Abstract
Background: The aims were to compare the consistency of epidermal growth factor receptor (EGFR) mutations in the plasma and tumor tissue of NSCLC patients, and to explore the prognostic significance of plasma EGFR mutation status in tyrosine kinase inhibitors (TKIs)-treated patients with tumor EGFR mutation. Methods: We evaluated EGFR gene (exons 18, 19, 20 and 21) mutation status in paired plasma and tumor tissue from 94 NSCLC patients before EGFR-TKIs treatments using the Scorpion amplification refractory mutation system (Scorpion-ARMS) method. Results: Our results demonstrated that the rates for EGFR mutations in 94 NSCLC patients were 20% (19/94, plasma samples) and 40% (38/94, tumor tissue samples), respectively. The consistency of EGFR mutations between plasma and tissue reached 80% (75/94, P<0.001). The sensitivity of tests using plasma samples was 50% (19/38) and the specificity was 100% (49/49) compared with tissue samples. 29 of the 38 patients were treated with TKIs. Among the 29 patients, 14 patients had EGFR mutations in both plasma and tumor tissue, and these patients had a significantly shorter overall survival (OS) than those with EGFR mutations in tumor tissue only by univariate analysis (P=0.019). Conclusions: Our data demonstrated the feasibility and potential utility of plasma cell-free DNA (cfDNA) as a source of specimens for EGFR mutation detection using the Scorpion ARMS method. Moreover, plasma EGFR mutation status before TKIs therapy might be of prognostic significance for TKIs-treated NSCLC patients with tumor tissue EGFR mutation.
Keywords: EGFR, plasma, cfDNA, prognosis, NSCLC
Introduction
Non-small cell lung cancer (NSCLC) is the most common histological subtype of lung cancer, which accounts for 80%of all histological subtypes [1]. Chemotherapy has been the primary treatment for NSCLC; however, the response rate has only been 17-22% [2]. Recently, tyrosine kinase inhibitor (TKIs), which targets epidermal growth factor receptor (EGFR), has become another treatment option for patients with NSCLC. Many studies have confirmed that activating EGFR gene mutations are effective markers for EGFR-TKIs sensitivity [3,4]. In Asia, 30-50% of NSCLC patients harbored mutant EGFR genes, among which the TKIs response rate was approximately 75%. In contrast, the response rate in patients with wild-type EGFR genes was only 10% [5-7]. The research indicated that patients with TKIs-sensitive EGFR mutations had longer progression-free survival (PFS) when receiving TKIs treatment instead of standard chemotherapy. Compared with chemotherapy, TKIs treatment was associated with fewer treatment-related adverse effects [8]. Therefore, detecting EGFR mutations is extremely important for predicting the responses to TKIs treatment in the first-line therapy for NSCLC. Currently, the routine sources of specimens for detecting EGFR mutations are formalin-fixed paraffin-embedded (FFPE) tissues from biopsy or from surgical resected specimen, where regions with a minimum of 50% of tumor cell penetration can be identified and enriched to avoid a false-negative result [9]. However, histological materials from biopsies are not always available because lung cancer is principally diagnosed at advanced stages. Previous reports have demonstrated that EGFR mutations could be detected in patient plasma, and the preliminary results seem promising [10-13]. Related technologies include Scorpion ARMS, sequencing, denaturing high-performance liquid chromatography (DHPLC), mutant-enriched PCR, and microfluidic digital PCR; however, the results obtained using these technologies appeared to vary. What’ more, as far as we know, the prognostic significance of plasma EGFR mutation status before TKIs therapy in TKIs-treated NSCLC patients with tumor tissue EGFR mutation hasn’t been reported. In our present study, we chose the Scorpion ARMS method, which used a Scorpion primer/probe in a real-time PCR setting and short probes that allow greater allelic specificity and a lower background [14]. Patients primitively diagnosed with NSCLC were recruited, and the EGFR mutation status in the paired plasma and tumor tissues of these patients was tested. We explored whether plasma could be another choice when tissue is unavailable for a patient with NSCLC, and analyzed the association between plasma EGFR mutation status before TKIs treatment and the outcome in TKIs-treated NSCLC patients with tissue EGFR mutation.
Materials and methods
Patients
Patients diagnosed with NSCLC by biopsy or by surgery at Peking Union Medical College Hospital from September 2013 to July 2014 were recruited. This study was approved by the institutional review board of Peking Union Medical College Hospital. Written informed consents for participation in the study were obtained from all participants.
FFPE and plasma samples
FFPE and paired plasma samples from 94 NSCLC patients were enrolled. Of the 94 FFPE specimens, 71 (75%) were from the primary tumor (7 from bronchial biopsy, 56 from percutaneous lung puncture, and 8 from surgery), 12 (13%) originated from lymph node metastasis, and 11 (12%) originated from distant metastasis (2 bone, 5 pleural effusion, 1 iliopsoas, 1 pelvic, 1 narrative (NAR), and 1 transbronchoscopic needle aspiration (TBNA). All FFPE specimens were histologically diagnosed and pathologically evaluated to confirm the diagnosis of NSCLC according to the 2004 World Health Organization (WHO) Classification. Six to eight sections (5 µm thick each) from qualified tissue blocks were mounted on slides. Then, sections were deparaffinized using xylene and rehydrated with gradient ethanol, and one of the slides was stained with hematoxylin-eosin. An experienced pathologist estimated the amount of tumor cells. A minimum of 100 tumor cells on each slide was used as the inclusion criterion based on the limit of Scorpion ARMS technology (approximately 1% sensitivity). Samples below this threshold were rejected. Nine-milliliter blood samples were collected in collection tubes containing EDTA before the start of EGFR-TKIs treatments. Centrifugation was performed at 3000 rpm for 10 min, and then the plasma was separated and stored at -80°C.
DNA extraction from plasma
An AmoyDx®Serum/Plasma Cell-free DNA Kit (Amoy Diagnostics, Xiamen, China) was used for DNA extraction. Two milliliters of plasma was consumed for each patient, and the extraction procedures were carefully performed according to the manufacturer’s manual.
DNA extraction from FFPE tissue sections
The slides were rinsed in water, and the selected tumor cells were carefully scraped from the slides into an Eppendorf tube with 180 μl of ATL lysis buffer with a dispensable sterile scalpel. Then, 20 μl of proteinase K was added, mixed and incubated at 56°C overnight. A QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) was used for extracting DNA from tumor tissue samples according to the manufacturer’s protocols.
DNA quality assessment and EGFR mutation detection
An EGFR RGQ PCR Kit and a Rotor-Gene Q Real-time PCR Platform (Qiagen, Hilden, Germany) were used for DNA quality assessment and for EGFR mutation detections, respectively. The cycling conditions for quality control (QC) runs and for mutation assays were as follows: 15 min incubation at 95°C, followed by 40 cycles of 95°C for 30 s, and 60°C for 1 min. Fluorescence was measured at 60°C. The data were interpreted according to the manufacturer’s instructions. Additionally, mutation assays were not performed when the corresponding QC run failed.
Statistical analyses
All experimental data were analyzed using SPSS 17 statistical software (Chicago, IL). McNemar’s test was used to assess the significance of the differences between the mutations detected in tissue and plasma samples. The degree of consistency was measured using the Kappa test. EGFR mutation in tissue samples were taken as the gold standard for the sensitivity and specificity measurements. We used the chi-square test or Fisher’s exact test to assess the relation between the EGFR gene mutation status and some clinical features. Progression-free survival (PFS) was defined as survival without disease progression or death and was calculated from the start of TKIs therapy until the first observation of disease progression or the last follow-up visit. Overall survival (OS) was calculated from the start of TKIs therapy until death or the last follow-up visit. Survival curves were constructed by the Kaplan-Meier method and compared using the log-rank test. The level of significance was defined as P<0.05 (two-tailed).
Results
Patients’ characteristics
The clinicopathological characteristics of the recruited patients were showed in Table 1. Smoking history was based on records from the patients’ first clinic visit, and a smoker was defined as a person who had smoked more than 100 cigarettes in his/her lifetime. Tumor stage was determined according to Goldstraw P et al. [15].
Table 1.
Patients’ clinicopathological characteristics
| Variable | No. of patients (%) |
|---|---|
| Overall | 94 (100) |
| Age | |
| Mean ± SDa | 58±11 |
| ≤60 | 56 (60) |
| >60 | 37 (40) |
| Gender | |
| Male | 61 (65) |
| Female | 33 (35) |
| Smoking history | |
| Smoker | 48 (51) |
| Never smoker | 46 (49) |
| Tumor type | |
| ADCb | 72 (77) |
| SCCc | 19 (20) |
| LCCd | 2 (2) |
| ADSCe | 1 (1) |
| Tumor stage | |
| IIA | 1 (1) |
| IIIA | 4 (4) |
| IIIB | 9 (10) |
| IV | 80 (85) |
| Tumor sample origin | |
| Tumor | 71 (75) |
| Nodes | 12 (13) |
| Metastasis | 11 (12) |
| Tumor sample source | |
| Biopsy | 78 (83) |
| Cytologyf | 5 (5) |
| Surgery | 11 (12) |
standard deviation;
adenocarcinoma;
squamous carcinoma;
large cell carcinoma;
adenosquamous carcinoma;
tumor cells of pleural effusion.
Comparison of EGFR mutation status observed in plasma samples and tissue samples
All histological specimens included in the analyses were valid for the Scorpion ARMS method. The plasma DNA samples from 4 patients failed in their QC runs, which were viewed as negative results. Among the 94 NSCLC patients, 19/94 (20%) had EGFR mutation-positive plasma samples, among which 17 harbored TKIs-sensitive mutations (deletions in exon 19 (12%), L858R (6%), G719X (1%), and 2 harbored partial-sensitive mutations (T790M and L858R (1%)). 38/94 (40%) were EGFR mutation-positive in tumor tissue samples, which contained deletions in exon 19 (20%), L858R (15%), G719X (2%), T790M and L858R (2%), and deletions in exon19 and G719X (1%). Figure 1 showed the EGFR T790M and L858R activated gene mutation in the plasma sample and the corresponding tissue sample. The mutation rates in tumor tissue and plasma samples were significantly different, as assessed by McNemar’s test (P<0.001) (Table 2). However, the degree of consistency for EGFR mutations in tumor tissue and plasma samples was high, with a coefficient of 0.54, as determined by the Kappa test (P<0.001). The overall concordance of EGFR mutation status between plasma and tissue samples was 80% (75/94), and the consistency between plasma and biopsy specimens was 81% (63/78). Compared with the tissue samples, the sensitivity and specificity of using plasma samples for EGFR mutation detection were 50% and 100%, respectively.
Figure 1.
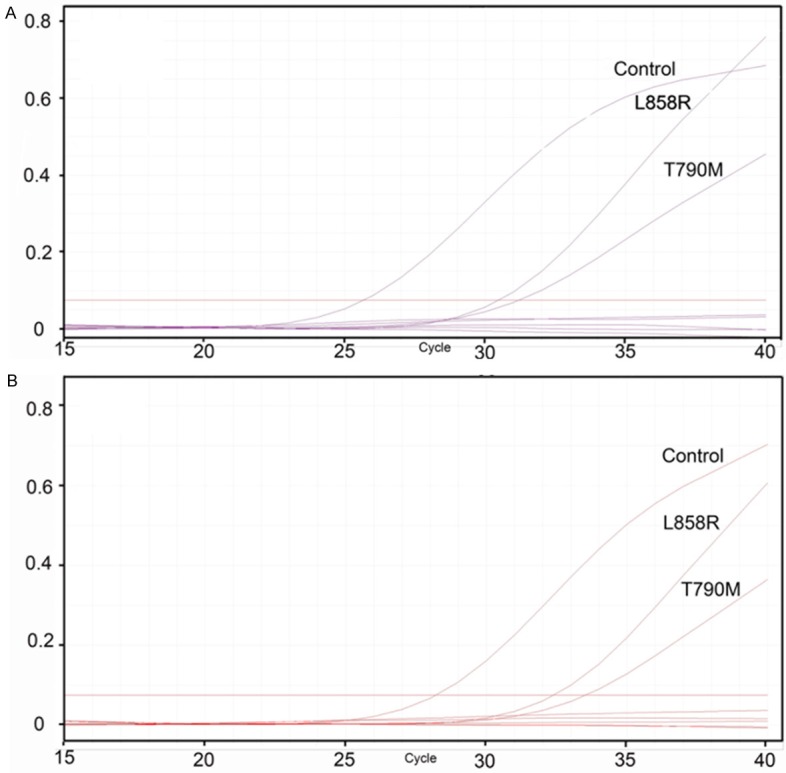
Amplification plot of EGFR T790M and L858R gene detected in paired plasma (A) and tissue sample (B). The horizontal line represents the threshold. Curves that crossing the threshold and the ΔCt calculated by specific gene control deducting the sample control is less than the confirmed scope (8.58 for L858R in exon 21 of EGFR and 6.38 for T790M) are considered positive for the specific gene. For L858R in exon 21 of EGFR, the sample’s ΔCt is 5.98 in the plasma sample and 4.82 in the corresponding tissue sample. For T790M, the sample’s ΔCt is 5.31 in the plasma sample and 5.53 in the corresponding tissue.
Table 2.
EGFR gene status detected in tumor tissue samples versus plasma samples in NSCLC patients
| Plasma | Tissue | |||
|---|---|---|---|---|
|
|
||||
| Mutation | Wild-type | Total | ||
| Mutation | 19 | 0 | 19 (20%) | |
| Wild-type | 19 | 56 | 75 (80%) | |
| Total | 38 (40%) | 56 (60%) | 94 (100%) | P<0.001 |
Correlation between EGFR mutation status and clinicopathological characteristics
The correlation between EGFR mutation status and patients’ clinicopathologic characteristics was summarized in Table 3. Tumor tissue EGFR mutations were more commonly detected in females, adenocarcinoma and never-smokers (P=0.013, P=0.001 and P<0.001, respectively), while plasma EGFR mutations were more frequent in adenocarcinoma and never-smokers (P=0.007 and P<0.001, respectively). There was a trend towards significance between plasma EGFR gene status and tumor stage in 94 NSCLC patients (P=0.093). 19 patients with EGFR plasma mutations were all found to have advanced-stage disease, whereas 61 of 75 patients without EGFR mutations in plasma samples had advanced-stage disease. Moreover, there was also a trend towards significance between plasma EGFR gene status and tumor stage in 38 NSCLC patients with tumor tissue EGFR mutation (P=0.053) (data not shown).
Table 3.
Association between EGFR mutation status and clinicopathological features
| Tumor tissue | P | Plasma | P | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Mutation | Wild-type | Mutation | Wild-type | |||
| Total | 38 | 56 | 19 | 75 | ||
| Age (years) | 0.666 | 0.603 | ||||
| ≤60 | 20 | 36 | 14 | 42 | ||
| >60 | 18 | 19 | 5 | 32 | ||
| Gender | 0.013 | 0.21 | ||||
| Male | 19 | 42 | 10 | 51 | ||
| Female | 19 | 14 | 9 | 24 | ||
| Smoking history | <0.001 | 0.001 | ||||
| Smoker | 10 | 38 | 6 | 42 | ||
| Never smoker | 28 | 18 | 13 | 33 | ||
| Tumor type | 0.001 | 0.007 | ||||
| adenocarcinoma | 36 | 36 | 19 | 53 | ||
| Non-adenocarcinoma | 2 | 20 | 0 | 22 | ||
| Tumor stage | 0.327 | 0.093 | ||||
| II-III | 4 | 10 | 0 | 14 | ||
| IV | 34 | 46 | 19 | 61 | ||
Treatment and outcome of patients with tumor tissue EGFR mutation
29 of 38 patients with in tumor tissue EGFR mutation were treated with TKIs (Table S1). 5 patients withdrawn TKIs because of financial infeasibility, 3 patients rejected any treatment and 1 was lost to follow-up. Among the 29 patients, 14 patients were found to have EGFR mutations in both tumor tissue and plasma samples.
Survival analysis in 29 TKIs-treated patients with tumor tissue EGFR mutation
The median follow-up for PFS and OS was 9.7 months (range, 0.5-14.0 months) and 10.3 months (range, 2.0-16.5 months), respectively.
We analyzed the influence of the following individual factors on survival: age, gender, smoking history, tumor type, lymph node metastasis, tumor stage, and plasma EGFR mutation status (Table 4). In univariate analysis, plasma EGFR mutation was a significant poor prognostic factor for OS (P=0.019) (Table 4; Figure 2A). It was also a potential poor prognostic factor for PFS (P=0.157) (Table 4; Figure 2B), although it did not reach statistical significance.
Table 4.
Univariate analyses for PFS and OS in 29 TKIs-treated NSCLC patients with tumor tissue EGFR mutation
| PFS | OS | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P | HR | 95% CI | P | |
| Age (years) | 0.036 | 0.117 | ||||
| ≤60 | 0.412 | 0.166-1.026 | 0.215 | 0.024-1.925 | ||
| >60 | Reference | Reference | ||||
| Gender | 0.677 | 0.704 | ||||
| Male | 1.192 | 0.492-2.887 | 1.395 | 0.233-8.366 | ||
| Female | Reference | Reference | ||||
| Smoking history | 0.477 | 0.491 | ||||
| Smoker | 1.388 | 0.528-3.650 | 1.817 | 0.303-10.882 | ||
| Never smoker | Reference | Reference | ||||
| Tumor type | 0.091 | 0.783 | ||||
| Adenocarcinoma | 0.197 | 0.023-1.685 | 21.242 | 0.000-4.270E+15 | ||
| Non-adenocarcinoma | Reference | Reference | ||||
| Lymph node metastasis | 0.191 | 0.414 | ||||
| Yes | 3.265 | 0.435-24.515 | 24.012 | 0.000-5.071E+06 | ||
| No | Reference | Reference | ||||
| Tumor stage | 0.981 | 0.505 | ||||
| II-III | 0.983 | 0.225-4.303 | 0.043 | 0.000-90661.512 | ||
| IV | Reference | Reference | ||||
| Plasma EGFR mutation | 0.157 | 0.019 | ||||
| Positive | 1.818 | 0.741-4.460 | 66.307 | 0.050-88082.845 | ||
| Negative | Reference | Reference | ||||
Figure 2.
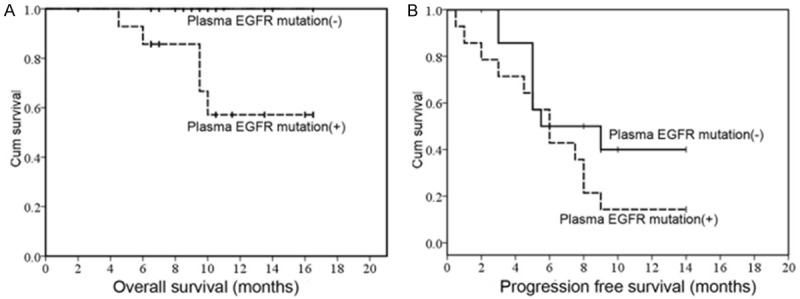
Univariate analyses of prognostic factors in 29 patients with EGFR-TKIs treatment (A). OS according to plasma EGFR mutation status. (B) PFS according to plasma EGFR mutation status.
Discussion
Our study demonstrated the possibility of using plasma samples as surrogates to tissue samples to determine the EGFR mutational status in patients with NSCLC. In 94 patient before the start of EGFR-TKIs treatments, we identified that 19 (20%) of plasma samples and 38 (40%) of tissue samples carried EGFR mutations using the Scorpion ARMS method. Generally, EGFR mutations are more common in females, adenocarcinoma and never-smokers [16], which was in concordance with the present study. The difference in our study was that the plasma EGFR mutations were not significantly related to the gender of the patients (Table 3). The lack of significance may be a result of the imbalanced ratio of genders (male: female =1.9:1).
Previous studies showed that compared with tissue, the detection of EGFR mutations in plasma/serum DNA using Scorpion ARMS method indicated a high specificity, which varied from 86% to 100% [17-19]. In our study, the specificity of tests using plasma samples was 100% (49/49) compared to the tissue samples, which implied that once the detection result was a positive one in a plasma sample, the physician could trust the result for treatment. In our study, the overall concordance and sensitivity of EGFR mutation status between plasma and tissue samples was 80% and 50%, respectively.
In previous studies, the concordances and sensitivity between the EGFR mutation statuses in the tissue and the plasma/serum samples using the Scorpion ARMS method was 66% to 93%, 43% to 86%, respectively [17-21]. The difference may be attributed to different sample types (plasma or serum), methods of cfDNA extractions, tissue detection methods and run conditions. In our research, we opted for plasma samples instead of serum for detecting EGFR mutations. First, patients with lung cancer have been confirmed to have higher DNA in plasma or serum samples than healthy volunteers, most likely originating from deciduous tumor cells or from circulating tumor cells (CTCs) [9]. Second, the serum samples contained a larger amount of genomic DNA [22], which might increase the risk of false-negative results. Liu et al. also used the Scorpion ARMS method to identify EGFR mutations in plasma samples and yielded a higher sensitivity of 68% [13]. Explanations could be that, for one thing, their study enrolled a higher percentage of adenocarcinoma patients; for another, they adopted a different DNA extraction kit (a QIAamp Circulating Nucleic Acid Kit) to ours, which might have improved the yielding of small DNA fragments [23]. Vallee A et al. increased sensitivity to 86% with the same EGFR mutation kit as ours by increasing the number of PCR cycles as well as the cut-off ΔCt value [17-21]. However, such practice exploited the performance characteristics of the kit and thus required further validation.
In our study, we showed the detection rate of plasma EGFR mutation using Scorpion ARMS method was 20% (19/94). Previous studies have demonstrated that the positive detection rate for EGFR mutations in plasma DNA using the other methods ranged from 14% to 46% [10,12,13,24-26]. Although the EGFR detection rate of the Scorpion ARMS method might be lower than other technologies, such as digital PCR, DHPLC, and peptide nucleic acid-locked nucleic acid (PNA-LNA) PCR clamp, this method allows for the identification of the 29 most common EGFR mutations clinically described thus far, out-competing the other above-mentioned methods. Additionally, of the 19 patients, 17 patients harbored TKIs-sensitive mutations and 2 harbored partial-sensitive mutations. Therefore, the Scorpion ARMS method can also be used to monitor the EGFR mutation status during the TKIs therapy. Last, those above-mentioned methods are faced with problems such as technical complexity, the cost of the tests and standardization. However, the Scorpion ARMS method utilized in this study is technically easier, less expensive and has been standardized. Most importantly, it is the only method certificated by FDA to detect EGFR mutations so far. According to the above discussion, our further study will try to employ the DNA extraction kit (a QIAamp Circulating Nucleic Acid Kit) and modified run conditions in order to explore whether the sensitivity of plasma EGFR mutation detection can be improved.
Although studies have tried their best to improve the detection sensitivity of plasma EGFR mutation, the highest sensitivity, to our knowledge, was 86% [17-21]. There might be two possible underlying explanations for the limited sensitivity of plasma EGFR mutation. One is that tumor cells carrying EGFR gene mutations have not entered the peripheral circulation [27]. The other is that the dilution of DNA derived from non-cancerous cells, such as inflamed cells, hinder the detection of mutations in plasma DNA samples [10]. Given this, tumor tissue EGFR mutation detection is necessary for plasma EGFR mutation-negative patients.
Several previous studies indicated that TKI-treated patients with plasma EGFR mutation had a significantly longer PFS than those without mutation [10,17,26]. However, these studies have not clarified the prognostic significance of plasma EGFR mutation status in TKI-treated patients with tumor tissue EGFR mutation. Our results showed, that the plasma EGFR mutations was a significant poor prognostic factor for OS in TKI-treated patients with tumor tissue EGFR mutation (P=0.019), and was a potential poor prognostic factor for PFS, though it did not reach statistical significance. Our results suggested that plasma EGFR mutation detection before TKI therapy might not only be an effective predictor of EGFR-TKI therapy, but also probably predict the prognosis of TKI-treated patients with tumor EGFR mutation. Rafael Rosell et al. [28] also showed that there was a significant association between serum EGFR L858R mutations and poor prognosis in erlotinib-treated patients with tumor tissue EGFR mutation. However, in that study, the serum EGFR mutation detection was performed during erlotinib therapy. Two possible explanations might account for our results. First, plasma EGFR mutation was prone to be correlated with advanced tumor stage and poor differentiation in NSCLC patients with tumor tissue EGFR mutation [24]. Consistent with this, our result also indicated that all the 19 cases with EGFR mutation in both plasma and tumor tissue was found to have advanced disease, and there was a trend towards significance between plasma EGFR gene status and tumor stage in NSCLC patients with tumor tissue EGFR mutation (P=0.053). Second, previous studies have demonstrated that plasma EGFR T790M mutation, which was correlated with poor prognosis, was more frequent in patients with plasma EGFR activating mutations than those without mutations [29,30]. T790M mutation might co-exist with sensitizing EGFR mutations at a low frequency in the plasma before EGFR-TKI therapy [29,31]. The T790M mutation frequency in the plasma cfDNA might fall below the limit of detection of Scorpion ARMS method in the present study. To verify the second explanation, we need to further evaluate plasma T790M mutation quantitatively in the patients with both plasma and tumor tissue EGFR mutation using a more sensitive method such as digital PCR. Our results revealed the prognostic significance of plasma EGFR mutation status before TKI treatment in patients with tumor tissue EGFR mutation, and thus clinically, it may be also important to detect the plasma EGFR mutation before TKIs treatment in patients with confirmed tumor tissue EGFR mutation. However, given the limitations of our study including small sample size and limited follow-up, the conclusions should be regarded with caution.
In conclusion, we demonstrated the feasibility and potential utility of EGFR mutations in cfDNA from plasma samples using the Scorpion ARMS method when tumor tissue is unavailable in NSCLC patients. Moreover, plasma EGFR mutation status before TKIs therapy might be of prognostic significance for TKIs-treated NSCLC patients with tumor tissue EGFR mutation.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Alberg AJ, Samet JM. Epidemiology of lung cancer. Chest. 2003;123:21s–49s. doi: 10.1378/chest.123.1_suppl.21s. [DOI] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH Eastern Cooperative Oncology Group. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Al Dayel F. EGFR mutation testing in non-small cell lung cancer (NSCLC) J Infect Public Health. 2012;5(Suppl 1):S31–34. doi: 10.1016/j.jiph.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Castro AS, Parente B, Goncalves I, Antunes A, Barroso A, Conde S, Neves S, Machado JC. Epidermal growth factor receptor mutation study for 5 years, in a population of patients with non-small cell lung cancer. Rev Port Pneumol. 2013;19:7–12. doi: 10.1016/j.rppneu.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Janne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J. Clin. Oncol. 2005;23:3227–3234. doi: 10.1200/JCO.2005.09.985. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 7.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 8.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, Porta R, Cobo M, Garrido P, Longo F, Moran T, Insa A, De Marinis F, Corre R, Bover I, Illiano A, Dansin E, de Castro J, Milella M, Reguart N, Altavilla G, Jimenez U, Provencio M, Moreno MA, Terrasa J, Munoz-Langa J, Valdivia J, Isla D, Domine M, Molinier O, Mazieres J, Baize N, Garcia-Campelo R, Robinet G, Rodriguez-Abreu D, Lopez-Vivanco G, Gebbia V, Ferrera-Delgado L, Bombaron P, Bernabe R, Bearz A, Artal A, Cortesi E, Rolfo C, Sanchez-Ronco M, Drozdowskyj A, Queralt C, de Aguirre I, Ramirez JL, Sanchez JJ, Molina MA, Taron M, Paz-Ares L, Pneumocancerologie GF, Toracica AIO. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 9.Ulivi P, Zoli W, Capelli L, Chiadini E, Calistri D, Amadori D. Target therapy in NSCLC patients: Relevant clinical agents and tumour molecular characterisation. Mol Clin Oncol. 2013;1:575–581. doi: 10.3892/mco.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, Wang X, Duan CJ, Wu NM, Guo ZQ, Liu YX, Liu HN, Wang YY, Wang J. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J. Clin. Oncol. 2009;27:2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 11.He C, Liu M, Zhou C, Zhang J, Ouyang M, Zhong N, Xu J. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer. 2009;125:2393–2399. doi: 10.1002/ijc.24653. [DOI] [PubMed] [Google Scholar]
- 12.Li XF, Ren RX, Ren SX, Chen XX, Cai WJ, Zhou F, Zhang YS, Su CX, Zhao C, Li JY, Cheng NN, Zhao MC, Zhou CC. Peripheral Blood for Epidermal Growth Factor Receptor Mutation Detection in Non-Small Cell Lung Cancer Patients. Transl Oncol. 2014;7:341–348. doi: 10.1016/j.tranon.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Lu Y, Zhu G, Lei Y, Zheng L, Qin H, Tang C, Ellison G, McCormack R, Ji Q. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol. 2013;66:1065–1069. doi: 10.1136/jclinpath-2013-201728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou SZ, Zhou M, Peng HY, Zeng AP, Yu QT, Song XQ. Comparison of ARMS and direct sequencing for detection of EGFR mutation and prediction of EGFR-TKI efficacy between surgery and biopsy tumor tissues in NSCLC patients. Med Oncol. 2014:31. doi: 10.1007/s12032-014-0926-3. [DOI] [PubMed] [Google Scholar]
- 15.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L, Canc IASL. The IASLC lung cancer staging project: Proposals for the revision of he TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 16.Chen ZY, Zhong WZ, Zhang XC, Su J, Yang XN, Chen ZH, Yang JJ, Zhou Q, Yan HH, An SJ, Chen HJ, Jiang BY, Mok TS, Wu YL. EGFR Mutation Heterogeneity and the Mixed Response to EGFR Tyrosine Kinase Inhibitors of Lung Adenocarcinomas. Oncologist. 2012;17:978–985. doi: 10.1634/theoncologist.2011-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura H, Kasahara K, Kawaishi M, Kunitoh H, Tamura T, Holloway B, Nishio K. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–3921. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- 18.Kimura H, Suminoe M, Kasahara K, Sone T, Araya T, Tamori S, Koizumi F, Nishio K, Miyamoto K, Fujimura M, Nakao S. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA) Br J Cancer. 2007;97:778–784. doi: 10.1038/sj.bjc.6603949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goto K, Ichinose Y, Ohe Y, Yamamoto N, Negoro S, Nishio K, Itoh Y, Jiang HY, Duffield E, McCormack R, Saijo N, Mok T, Fukuoka M. Epidermal Growth Factor Receptor Mutation Status in Circulating Free DNA in Serum From IPASS, a Phase III Study of Gefitinib or Carboplatin/ Paclitaxel in Non-small Cell Lung Cancer. J Thorac Oncol. 2012;7:115–121. doi: 10.1097/JTO.0b013e3182307f98. [DOI] [PubMed] [Google Scholar]
- 20.Kuang Y, Rogers A, Yeap BY, Wang L, Makrigiorgos M, Vetrand K, Thiede S, Distel RJ, Janne PA. Noninvasive Detection of EGFR T790M in Gefitinib or Erlotinib Resistant Non-Small Cell Lung Cancer. Clin Cancer Res. 2009;15:2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vallee A, Marcq M, Bizieux A, El Kouri C, Lacroix H, Bennouna J, Douillard JY, Denis MG. Plasma is a better source of tumor-derived circulating cell-free DNA than serum for the detection of EGFR alterations in lung tumor patients. Lung Cancer. 2013;82:373–374. doi: 10.1016/j.lungcan.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Board RE, Williams VS, Knight L, Shaw J, Greystoke A, Ranson M, Dive C, Blackhall FH, Hughes A. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann N Y Acad Sci. 2008;1137:98–107. doi: 10.1196/annals.1448.020. [DOI] [PubMed] [Google Scholar]
- 23.Yuan HH, Zhu ZZ, Lu YC, Liu F, Zhang WY, Huang G, Zhu GS, Jiang B. A Modified Extraction Method of Circulating Free DNA for Epidermal Growth Factor Receptor Mutation Analysis. Yonsei Med J. 2012;53:132–137. doi: 10.3349/ymj.2012.53.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Han RB, Zhao J, Wang J, Yang F, Zhong W, Zhang L, Li LY, Wang MZ. Comparison of Epidermal Growth Factor Receptor Mutation Statuses in Tissue and Plasma in Stage I-IV Non-Small Cell Lung Cancer Patients. Respiration. 2013;85:119–125. doi: 10.1159/000338790. [DOI] [PubMed] [Google Scholar]
- 25.Yung TKF, Chan KCA, Mok TSK, Tong J, To KF, Lo YMD. Single-Molecule Detection of Epidermal Growth Factor Receptor Mutations in Plasma by Microfluidics Digital PCR in Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 26.Chen YM, Fan WC, Tseng PC, Tsai CM, Chou TY, Wu CH, Chou KT, Lee YC, Perng RP, Whang-Peng J. Plasma epidermal growth factor receptor mutation analysis and possible clinical applications in pulmonary adenocarcinoma patients treated with erlotinib. Oncol Lett. 2012;3:713–717. doi: 10.3892/ol.2011.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack PC, Holland WS, Burich RA, Sangha R, Sobs LJ, Li YJ, Beckett LA, Lara PN, Davies AM, Gandara DR. EGFR Mutations Detected in Plasma Are Associated with Patient Outcomes in Erlotinib Plus Docetaxel-Treated Non-small Cell Lung Cancer. J Thorac Oncol. 2009;4:1466–1472. doi: 10.1097/JTO.0b013e3181bbf239. [DOI] [PubMed] [Google Scholar]
- 28.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, Paz-Ares L, Bover I, Garcia-Campelo R, Moreno MA, Catot S, Rolfo C, Reguart N, Palmero R, Sanchez JM, Bastus R, Mayo C, Bertran-Alamillo J, Molina MA, Sanchez JJ, Taron M, Grp SLC. Screening for Epidermal Growth Factor Receptor Mutations in Lung Cancer. N Engl J Med. 2009;361:958–U938. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 29.He C, Zheng L, Xu Y, Liu M, Li Y, Xu J. Highly sensitive and noninvasive detection of epidermal growth factor receptor T790M mutation in non-small cell lung cancer. Clin Chim Acta. 2013;425:119–124. doi: 10.1016/j.cca.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Chen R, Wang S, Zhong J, Wu M, Zhao J, Duan J, Zhuo M, An T, Wang Y, Bai H, Wang J. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One. 2014;9:e110780. doi: 10.1371/journal.pone.0110780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Janne PA, Joshi VA, McCollum D, Evans TL, Muzikansky A, Kuhlmann GL, Han M, Goldberg JS, Settleman J, Iafrate AJ, Engelman JA, Haber DA, Johnson BE, Lynch TJ. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J. Clin. Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


