezzo NM, Silva AE. Helicobacter pylori infection: host immune response, implications on gene expression and microRNAs. World J Gastroenterol. 2014;20:1424–37. doi: 10.3748/wjg.v20.i6.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsujiura M, Komatsu S, Ichikawa D, Shiozaki A, Konishi H, Takeshita H, Moriumura R, Nagata H, Kawaguchi T, Hirajima S, Arita T, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–9. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou H, Guo JM, Lou YR, Zhang XJ, Zhong FD, Jiang Z, Cheng J, Xiao BX. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J Mol Med (Berl) 2010;88:709–17. doi: 10.1007/s00109-010-0617-2. [DOI] [PubMed] [Google Scholar]
- 12.Cai H, Yuan Y, Hao YF, Guo TK, Wei X, Zhang YM. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol. 2013;30:452. doi: 10.1007/s12032-012-0452-0. [DOI] [PubMed] [Google Scholar]
- 13.Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17:26–33. doi: 10.1007/s10120-013-0259-5. [DOI] [PubMed] [Google Scholar]
- 14.Kosuke N, Oguma H, Yamamoto M. Early gastric cancer with lymph node metastasis. Ann Surg. 2011;253:840–1. doi: 10.1097/10.1097/SLA.0b013e318211d91b. author reply 841. [DOI] [PubMed] [Google Scholar]
- 15.Kim BS, Cho SW, Min SK, Lee BH. Differences in prognostic factors between early and advanced gastric cancer. Hepatogastroenterology. 2011;58:1032–40. [PubMed] [Google Scholar]
- 16.Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143:35–47. e2. doi: 10.1053/j.gastro.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Ishiguro H, Kimura M, Takeyama H. Role of microRNAs in gastric cancer. World J Gastroenterol. 2014;20:5694–9. doi: 10.3748/wjg.v20.i19.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu S, Liu Y, Wang J, Guo Z, Zhang Q, Yu F, Zhang Y, Huang K, Li Y, Song E, Zheng XL, Xiao H. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab. 2012;97:2084–92. doi: 10.1210/jc.2011-3059. [DOI] [PubMed] [Google Scholar]
- 19.Wu Q, Wang C, Lu Z, Guo L, Ge Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin Chim Acta. 2012;413:1058–65. doi: 10.1016/j.cca.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 20.Hennessey PT, Sanford T, Choudhary A, Mydlarz WW, Brown D, Adai AT, Ochs MF, Ahrendt SA, Mambo E, Califano JA. Serum microRNA biomarkers for detection of non-small cell lung cancer. PLoS One. 2012;7:e32307. doi: 10.1371/journal.pone.0032307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a sevenmicroRNA signature. Gut. 2010;59:579–85. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Huang SK, Zhao M, Yang M, Zhong JL, Gu YY, Peng H, Che YQ, Huang CZ. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One. 2014;9:e87451. doi: 10.1371/journal.pone.0087451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, Zhang Y, Chen X, Wang J, Zen K, Zhang CY, Zhang C. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One. 2014;9:e92292. doi: 10.1371/journal.pone.0092292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, Xiao C, Hu WY, Han J, Sun P. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70:8547–57. doi: 10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]


