Abstract
Background: Many studies have suggested a relationship between human papillomavirus (HPV) infection and the risk of esophageal squamous cell carcinoma (ESCC). However, findings are inconclusive, potentially because of geographic heterogeneity and variations in detection methods. Objectives: We sought to further investigate the prevalence of HPV with a new detection method, the MassARRAY Sequenom technique, in esophageal squamous cell carcinomas occurring in patients belonging to Kazakh populations in Xinjiang, China. Study design: In the present study, a novel genotyping method for detecting 30 HPV genotypes, specifically by genotyping both the HPV E6 and L1 genes with multiplex PCR using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (PCR-MS) was first adopted to evaluate HPV genotypes in 89 esophageal cancer samples and 49 matched adjacent normal esophageal tissues. Results: Six HPV genotypes (HPV6, HPV16, HPV33, HPV39, HPV51, and HPV82) were present in at least 51.7% of the esophageal carcinoma tissues, which was significantly greater than 28.6% prevalence among controls (P < 0.05). HPV16 was the most common of all the genotypes investigated (HPV16 prevalence in carcinoma tissue: 49.4%; odds ratio 3.02, 95% confidence interval 1.39-6.53). HPV-positive ESCC patients were generally younger than HPV-negative patients (P = 0.04). In addition, HPV infection was more common in cases of well-differentiated and shallower invasive depth. Conclusions: Based on this new detection method, our findings reiterate the possibility that HPV infection (especially HPV16) may be involved in the etiology of esophageal carcinoma in the Kazakh populations and that HPV E6 gene positivity may be associated with prognosis of patients.
Keywords: Human papillomavirus, HPV genotyping, MassARRAY Sequenom technique, esophageal squamous cell carcinoma
Introduction
Esophageal carcinoma (EC) is one of the top eight most common malignant tumors worldwide [1], with an estimated 500,000 new cases diagnosed and 400,000 deaths annually [2]. Kazakh population is a nomadic tribe and mainly residing in Xinjiang, the northwest of China. A hospital-based retrospective study during a 10-year period showed that the Kazakh population has a high incidence of esophageal squamous cell carcinoma (ESCC) [1,2]. The mortality rate in this population reaches 155.9/100,000, which is considerably higher than the average Chinese rate of 15.2/100,000 [3]. Epidemiological studies revealed that excessive alcohol consumption, tobacco use, micronutrient deficiency, and dietary exposure to potential carcinogens all increase the risk of esophageal carcinoma [4,5]. However, the exact etiology and mechanisms remain unclear.
Human papillomaviruses (HPV) are small, non-enveloped double-stranded DNA viruses, comprising early (E) and late (L) genes, and an untranslated long control region. Studies have reported relationships between HPV infection and non-genital cancers, including head and neck malignancies, and ESCC [6-8]. However, subsequent studies have found highly varying rates of HPV infection in ESCC, ranging from 0% to 100% [3,9-14]. These inconsistent findings may derive from different geographic regions [15,16], different sensitivities and specificities of the detection methods [17-19]. Viral load is generally lower in ESCC than in cervical cancer specimen [20]. Moreover, viral load in ESCC formalin-fixed paraffin-embedded (FFPE) tissues is lower than that in fresh specimen, as HPV genes may degrade in FFPE tissues [21-24]. Therefore, there is an outstanding need for more efficient methods of detecting HPV in ESCC FFPE tissues, which could clarify the relationship between HPV and ESCC.
Currently, the accurate detection of HPV mostly relies on the detection of viral nucleic acids. Of these detection systems, PCR-based amplification techniques were most widely used. Generally, PCR amplification is performed using primers that mostly target the highly conserved L1 region. Then, the amplification products are analyzed by either real-time PCR, reverse hybridization analysis, sequencing, or other methods. Despite their widespread use, these PCR-based methods have some common limitations, such as that PCR sequencing lacks of the ability to genotype multiple HPV genotypes in a single sample, whereas the hybridization method has poor sensitivity although it could detect multiple-type infections. Moreover, the L1 region may be lost during integration of the viral genome in cancers [21,25]. In such cases, HPV may not be detected by genotyping HPV L1.
Recent studies have noted that the PCR-based mass spectrometry system (PCR-MS) showed high specificities, high sensitivities, and high throughput for detecting HPV in cervical cancer tissues [26-28]. In the present study, we adopted the PCR-MS method targeting both E6 and L1 genes, to detect HPV in ESCC FFPE specimens from patients belonging to Kazakh populations. We sought to further investigate the prevalence of HPV in ESCC occurring in this population.
Materials and methods
Subjects
Eighty-nine Kazakh patients with primary ESCC (51 males and 38 females) collected from the First Affiliated Hospital of Shihezi University School of Medicine and Xinjiang Yili Prefecture Friendship Hospital between 2000-2012 were enrolled. Of the 89 ESCC specimens, 49 matched adjacent normal esophageal tissues were available as controls. Another twenty cervical squamous cell carcinoma (CSCC) specimens from Xinjiang Uighur patients which were HPV positive (detected by HPV PCR-reverse dot blot) were served as positive controls. All samples were fixed in 10% buffered formalin after surgical resection, routinely processed, and embedded in paraffin. Research protocol utilized in this study was approved by the Medical Ethics and Human Clinical Trial Committee of Shihezi University School of Medicine, and all recruited subjects were enrolled with written informed consent.
DNA preparation and quality control
The FFPE samples were cut into 5-μm slices. 10-15 sections per sample were collected for genomic DNA extraction using QIAamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer’s instructions. Methods to minimize the possibility of cross-contamination were used as previously described [3,29]. Briefly, DNA extraction, PCR amplification, and products detection were carried out in separated spaces. During FFPE tissues sectioning, the blade was disinfected with 75% medicinal alcohol before cutting of each sample, and paraffin-only samples were cut to serve as no contamination control for every 5 samples. All DNA samples were quantified and diluted to approximately 50 ng/μl. The quality of the prepared DNA was validated by amplifying of human β-globin (HBB) as internal control. DNA of sufficient quality was chosen for further study.
HPV PCR-reverse dot blot (PCR-RDB)
We used a commercial HPV genotyping DNA chip (Decipher Bioscience Shenzhen Ltd.) which is able to identify 23 HPV subtypes, including 18 high-risk types and 5 low-risk types in one reaction according to the manufacturer’s instructions and previous study [30]. Briefly, PCR amplification was designed for HPV L region (primer set MY09/11). The type-specific biotinylated probes were used to test the subtype-specific sequences. Amplicons hybridized to probes were detected using streptavidin horseradish peroxidase-mediated color precipitation. If a blue spot appeared at a certain position on the membrane, its presence suggested that the corresponding HPV type was contained in the specimen.
HPV detection and typing with MassARRAY MALDI-TOF MS
PCR-MS was performed by using the Sequenom MassARRAY matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) system (Sequenom Inc.) according to the manufacturer’s instructions and a previously described protocol [28]. Briefly, this method consists of a three-step process: a multiplex primary PCR, primer extension, and MALDI-TOF MS separation of products on a matrix-loaded silicon chip array. Primary PCR amplification which is a high-throughput 61-plex analysis of 30 distinct HPV genotypes for two HPV genes, E6 and L1, coupled with HBB as a DNA extraction quality control, was performed in 384-well microplates using MasScan_HPV Primer Mix (Sequenom Inc.). Primary PCR mixes were dephosphorylated with shrimp alkaline phosphatase, and then the single-base extension reaction was performed with the iPLEX Pro Reagent kit. After desalting by the addition of Clean Resin, each iPLEX product was transferred onto SpectroChip. Then data were collected by the use of SpectroAcquire and analyzed with Mass-ARRAY Typer 4.1.0 software.
Positive controls (DNA extracted from HPV16+ SiHa cells and HPV18+ HeLa cells), negative controls (DNA extracted from HPV- HEK293 cells and paraffin-only samples collected during sectioning) and blank control (water) were used in this study. To confirm the repeatability of this new assay, 20 out of these 89 ESCC samples were randomly selected and tested again.
Statistical analysis
Pearson’s chi-square test was used to compare HPV infection in cases and controls, and groups of ESCC samples defined by clinicopathological parameters. The statistical analyses were performed using SPSS 17.0. All reported P-values are 2-sided. Values of P < 0.05 were considered as statistically significant.
Results
Consistency between PCR-MS and PCR-RDB of detecting HPV in CSCC tissues
Twenty cervical cancer samples which were HPV16-positvie detected by PCR-RDB were served as positive controls. PCR-MS showed good consistency with PCR-RDB as consistent genotypes were identified in 19 of the 20 (95.0%) cervical cancer samples (including 18 HPV16 single infection and 1 HPV16 and HPV58 double infections). The remaining case was identified as HPV16 and HPV35 co-infection by PCR-RDB but was detected as HPV35 single infection by PCR-MS (Table 1; Figure 1). In all CSCC samples, both E and L probes could be used in the detection of HPV16.
Table 1.
A high consistency in HPV genotypes detected by PCR-RDB and PCR-MS in cervical cancer
| Sample ID | HPV type determined by | |
|---|---|---|
|
| ||
| PCR-RDB | PCR-MS | |
| 1 | 16 | 16ELa |
| 2 | 16 | 16EL |
| 3 | 16 | 16EL |
| 4 | 16 | 16EL |
| 5 | 16/35 | 35EL |
| 6 | 16 | 16EL |
| 7 | 16 | 16EL |
| 8 | 16/58 | 16EL/58L |
| 9 | 16 | 16EL |
| 10 | 16 | 16EL |
| 11 | 16 | 16EL |
| 12 | 16 | 16EL |
| 13 | 16 | 16EL |
| 14 | 16 | 16EL |
| 15 | 16 | 16EL |
| 16 | 16 | 16EL |
| 17 | 16 | 16EL |
| 18 | 16 | 16EL |
| 19 | 16 | 16EL |
| 20 | 16 | 16EL |
E: HPV E6 probe positive; L: HPV L1 probe positive.
Figure 1.
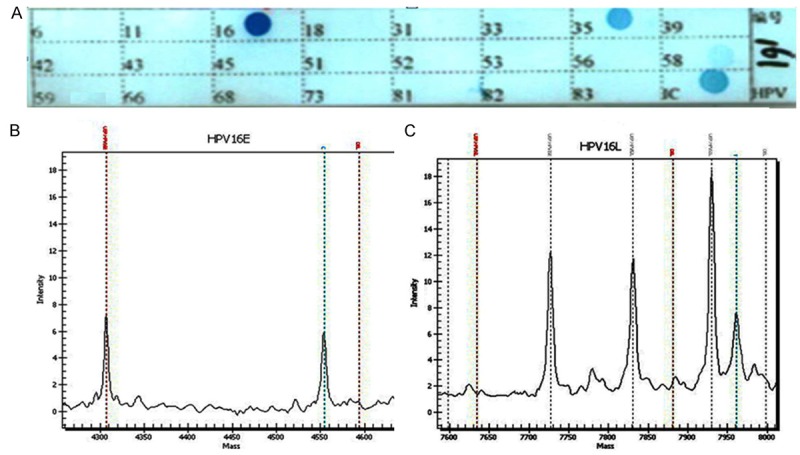
HPV genotyping using PCR-RDB and PCR-MS in cervical carcinoma. A. Representative examples of PCR-RDB (23 HPV types). Once the HPV gene chip procedure was completed, the different HPV types were indicated by blue spots on the HPV gene chip. IC: Internal control. B, C. Representative examples of PCR-MS: HPV16 type positive by genotyping both the E and L genes.
HPV genotyping using PCR-MS by targeting E6 and L1 regions in ESCC samples
89 ESCC and 49 control samples from Kazakh patients were analyzed using the multiplex PCR-MS by genotyping E6 regions (E probe) and L1 regions (L) (Figure 2). Case and control groups had similar distribution of age and sex (P > 0.05 for both) (Table 2). Results indicated that 46 of the 89 (51.7%) ESCC cases harbored HPV infection, including that 36 cases were E probe positive, 20 cases were L probe positive, and 10 cases were E and L double positive. 14 of the 49 (28.6%) control samples also harbored HPV infection, including that 10 controls were E probe positive, 11 controls were L probe positive, and 7 controls were double positive. Among the 89 ESCC samples, 44 cases were single-type infection and 2 cases were double infections. Whereas in the 49 control samples, 12 cases were single-type infection and 2 cases were double infections. The overall detection rate of HPV infection was significantly greater in the ESCC samples than that in the control samples (51.7% vs. 28.6%, P = 0.004, odds ratio [OR] 2.67, 95% confidence interval [CI] 1.27-5.64) (Table 3).
Figure 2.
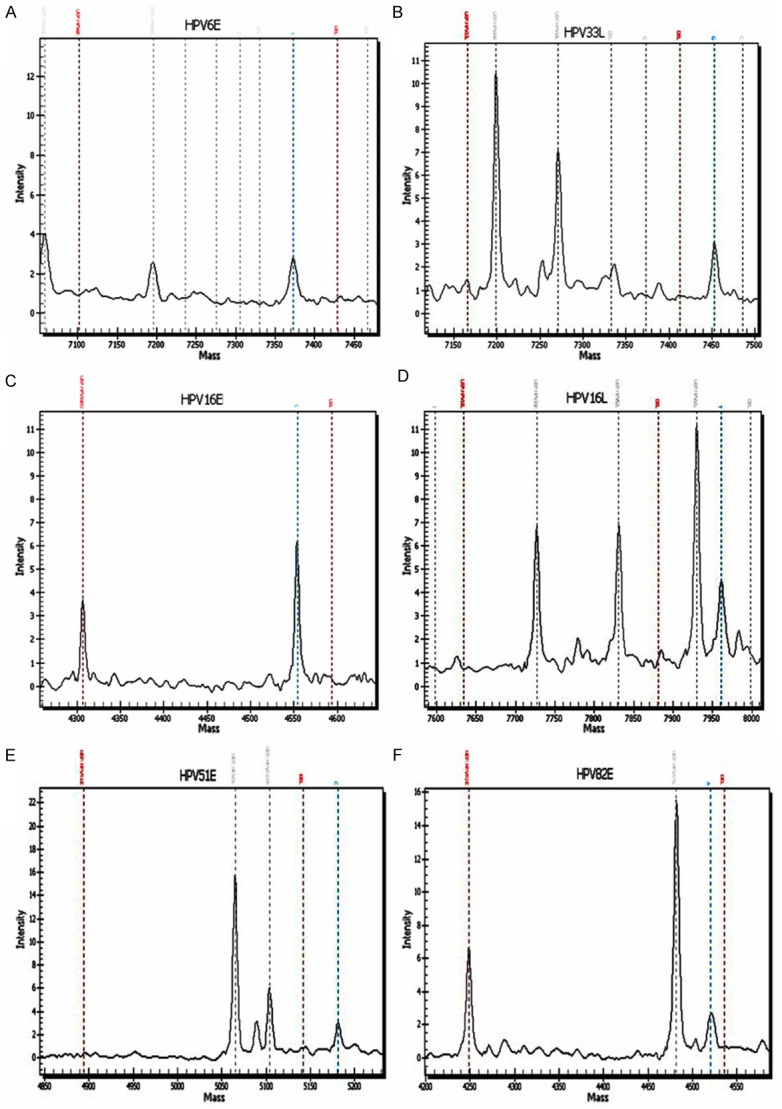
HPV genotyping using PCR-MS in esophageal carcinoma. Representative examples of PCR-MS: HPV6 positive by genotyping the E gene (A), HPV33 positive by genotyping the L gene (B). HPV16 positive by genotyping the E gene (C). HPV16 positive by genotyping the L gene (D). HPV51 and HPV82 positive by genotyping the E gene (E and F).
Table 2.
Distribution of age and sex of Kazakh subjects
| Characteristics | Cases (n = 89) N (%) | Control (n = 49) N (%) | P |
|---|---|---|---|
| Age (years) | |||
| ≤ 60 | 39 (43.8) | 25 (51.0) | 0.42 |
| > 60 | 50 (56.2) | 24 (49.0) | |
| Gender | |||
| Female | 38 (42.7) | 28 (57.1) | 0.104 |
| Male | 51 (57.3) | 21 (42.9) |
Table 3.
HPV infection rates in Kazakh esophageal carcinoma patients and control samples analyzed using unconditional logistic regression
| Types | Cases (n = 89) N (%)a | Control (n = 49) N (%)a | χ2 | OR (95% CI) |
|---|---|---|---|---|
| Total | 46 (51.7) | 14 (28.6) | 6.87* | 2.67 (1.27-5.64) |
| High-risk | 46 | 13 | 8.17* | 2.96 (1.39-6.32) |
| HPV16 | 44 (49.4) | 12 (24.5) | 8.16* | 3.02 (1.39-6.53) |
| HPV33 | N | 1 (2.0) | ||
| HPV39 | 1 (1.1) | N | ||
| HPV51 | 1 (1.1) | 1 (2.0) | 0.19 | 0.55 (0.03-8.92) |
| HPV82 | 2 (2.2) | 1 (2.0) | 0.01 | 1.10 (0.10-12.49) |
| Low-risk | 0 | 1 (2.0) | ||
| HPV6 | N | 1 (2.0) |
P < 0.05.
The total number of HPV positives was smaller than the total number of HPV gene-type positives due to the multiple infections.
1 co-infection with HPV16 and HPV82, and 1 co-infection with HPV16 and HPV39 were confirmed among all the cases. 1 co-infection with HPV16 and HPV51 and 1 co-infection with HPV16 and HPV33 were confirmed among all the controls. N: negative.
A total of six HPV genotypes was detected in these samples, including 5 high-risk types (HPV16, 33, 39, 51, 82) and 1 low-risk type (HPV6) (Table 3). In both the ESCC and control samples, high-risk HPV16 was the predominant genotype (44 of 89 ESCC (49.4%), and 12 of 49 control samples (24.5%), respectively), followed by HPV82, which was identified in two ESCC cases and one control samples (2.2% of all positive samples and 2% of control samples). Besides that, high-risk HPV39 and 51 were detected in one ESCC case respectively, whereas high-risk HPV33, 51 and low-risk HPV6 were detected in one control sample respectively. The detection of high-risk HPV DNA (OR 2.96, 95% CI 1.39-6.32) and the detection of HPV16 specifically (OR 3.02, 95% CI 1.39-6.53) both exhibited strong positive associations with the risk of esophageal carcinoma in the Kazakh cohort.
Association between HPV infection and the clinical characteristics of ESCC patients
To evaluate whether HPV infection could influence esophageal malignant progression, we analyzed associations between HPV infection and the clinical characteristics of patients. Specifically, HPV infection was more prevalent in younger patients (≤ 60, 64.1% vs. 42.0%, P = 0.04). HPV infection rates were correlated to the degree of differentiation as the rates were higher in well-differentiated (67.5%) ESCC than they were in moderately differentiated (55.2%) or poorly differentiated (33.3%) ESCC, although statistical significance was not achieved (P = 0.20), which might due to the small sample size (Table 4). Moreover, HPV infection was more common in cases with invasion to the muscular layer than in cases with invasion to the fibrous layer especially identified by genotyping HPV E6 single (54.8% vs. 32.8%, P = 0.04). However, no significant correlation was found between HPV infection status and other clinical parameters (P > 0.05).
Table 4.
Association between HPV infection and the clinical characteristics of ESCC
| Factor | N (%) | HPV n (%) | P | HPV E6 n (%) | P | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| +a | - | +b | - | ||||
| Total | 89 (100) | 46 (51.7) | 43 (48.3) | 36 (40.4) | 53 (59.6) | ||
| Age (years) | |||||||
| ≤ 60 | 39 (43.8) | 25 (64.1) | 14 (35.9) | 0.04* | 21 (53.8) | 18 (46.2) | 0.02* |
| >60 | 50 (56.2) | 21 (42.0) | 29 (58.0) | 15 (30.0) | 35 (70.0) | ||
| Gender | |||||||
| Female | 38 (42.7) | 17 (44.7) | 21 (55.3) | 0.26 | 12 (31.6) | 26 (68.4) | 0.14 |
| Male | 51 (57.3) | 29 (56.9) | 22 (43.1) | 24 (47.1) | 27 (52.9) | ||
| Anatomic location | |||||||
| Upper + Middle | 51 (57.3) | 25 (49.0) | 26 (51.0) | 0.56 | 20 (39.2) | 31 (60.8) | 0.78 |
| Lower | 38 (42.7) | 21 (55.3) | 17 (44.7) | 16 (42.1) | 22 (57.9) | ||
| Differentiation | |||||||
| Well-differentiated | 13 (14.6) | 8 (61.5) | 5 (38.5) | 0.20 | 8 (61.5) | 5 (38.5) | 0.09 |
| Mild-differentiated | 58 (65.2) | 32 (55.2) | 26 (44.8) | 24 (41.4) | 34 (58.6) | ||
| Poorly-differentiated | 18 (20.2) | 6 (33.3) | 12 (66.7) | 4 (22.2) | 14 (77.8) | ||
| Tumor invasive depth | |||||||
| Muscularis layer | 31 (34.8) | 20 (64.5) | 11 (35.5) | 0.08 | 17 (54.8) | 14 (45.2) | 0.04* |
| Fibrous layer | 58 (65.2) | 26 (44.8) | 32 (55.2) | 19 (32.8) | 39 (67.2) | ||
| Lymph node metastasis | |||||||
| No | 46 (51.7) | 26 (56.5) | 20 (43.5) | 0.35 | 21 (45.7) | 25 (54.3) | 0.30 |
| Yes | 43 (48.3) | 20 (46.5) | 23 (53.5) | 15 (34.9) | 28 (65.1) | ||
HPV E probe or L probe positive.
HPV E probe single positive.
P<0.05.
Discussion
Appropriate HPV screening and treatment are important means of preventing HPV-related cancers, especially for medically underserved populations, such as Kazakh populations living in Xinjiang. In recent years, many reports have been published on the topic of HPV infection in esophageal carcinoma [31]. However, these reports have been limited by the shortcomings of existing methods [32]. Peng et al [28] provided the first description of a novel method (PCR-MS) for HPV detection, in which short regions (60-120 bp) of dual genes (E6 and L1) were targeted to distinguish each HPV.
In our study, the new high-sensitive assay was first adopted for HPV detection in ESCC samples. Using the PCR-MS method, we found a 51.7% HPV infection rate in cases of ESCC from the Xinjiang Kazakh population. This rate is higher than other studies that were also conducted in the Xinjiang region. A rate of 31.7% was identified in our previous study, in which a HPV gene chip was used [30], and a rate of 30% was noted in a study that used the INNO-LiPA HPV Genotyping v2 test [33]. These discrepancies can be explained by the different types of PCR that were used in each study, the previous studies used consensus PCR targeting the L1 region with either MY09/11 primer set (in the gene chip assay) or SPF10 primers (in the INNO-LiPA assay). In contrast, both the E6 and L1 genes were genotyped in the present study. Indeed, the dual-gene detection of PCR-MS could improve the detection rate of HPV in ESCC FFPE tissues. When targeting the HPV L1 gene alone, HPV infection was only detected in 22.5% of the ESCC samples, whereas when HPV E6 gene was also targeted, HPV infection was detected in 51.7% of these samples, constituting a significantly greater positive rate. This difference suggests that the HPV L1 gene may be lost during the process of integration in esophageal cancer as similar phenomena have been reported in cervical cancers [21,25]. Moreover, it is potentially explained by the lower HPV viral load in ESCC tissues, as compared with CSCC tissues [11,22,34,35]. Extremely low levels of HPV DNA load in ESCC tissues (even 0.01 copies per cell) may not meet the minimum threshold of detection for current methods.
The rate of HPV detection has also been re-ported to differ by geographic region [36]. Particularly, HPV has been detected more frequently in regions with high incidences of esophageal cancer, particularly in China and South Africa [14-16]. Rates of HPV detection were extremely low (or even zero) in studies from other geographic regions, including Poland [37], Southern Iran [38], and the United States [39]. It was suspected that cross-contamination of specimens could lead to false-positive results [40,41]. However, in the present assay, methods of contamination control were adopted during FFPE tissues sectioning. Moreover, most of the L1 probes adopted in the PCR-MS assay were excluded from the MY region. Thus, this method could eliminate false-positive results caused by widespread contamination of generally used MY-PCR amplicon. In consideration of the differences of reported HPV detection rates, our findings support the hypothesis that ESCC might have different etiologies in low- and high-incidence geographic regions, specifically because HPV only plays an important role in the latter [36].
High detection rates of HPV16 not only exist in Kazakh esophageal cancer but also in normal esophagus. These results suggest that HPV16 infection may be a common phenomenon in Kazakh, where certain living habits (e.g. peppery food, hot salted milk tea, crusty pancakes, and unsanitary drinking water) may lead to damage to the esophagus and infection with high-risk HPV genotypes. Similar observation was also revealed in previous studies [3,22,29-31]. Studies linking HPV with oncogenesis in Kazakh cohort have shown that HPV infection was associated with p53 codon 72 polymorphism [42,43], HLA-DRB1*1501 and HLA-DQB1*0301 alleles [3], overexpression of HLA-G [29], heterozygote of PLCE1 rs2274223 [30], thereby contributing to ESCC oncogenesis. We found a higher detection rate in younger patients and well-differentiated ESCC, suggesting that HPV may be an essential component of the early stages of ESCC carcinogenesis in the Kazakh population. When genotyping the HPV E6 gene, HPV was more commonly detected in cases with invasion to the muscularis layer than in cases with invasion to the fibrous layer. These connections might partially contribute to the better prognosis among patients with HPV-positive ESCC in northern China [44]. However, additional studies are necessary to provide direct connections between HPV infection and ESCC carcinogenesis.
In conclusion, our study shows that the PCR-MS method is a very sensitive assay for the detection of HPV infection in FFPE tissue samples of ESCC. Using this dual-gene detection method, we found that a high rate of HPV infection was correlated with the risk of ESCC in Kazakh cohort. However, there are several limitations of this study such as the relatively small number of samples and the loss of other epidemiology information which could help us to assess the interaction between high-risk HPV infection and other environment factors in ESCC carcinogenesis.
Acknowledgements
This work was supported by the grants from the Ministry of Science and Technology of China (2010DFB34100 and 2012AA02A503) and the National Natural Science Foundation of China (No.81460416, 81160301, 81360358, 81260301).
Disclosure of conflict of interest
None.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J. Clin. Oncol. 2006;24:2137–50. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Hu J, Li L, Pang L, Chen Y, Yang L, Liu C, Zhao J, Chang B, Qi Y, Liang W, Li F. HLA-DRB1*1501 and HLA-DQB1*0301 alleles are positively associated with HPV16 infection-related Kazakh esophageal squamous cell carcinoma in Xinjiang China. Cancer Immunol Immunother. 2012;61:2135–41. doi: 10.1007/s00262-012-1281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou XN, Taylor PR, Mark SD, Chao A, Wang W, Dawsey SM, Wu YP, Qiao YL, Zheng SF. Seasonal variation of food consumption and selected nutrient intake in Linxian, a high risk area for esophageal cancer in China. Int J Vitam Nutr Res. 2002;72:375–82. doi: 10.1024/0300-9831.72.6.375. [DOI] [PubMed] [Google Scholar]
- 5.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Zandberg DP, Bhargava R, Badin S, Cullen KJ. The role of human papillomavirus in nongenital cancers. CA Cancer J Clin. 2013;63:57–81. doi: 10.3322/caac.21167. [DOI] [PubMed] [Google Scholar]
- 7.Suzuk L, Noffsinger AE, Hui YZ, Fenoglio-Preiser CM. Detection of human papillomavirus in esophageal squamous cell carcinoma. Cancer. 1996;78:704–10. doi: 10.1002/(SICI)1097-0142(19960815)78:4<704::AID-CNCR2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 8.Liyanage SS, Rahman B, Gao Z, Zheng Y, Ridda I, Moa A, Newall AT, Seale H, Li Q, Liu JF, Macintyre CR. Evidence for the aetiology of human papillomavirus in oesophageal squamous cell carcinoma in the Chinese population: a meta-analysis. BMJ Open. 2013;3:e003604. doi: 10.1136/bmjopen-2013-003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peixoto Guimaraes D, Hsin Lu S, Snijders P, Wilmotte R, Herrero R, Lenoir G, Montesano R, Meijer CJ, Walboomers J, Hainaut P. Absence of association between HPV DNA, TP53 codon 72 polymorphism, and risk of oesophageal cancer in a high-risk area of China. Cancer Lett. 2001;162:231–5. doi: 10.1016/s0304-3835(00)00643-1. [DOI] [PubMed] [Google Scholar]
- 10.Chang F, Syrjanen S, Shen Q, Cintorino M, Santopietro R, Tosi P, Syrjänen K. Evaluation of HPV, CMV, HSV and EBV in esophageal squamous cell carcinomas from a high-incidence area of China. Anticancer Res. 2000;20:3935–40. [PubMed] [Google Scholar]
- 11.Shuyama K, Castillo A, Aguayo F, Sun Q, Khan N, Koriyama C, Akiba S. Human papillomavirus in high- and low-risk areas of oesophageal squamous cell carcinoma in China. Br J Cancer. 2007;96:1554–9. doi: 10.1038/sj.bjc.6603765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonsson A, Nancarrow DJ, Brown IS, Green AC, Drew PA, Watson DI, Hayward NK, Whiteman DC Australian Cancer Study. High-risk human papillomavirus in esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2010;19:2080–7. doi: 10.1158/1055-9965.EPI-10-0033. [DOI] [PubMed] [Google Scholar]
- 13.Guo F, Liu Y, Wang X, He Z, Weiss NS, Madeleine MM, Liu F, Tian X, Song Y, Pan Y, Ning T, Yang H, Shi X, Lu C, Cai H, Ke Y. Human papillomavirus infection and esophageal squamous cell carcinoma: a case-control study. Cancer Epidemiol Biomarkers Prev. 2012;21:780–5. doi: 10.1158/1055-9965.EPI-11-1206. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Li J, Diao M, Cai Z, Yang J, Zeng Y. Statistical analysis of human papillomavirus in a subset of upper aerodigestive tract tumors. J Med Virol. 2013;85:1775–85. doi: 10.1002/jmv.23662. [DOI] [PubMed] [Google Scholar]
- 15.Schafer G, Kabanda S, van Rooyen B, Marusic MB, Banks L, Parker MI. The role of inflammation in HPV infection of the Oesophagus. BMC Cancer. 2013;13:185. doi: 10.1186/1471-2407-13-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardefeldt HA, Cox MR, Eslick GD. Association between human papillomavirus (HPV) and oesophageal squamous cell carcinoma: a meta-analysis. Epidemiol Infect. 2014;142:1119–37. doi: 10.1017/S0950268814000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liyanage SS, Segelov E, Garland SM, Tabrizi SN, Seale H, Crowe PJ, Dwyer DE, Barbour A, Newall AT, Malik A, Macintyre CR. Role of human papillomaviruses in esophageal squamous cell carcinoma. Asia Pac J Clin Oncol. 2013;9:12–28. doi: 10.1111/j.1743-7563.2012.01555.x. [DOI] [PubMed] [Google Scholar]
- 18.Poljak M, Cerar A, Seme K. Human papillomavirus infection in esophageal carcinomas: a study of 121 lesions using multiple broad-spectrum polymerase chain reactions and literature review. Human Pathol. 1998;29:266–71. doi: 10.1016/s0046-8177(98)90046-6. [DOI] [PubMed] [Google Scholar]
- 19.Syrjanen KJ. HPV infections and oesophageal cancer. J Clin Pathol. 2002;55:721–8. doi: 10.1136/jcp.55.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Si HX, Tsao SW, Poon CS, Wang LD, Wong YC, Cheung AL. Viral load of HPV in esophageal squamous cell carcinoma. Int J Cancer. 2003;103:496–500. doi: 10.1002/ijc.10865. [DOI] [PubMed] [Google Scholar]
- 21.Corden SA, Sant-Cassia LJ, Easton AJ, Morris AG. The integration of HPV-18 DNA in cervical carcinoma. Mol Pathol. 1999;52:275–82. doi: 10.1136/mp.52.5.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D, Zhang Q, Zhou L, Huo L, Zhang Y, Shen Z, Zhu Y. Comparison of prevalence, viral load, physical status and expression of human papillomavirus-16, -18 and -58 in esophageal and cervical cancer: a case-control study. BMC Cancer. 2010;10:650. doi: 10.1186/1471-2407-10-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Si HX, Tsao SW, Poon CS, Wong YC, Cheung AL. Physical status of HPV-16 in esophageal squamous cell carcinoma. J Clin Virol. 2005;32:19–23. doi: 10.1016/j.jcv.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Zehbe I, Rylander E, Edlund K, Wadell G, Wilander E. Detection of human papillomavirus in cervical intra-epithelial neoplasia, using in situ hybridization and various polymerase chain reaction techniques. Virchows Arch. 1996;428:151–7. doi: 10.1007/BF00200657. [DOI] [PubMed] [Google Scholar]
- 25.Noffsinger AE, Suzuk L, Hui YZ, Gal AA, Fenoglio-Preiser CM. Differential sensitivities of E6 type-specific and L1 consensus primers in the detection of human papillomavirus in anal carcinoma. Mod Pathol. 1995;8:509–14. [PubMed] [Google Scholar]
- 26.Soderlund-Strand A, Dillner J, Carlson J. High-throughput genotyping of oncogenic human papilloma viruses with MALDI-TOF mass spectrometry. Clin Chem. 2008;54:86–92. doi: 10.1373/clinchem.2007.092627. [DOI] [PubMed] [Google Scholar]
- 27.Yi X, Li J, Yu S, Zhang A, Xu J, Yi J, Zou J, Nie X, Huang J, Wang J. A new PCR-based mass spectrometry system for high-risk HPV, part I: methods. Am J Clin Pathol. 2011;136:913–9. doi: 10.1309/AJCPWTZDT0Q7DOVI. [DOI] [PubMed] [Google Scholar]
- 28.Peng J, Gao L, Guo J, Wang T, Wang L, Yao Q, Zhu H, Jin Q. Type-specific detection of 30 oncogenic human papillomaviruses by genotyping both E6 and L1 genes. J Clin Microbiol. 2013;51:402–8. doi: 10.1128/JCM.01170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J, Li L, Liu Y, Chen Y, Liu C, Liang W, Zhao J, Zou H, Cui X, Qi Y Li L, Feng L. Overexpression of HLA-G Is positively associated with Kazakh esophageal squamous cell carcinoma in Xinjiang, China. Viral Immunol. 2013;26:180–4. doi: 10.1089/vim.2012.0085. [DOI] [PubMed] [Google Scholar]
- 30.Cui X, Chen Y, Liu L, Li L, Hu J, Yang L, Liang W, Li F. Heterozygote of PLCE1 rs2274223 increases susceptibility to human papillomavirus infection in patients with esophageal carcinoma among the Kazakh populations. J Med Virol. 2014;86:608–17. doi: 10.1002/jmv.23775. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Tian X, Liu F, Zhao Y, Sun M, Chen D, Lu C, Wang Z, Shi X, Zhang Q, Zhang D, Shen Z, Li F, Harris CC, Cai H, Ke Y. Detection of HPV DNA in esophageal cancer specimens from different regions and ethnic groups: a descriptive study. BMC Cancer. 2010;10:19. doi: 10.1186/1471-2407-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molijn A, Kleter B, Quint W, van Doorn LJ. Molecular diagnosis of human papillomavirus (HPV) infections. J Clin Virol. 2005;32(Suppl 1):S43–51. doi: 10.1016/j.jcv.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Lu XM, Monnier-Benoit S, Mo LZ, Xu SY, Pretet JL, Liu Z, Vuitton DA, Mougin C. Human papillomavirus in esophageal squamous cell carcinoma of the high-risk Kazakh ethnic group in Xinjiang, China. Eur J Surg Oncol. 2008;34:765–70. doi: 10.1016/j.ejso.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Liu T, Liu Q, Liang M, Zheng S, Li XL, Lu X, Sheyhidin I. Viral load of HPV 16/18 in esophageal squamous cell carcinoma in three ethnic groups living in Xinjiang Autonomous Region, China. Mol Biol Rep. 2013;40:2045–52. doi: 10.1007/s11033-012-2263-y. [DOI] [PubMed] [Google Scholar]
- 35.Zhang QY, Zhang DH, Shen ZY, Xu LY, Li EM, Au WW. Infection and integration of human papillomavirus in esophageal carcinoma. Int J Hyg Environ Health. 2011;214:156–61. doi: 10.1016/j.ijheh.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Syrjanen K. Geographic origin is a significant determinant of human papillomavirus prevalence in oesophageal squamous cell carcinoma: systematic review and meta-analysis. Scand J Infect Dis. 2013;45:1–18. doi: 10.3109/00365548.2012.702281. [DOI] [PubMed] [Google Scholar]
- 37.Dabrowski A, Kwasniewski W, Skoczylas T, Bednarek W, Kuzma D, Gozdzicka-Jozefiak A. Incidence of human papilloma virus in esophageal squamous cell carcinoma in patients from the Lublin region. World J Gastroenterol. 2012;18:5739–44. doi: 10.3748/wjg.v18.i40.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noori S, Monabati A, Ghaderi A. The prevalence of human papilloma virus in esophageal squamous cell carcinoma. Iran J Med Sci. 2012;37:126–33. [PMC free article] [PubMed] [Google Scholar]
- 39.El-Serag HB, Hollier JM, Gravitt P, Alsarraj A, Younes M. Human papillomavirus and the risk of Barrett’s esophagus. Dis Esophagus. 2013;26:517–21. doi: 10.1111/j.1442-2050.2012.01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel K, Mining S, Wakhisi J, Gheit T, Tommasino M, Martel-Planche G, Hainaut P, Abedi-Ardekani B. TP53 mutations, human papilloma virus DNA and inflammation markers in esophageal squamous cell carcinoma from the Rift Valley, a high-incidence area in Kenya. BMC Res Notes. 2011;4:469. doi: 10.1186/1756-0500-4-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haeri H, Mardany O, Asadi-Amoli F, Shahsiah R. Human papilloma virus and esophageal squamous cell carcinoma. Acta Med Iran. 2013;51:242–5. [PubMed] [Google Scholar]
- 42.Lu XM, Zhang YM, Lin RY, Liang XH, Zhang YL, Wang X, Zhang Y, Wang Y, Wen H. p53 polymorphism in human papillomavirus-associated Kazakh’s esophageal cancer in Xinjiang, China. World J Gastroenterol. 2004;10:2775–8. doi: 10.3748/wjg.v10.i19.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar J, Dominguez E, Li G, Kusewitt DF, Johnson DG. Modeling gene-environment interactions in oral cavity and esophageal cancers demonstrates a role for the p53 R72P polymorphism in modulating susceptibility. Mol Carcinogene. 2014;53:648–58. doi: 10.1002/mc.22019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao F, Han H, Zhang F, Wang B, Ma W, Wang Y, Sun G, Shi M, Ren Y, Cheng Y. HPV infection in esophageal squamous cell carcinoma and its relationship to the prognosis of patients in northern China. ScientificWorldJournal. 2014;2014:804738. doi: 10.1155/2014/804738. [DOI] [PMC free article] [PubMed] [Google Scholar]


