Abstract
CYP2C19 is a highly polymorphic gene and CYP2C19 enzyme results in broad inter-individual variability in response to certain clinical drugs, while little is known about the genetic variation of CYP2C19 in Li Chinese population. The aim of this study was to identify different CYP2C19 mutant alleles and determine their frequencies, along with genotype frequencies, in the Li Chinese population. We used DNA sequencing to investigate promoter, exons, introns, and 3’UTR of the CYP2C19 gene in 100 unrelated healthy Li individuals from Hainan Province, China. We also used SIFT and PolyPhen-2 to predict the protein function of the non-synonymous mutation in CYP2C19 coding regions. We identified 22 different CYP2C19 polymorphisms in the Li Chinese population, including three novel variants (-254A > G, 17807T > C and 58025C > T). The allele frequencies of CYP2C19*1A, *1B, *2A and *3A were 50%, 24%, 24.5%, and 1.5%, respectively. The most common genotype combinations were *1A/*1B (48%) and *1A/*2A (49%). Additionally, the mutation Ala161Pro was predicted to be intolerant and possibly damaging by SIFT and PolyPhen-2, respectively. Our results shed new light on CYP2C19 polymorphisms in Li individuals, which may help to optimize pharmacotherapy effectiveness by providing personalized medicine to this ethnic group.
Keywords: Genetic polymorphism, CYP2C19, Li Chinese population, ethnic groups
Introduction
The cytochrome P450 (CYP450) superfamily is a large and diverse group of enzymes, mainly localized in the endoplasmic reticulum, that metabolize many common therapeutic drugs [1]. CYP2C19 is a member of the CYP2C subfamily of cytochromes P450 and involved in the metabolism of a range of clinically important compounds [2]. These compounds include certain tricyclic antidepressants (e.g., amitriptyline, clomipramine and imipramine), anticonvulsant drugs (e.g., phenytoin and diazepam), antiulcer drugs (e.g., lansoprazole, omeprazole and rabeprazole), benzodiazepines (e.g. quazepam, diazepam and unitrazepam) and specific b-adrenoceptor blockers [3-6].
CYP2C19 is a highly polymorphic gene and genetic variants in the CYP2C19 might cause changes to the enzyme, thus giving rise to different enzymatic activities and resulting in great intra- and inter-population differences in therapeutic outcomes and adverse drug reactions [7]. To date, at least 34 alleles of CYP2C19 have been identified. Among them, CYP2C19*2 and CYP2C19*3 are the most prevalent alleles and have been associated with decreased metabolism of the substrates (drugs); by contrast, CYP2C19*17 is less studied and showed increased gene expression and enzyme activity [8]. Previous studies had demonstrated significant inter-individual and inter-ethnic differences in the frequencies of CYP2C19 alleles and genotypes [9].
The population of China consists of Han Chinese and 55 ethnic minorities currently recognized by the People’s Republic of China. Li is one of the most ancient ethnic groups, having their own spoken and written language. Li population, living mainly in Hainan Island, is geographically isolated from other ethnic groups in the region. To our knowledge, no genotype information on CYP2C19 mutants in this population is available. We systematically screened the whole CYP2C19 genes of 100 healthy, unrelated Li people for polymorphisms and compared their allelic frequencies with previous observations of other ethnic groups, hoping to offer recommendations pertaining to the drug substrates of CYP2C19 in the Li population.
Materials and methods
Subjects
One hundred healthy, unrelated Li Chinese (50 males and 50 females) were recruited between March 2013 and October 2014 from Hainan Provincial People’s Hospital. All participants were Li Chinese residing in the Hainan province, and they had at least three generations of Li paternal ancestry. Subjects with any type of medical illness, organ transplant, drug or alcohol addiction, and pregnant females were excluded from the study. These exclusion criteria were used to minimize controllable factors that may have influenced genetic variation in the genes of interest.
The purpose and experimental procedures of the study were explained to all participants, and written informed consent was obtained from all individuals prior to sample donation. The study protocol was performed in accordance with the Declaration of Helsinki and was approved by The Ethics Committees of Hainan Provincial People’s Hospital.
PCR and DNA sequencing
A blood sample (5 mL) was taken from each subject into an EDTA tube and genomic DNA was extracted using the GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag Ltd.) according to the manufacturer’s instructions. Primers for PCR were designed to amplify the 5’ flanking regions, all exons, and all introns of the CYP2C19 gene, and their sequences are provided in Table 1. Polymerase chain reaction (PCR) for all single nucleotide polymorphisms (SNPs) was performed in 10 μL reactions with 5 μL HotStar Taq Master Mix, 1 μL of template DNA, 0.5 μL each primer (5 μM) and 3 μL deionized water. Thermal cycling conditions were as follows: a initial denaturation step of 15 min at 95°C, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55-64°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 3 min. The PCR products were sequenced using the ABI PrismBigDye Terminator Cycle Sequencing Kit version 3.1 (Applied Biosystems) on an ABI Prism3100 sequencer (Applied Biosystems).
Table 1.
Primers used to amplify regions of CYP2C19
| Primer name | Primer Sequence (5’-3’) | PCR product size (bp) |
|---|---|---|
| Promoter_F | GCCTGTTTTATGAACAGGATGA | 918 |
| Promoter_R | TAAGACAACCGTGAGCTTGC | |
| Exon1_F | ACAGAGTGGGCACTGGGACGA | 844 |
| Exon1_R | GGTCCTAAACCCACAGCTGCTTCC | |
| Exon2_3_F | TTGTCTGACCATTGCCTTGA | 833 |
| Exon2_3_R | TCTCAGCTTCAAACCCTGCT | |
| Exon4_F | CCCCAACTATTCTCACCCTTT | 916 |
| Exon4_R | AAAGTGTGAATTGAAGGACAAGC | |
| Exon5_F | TCAGGTTGTGCAAACTCTTTT | 908 |
| Exon5_R | CCTTCACTCACTTTTTGATGGA | |
| Exon6_F | ATGTTGGTAAGTATACAATGTGAGT | 386 |
| Exon6_R | TCACACCATTAAATTGGGACAGA | |
| Exon7_F | TTTTGATTGGAAATTTTAGTCCATT | 921 |
| Exon7_R | TCAGTTCTTTCCAAACTGACCT | |
| Exon8_F | GTCACTGGCCTTAAGCTCATGCCT | 718 |
| Exon8_R | CCCAGCCTAGGGGGTGAGGG | |
| Exon9_F | TGAGAGTAGGGGAGGTGAAGA | 907 |
| Exon9_R | GATGACGGGTCAGAAGAAGC | |
| 3’-UTR_F | ACGGATTTGTGTGGGAGAGGGC | 674 |
| 3’-UTR_R | AATGCTCAGCCAAAATAGCTTCCCT |
Data analysis
Sequencher 4.10.1 (http://www.genecodes.com/) software was used to initially analyze the sequences including manual curation, fragment assembly, and mutation detection. We named the CYP2C19 variants based on the nucleotide reference sequence NG_008384.2 and CYP allele nomenclature (http://www.cypalleles.ki.se/). Allelic frequency comparisons between Li Chinese population and other populations were performed using the Chi-squared test with a significance level set at P = 0.05 [10]. HAPLOVIEW 4.1 (http://broad.mit.edu/mpg/haploview) was used to assess linkage disequilibrium (LD) and Hardy-Weinberg equilibrium for each genetic variant [11]. Haplotypes were constructed from the selected SNPs and haplotype frequencies were derived for the Li population.
Transcriptional prediction
We analyzed non-synonymous SNPs in the CYP2C19 coding regions to predict the corresponding protein function. Two algorithms, SIFT (Sorting Intolerant From Tolerant, http://sift.bii.a-star.edu.sg/) and PolyPhen-2 (Polymorphism Phenotyping v2, http://genetics.bwh.harvard.edu/pph2/), were used to perform the functional prediction of non-synonymous SNPs [12]. Each variant was given a score based on the impact of its mutation on protein function. The SIFT divided results into four categories based on these scores: tolerant (0.201-1.00), borderline (0.101-0.20), potentially intolerant (0.051-0.10) and intolerant (0.00-0.05). PolyPhen-2 results were divided into three categories: benign, possibly damaging and probably damaging.
Results
Genetic variants
We sequenced CYP2C19 from our study subjects and successfully identified a total of 22 CYP2C19 polymorphisms in this population. Three of the polymorphisms had not previously been reported in either the NCBI database or the Human Cytochrome P450 Allele Nomenclature Committee tables (Table 2). -254A > G was in the promoter region, 17807T > C was a synonymous mutation in exon 4, and 58025C > T was in the intron 6.
Table 2.
Frequency distribution of CYP2C19 polymorphisms in 100 Li subjects
| NO. | SNP | Position | Nucleotide change | Region | Allele | Frequencies | Amino-acid effect | |
|---|---|---|---|---|---|---|---|---|
| 1 | rs190944530 | -283 | G > T | Promoter | 9/100 | 9% | No translated | |
| 2 | -254 | A > G | Promoter | Novel 1 | 1/100 | 1% | No translated | |
| 3 | rs17885098 | 99 | C > T | Exon 1 | 91/94 | 96.80% | Pro33Proa | |
| 4 | rs17878649 | 12306 | G > A | Intron 1 | 1/100 | 1% | No translated | |
| 5 | rs145328984 | 12401 | C > T | Exon 2 | 1/100 | 1% | Arg73Cysb | |
| 6 | rs12769205 | 12662 | A > G | Intron 2 | 55/100 | 55% | No translated | |
| 7 | rs181297724 | 12834 | G > C | Exon 3 | 3/99 | 3.03% | Ala161Prob | |
| 8 | 17807 | T > C | Exon 4 | Novel 2 | 1/98 | 1.02% | Asp165Aspa | |
| 9 | rs4986893 | 17948 | G > A | Exon 4 | CYP2C19*3A | 3/98 | 3.06% | Trp212Terb |
| 10 | rs184151290 | 18074 | C > T | Intron 4 | 1/98 | 1.02% | No translated | |
| 11 | rs7088784 | 18911 | A > G | Intron 4 | 5/100 | 5% | No translated | |
| 12 | rs4244285 | 19154 | G > A | Exon 5 | CYP2C19*2A | 53/100 | 53% | Pro227Proa |
| 13 | rs12571421 | 19520 | A > G | Intron 5 | 53/100 | 53% | No translated | |
| 14 | 58025 | C > T | Intron 6 | Novel 3 | 5/100 | 5% | No translated | |
| 15 | rs28399513 | 79936 | T > A | Intron 6 | 53/100 | 53% | No translated | |
| 16 | rs3758580 | 80160 | C > T | Exon 7 | CYP2C19*2A | 53/100 | 53% | Val330Vala |
| 17 | rs3758581 | 80161 | A > G | Exon 7 | 100/100 | 100% | Ile331Valb | |
| 18 | rs4917623 | 87106 | C > T | Intron 7 | 61/94 | 64.89% | No translated | |
| 19 | rs17886522 | 87313 | A > C | Exon 8 | CYP2C19*3A | 3/100 | 3% | Gly417Glya |
| 20 | rs17882572 | 87594 | G > T | Intron 8 | 3/100 | 3% | No translated | |
| 21 | rs17885052 | 87620 | A > T | Intron 8 | 5/100 | 5% | No translated | |
| 22 | rs191493794 | 90647 | C > T | 3’-UTR | 5/100 | 5% | No translated | |
Synonymous mutations;
non-synonymous mutations.
Allele frequency and genotype frequency
Four CYP2C19 alleles were detected in the Li study group (Table 3). The CYP2C19*1A allele had the highest frequency (50%), followed by the CYP2C19*1B allele (24.00%), and the CYP2C19*2A allele (24.50%). The last allele, CYP2C19*3A, was relatively rare with frequencies of only 1.50%.
Table 3.
Allele and genotype frequencies of CYP2C19 in Li population
| Total (N = 100) | Frequency | Phenotype | ||
|---|---|---|---|---|
| Allele | *1A | 100 | 50.00% | Normal |
| *1B | 48 | 24.00% | Normal | |
| *2A | 49 | 24.50% | None | |
| *3A | 3 | 1.50% | None | |
| Genotype | *1A/*1B | 48 | 48.00% | Normal enzyme activity |
| *1A/*2A | 49 | 49.00% | Decreased enzyme activity | |
| *1A/*3A | 3 | 3.00% | Decreased enzyme activity |
We also detected three CYP2C19 genotypes, with a frequency range from 3.00% to 49.00% in this Li population. Individuals with the wild-type *1/*1 genotype have normal enzyme activity, and this genotype was the relatively prevalent (48.00%) in our study group. Other identified genotypes included the heterozygous genotype *1/*2 (49.00%) and *1/*3 (3.00%), which leads to decreased enzyme activity. According to Haploview analysis, all allele and genotype frequencies (Table 3) were in Hardy-Weinberg equilibrium.
Inter-population comparisons
We further compared CYP2C19 allele frequencies between our data and previously published data from different countries and ethnic groups in east Asia [13-16], south Asia [17], Europe [18-22] and Africa [23-25] (Table 4). Our results showed that the frequency of the wild-type allele, CYP2C19*1, in our study group was significantly lower (P < 0.01) than in Caucasian populations, but was highest in Asian groups. Furthermore, the frequencies of CYP2C19*2 and CYP2C19*3 were significantly higher (P < 0.05) among those of Chinese descent compared with Caucasians. Additionally, we found no significant differences between Li Chinese and Africans.
Table 4.
Allele frequencies of CYP2C19 in different populations
| Populations | Total Number | Allele frequency (%) | References | ||
|---|---|---|---|---|---|
|
| |||||
| CYP2C19*1 | CYP2C19*2 | CYP2C19*3 | |||
| Asians | |||||
| Chinese Li | 100 | 74.00 | 24.50 | 1.50 | Present study |
| Chinese Han | 100 | 67.50 | 25.50 | 2.00 | [13] |
| Chinese Dai | 193 | 66.30 | 30.30 | 3.40 | [14] |
| Japanese | 140 | 53.90** | 35.00 | 11.10** | [15] |
| Korean | 103 | 67.00 | 21.00 | 12.00** | [16] |
| Vietnamese | 90 | 62.00 | 24.00 | 14.00** | [17] |
| Thai | 121 | 59.90* | 35.10 | 5.00 | [17] |
| Caucasians | |||||
| Swedish | 175 | 76.60 | 23.10 | 0.30 | [18] |
| Russian | 290 | 88.30** | 11.40** | 0.30 | [19] |
| Italian | 360 | 88.90** | 11.10** | 0.00* | [20] |
| Bolivian | 778 | 92.10** | 7.80** | 0.10** | [21] |
| Faroese | 312 | 97.10** | 2.90** | 0.00* | [22] |
| Africans | |||||
| Tanzanian | 251 | 81.50 | 17.90 | 0.60 | [23] |
| Ethiopian | 114 | 84.00 | 14.00 | 2.00 | [24] |
| Zimbabwean | 84 | 86.90* | 13.10 | 0.00 | [25] |
P < 0.01, compared with the data of the present study;
P < 0.05, compared with the data of the present study.
Linkage disequilibrium analysis
We performed LD analysis using Haploview with confidence intervals to define LD blocks (Figure 1). The extent of LD for each pair of SNPs was measured by the D’ value, which was most accurate when minor allele frequencies (MAFs) were greater than 5%. Haplotype analysis identified one LD blocks within CYP2C19, and very strong linkage was found between rs4244285, rs12571421, novel variant 58025C > T, rs28399513 and rs3758580.
Figure 1.
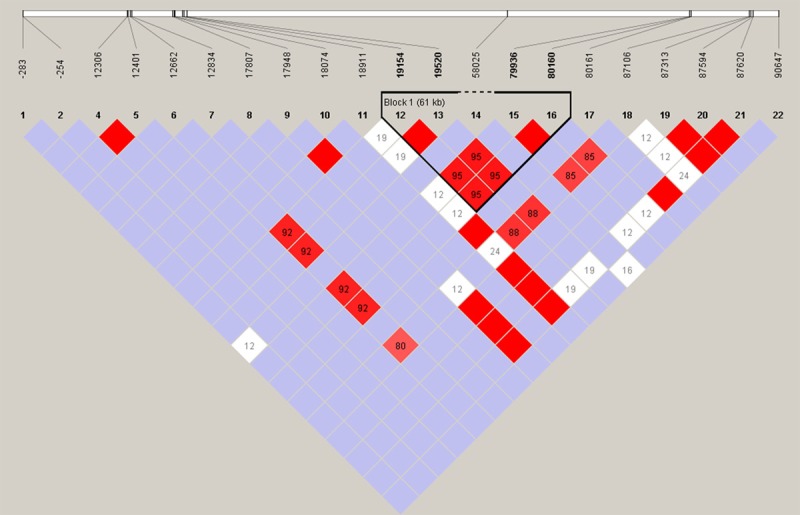
Linkage disequilibrium analysis of CYP2C19. LD is displayed by standard color schemes, with bright red for very strong LD (LOD > 2, D’ = 1), pink red (LOD > 2, D’ < 1) and blue (LOD < 2, D’ = 1) for intermediate LD, and white (LOD < 2, D’ < 1) for no LD.
Predicted protein function of the non-synonymous mutation
We identified four non-synonymous mutation of CYP2C19 in our study group, Arg73Cys, Ala161Pro, Trp212Ter and Ile331Val. Trp212Ter was excluded because it is a termination of protein sequence. Analysis using SIFT of the Arg73Cys and Ala161Pro variants indicated that they were intolerant (score = 0.01), while the variant Ile331Val was identified as tolerant (score = 0.29). PolyPhen-2 results for the Arg73Cys and Ile331Val revealed that both mutations were benign; while Ala161Pro was identified as possibly damaging. PolyPhen-2 utilized two models (HumDiv and HumVar), in which the latter is more rigorous in its false discovery rate. So the HumVar dataset was usually used to predict protein function (Figure 2). The protein function prediction results from SIFT and PolyPhen-2 analysis of the Arg73Cys were inconsistent. However, the protein function prediction results of the Ala161Pro and Ile331Val variants were highly consistent.
Figure 2.
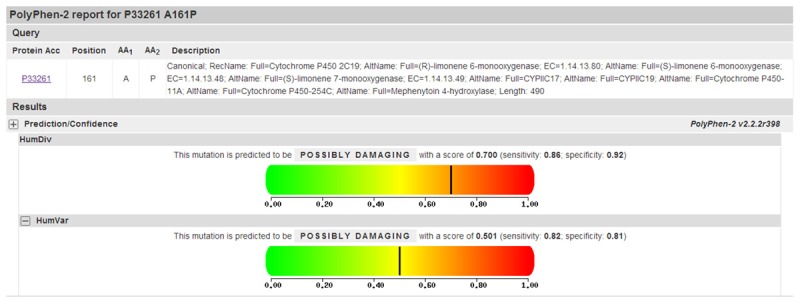
PolyPhen-2 prediction of functional change resulting from an amino acid mutation at position 161.
Discussion
Genetic polymorphisms in CYP2C19 are highly involving in the metabolism of many clinically prescribed drugs and may give rise to important inter-individual and inter-ethnic differences in patient responsiveness and adverse drug reactions [26]. All of these years, several study has determined CYP2C19 genetic polymorphisms in Han Chinese populations, few studies to date have focused on ethnic minorities in China, especially Li Chinese. We identified 22 genetic variants including three novel polymorphisms, four alleles, and three genotypes of CYP2C19 in our study Li Chinese population, and compared these data with previous observations of other ethnic groups. Therefore, our results provide a better understanding of CYP2C19 polymorphisms and a potential database for promoting personalized medicine in Li Chinese population.
The frequency of the wild-type CYP2C19 allele (CYP2C19*1) in the Li Chinese study population was significantly lower (P < 0.01) than in Caucasian populations, which was consistent with findings in previous studies on the Asian populations [27,28]. CYP2C19*2 and CYP2C19*3 have been determined as null alleles and resulting in the total absence of enzyme activity and *1/*2, *1/*3 genotypes have been associated with decreased enzyme activity in previous studies [29]. The occurrence of CYP2C19*2 in the Li subjects in our study was significantly higher (P < 0.01) than that reported for Caucasians, which suggested the pharmacological or toxicological properties of medications that are metabolized by CYP2C19 are likely to differ between Li Chinese and Caucasian populations. Interestingly, the allele frequency of CYP2C19 in our Li group has no significant differences compared with Africans, which may relate to the similarity of their residences. Hainan Province is in the southernmost point of China, and belongs to tropical and subtropical zones, which was consistent with most part of African territory.
Recent studies have shown that CYP2C19 polymorphisms have caused a diverse responsiveness to clopidogrel [30]. The risk of cardiovascular events is increased in patients who are PM (poor metabolizer, carrying at least one CYP2C19*2 allele) despite patients receiving adequate doses of an antiplatelet agent, clopidogrel [31]. In our current study, we determined that the allele *2 and *3 are common genetic variant in the Li Chinese population, and individuals who are homozygous carriers for the *2 or *3 allele show decreased enzyme activity compared to the wild type. So clinical treatment used clopidogrel should be more carefully in Li Chinese population.
Analysis of genetic variants in the coding region revealed variant Ala161Pro will affect the protein structure and function, and the results of SITF and PolyPhen-2 were highly consistent. However, the protein prediction results of Arg73Cys from the SIFT and PolyPhen-2 were inconsistent. The prediction accuracy of SIFT and PolyPhen-2 is 63% and 75%, while the false positive rate is 19% and 9%, respectively [12,32]. Therefore, the results identified here should be confirmed by other means in further studies.
In summary, our results provide a basic profile of CYP2C19 polymorphisms in the Li Chinese population, and future studies will use a larger sample size of Li Chinese, leading to the enhanced application of personalized medicine in this population.
Acknowledgements
This work is supported by Special Project of Science and Technology for Hainan Province Social Development (No. SF201402) and Applied Technology Research and Development and Demonstration Projects of Hainan Province (No. ZDXM2014119).
Disclosure of conflict of interest
None.
References
- 1.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Gerbal-Chaloin S, Pascussi JM, Pichard-Garcia L, Daujat M, Waechter F, Fabre JM, Carrère N, Maurel P. Induction of CYP2C genes in human hepatocytes in primary culture. Drug Metab Dispos. 2001;29:242–251. [PubMed] [Google Scholar]
- 3.Andersson T, Holmberg J, Walan A. Pharmacokinetics and effect on caffeine metabolism of the proton pump inhibitors, omeprazole, lansoprazole, and pantoprazole. Br J Clin Pharmacol. 1998;45:369–375. doi: 10.1046/j.1365-2125.1998.t01-1-00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardiner SJ, Begg EJ. Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacol Rev. 2006;58:521–590. doi: 10.1124/pr.58.3.6. [DOI] [PubMed] [Google Scholar]
- 5.Onof S, Hatanaka T, Miyazawa S, Tsutsui M, Aoyama T, Gonzalez F, Satoh T. Human liver microsomal diazepam metabolism using cDNA-expressed cytochrome P450s: role of CYP2B6, 2C19 and the 3A subfamily. Xenobiotica. 1996;26:1155–1166. doi: 10.3109/00498259609050260. [DOI] [PubMed] [Google Scholar]
- 6.Paveliu MS, Bengea S, Paveliu FS. Individualized drug response related to genetic variations of cytochrome P450 isoforms and other enzymes. Farmacia. 2010;58:245–254. [Google Scholar]
- 7.Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol. 2001;52:349–355. doi: 10.1046/j.0306-5251.2001.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang M, Tybring G, Dahl ML, Lindh JD. Impact of cytochrome P450 2C19 polymorphisms on citalopram/escitalopram exposure: a systematic review and meta-analysis. Clin Pharmacokinet. 2014;53:801–811. doi: 10.1007/s40262-014-0162-1. [DOI] [PubMed] [Google Scholar]
- 9.Hu LM, Dai DP, Hu GX, Yang JF, Xu RA, Yang LP, Qian JC, Ge RS, Cai JP. Genetic polymorphisms and novel allelic variants of CYP2C19 in the Chinese Han population. Pharmacogenomics. 2012;13:1571–1581. doi: 10.2217/pgs.12.141. [DOI] [PubMed] [Google Scholar]
- 10.Adamec C. Example of the use of the nonparametric test. Test X2 for comparison of 2 independent examples. Cesk Zdrav. 1964;12:613. [PubMed] [Google Scholar]
- 11.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 12.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Q, Yu X, Lin H, Wang L, Yun Q, Hu S, Wang D. Genetic polymorphism, linkage disequilibrium, haplotype structure and novel allele analysis of CYP2C19 and CYP2D6 in Han Chinese. Pharmacogenomics J. 2009;9:380–394. doi: 10.1038/tpj.2009.31. [DOI] [PubMed] [Google Scholar]
- 14.He N, Yan FX, Huang SL, Wang W, Xiao ZS, Liu ZQ, Zhou HH. CYP2C19 genotype and S-mephenytoin 4’-hydroxylation phenotype in a Chinese Dai population. Eur J Clin Pharmacol. 2002;58:15–18. doi: 10.1007/s00228-002-0425-x. [DOI] [PubMed] [Google Scholar]
- 15.Kimura M, Ieiri I, Mamiya K, Urae A, Higuchi S. Genetic polymorphism of cytochrome P450s, CYP2C19, and CYP2C9 in a Japanese population. Ther Drug Monit. 1998;20:243–247. doi: 10.1097/00007691-199806000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Roh HK, Dahl ML, Tybring G, Yamada H, Cha YN, Bertilsson L. CYP2C19 genotype and phenotype determined by omeprazole in a Korean population. Pharmacogenetics. 1996;6:547–551. doi: 10.1097/00008571-199612000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Yamada S, Onda M, Kato S, Matsuda N, Matsuhisa T, Yamada N, Miki M, Matsukura N. Genetic differences in CYP2C19 single nucleotide polymorphisms among four Asian populations. J Gastroenterol. 2001;36:669–672. doi: 10.1007/s005350170029. [DOI] [PubMed] [Google Scholar]
- 18.Chang M, Dahl ML, Tybring G, Gotharson E, Bertilsson L. Use of omeprazole as a probe drug for CYP2C19 phenotype in Swedish Caucasians: comparison with S-mephenytoin hydroxylation phenotype and CYP2C19 genotype. Pharmacogenetics. 1995;5:358–363. doi: 10.1097/00008571-199512000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Gaikovitch EA, Cascorbi I, Mrozikiewicz PM, Brockmöller J, Frötschl R, Köpke K, Gerloff T, Chernov JN, Roots I. Polymorphisms of drug-metabolizing enzymes CYP2C9, CYP2C19, CYP2D6, CYP1A1, NAT2 and of P-glycoprotein in a Russian population. Eur J Clin Pharmacol. 2003;59:303–312. doi: 10.1007/s00228-003-0606-2. [DOI] [PubMed] [Google Scholar]
- 20.Scordo MG, Caputi AP, D’Arrigo C, Fava G, Spina E. Allele and genotype frequencies of CYP2C9, CYP2C19 and CYP2D6 in an Italian population. Pharmacol Res. 2004;50:195–200. doi: 10.1016/j.phrs.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Bravo-Villalta HV, Yamamoto K, Nakamura K, Bayá A, Okada Y, Horiuchi R. Genetic polymorphism of CYP2C9 and CYP2C19 in a Bolivian population: an investigative and comparative study. Eur J Clin Pharmacol. 2005;61:179–184. doi: 10.1007/s00228-004-0890-5. [DOI] [PubMed] [Google Scholar]
- 22.Halling J, Petersen MS, Damkier P, Nielsen F, Grandjean P, Weihe P, Lundgren S, Lundblad MS, Brøsen K. Polymorphism of CYP2D6, CYP2C19, CYP2C9 and CYP2C8 in the Faroese population. European J Clin Pharmacol. 2005;61:491–497. doi: 10.1007/s00228-005-0938-1. [DOI] [PubMed] [Google Scholar]
- 23.Herrlin K, Massele AY, Jande M, Alm C, Tybring G, Abdi YA, Wennerholm A, Johansson I, Dahl ML, Bertilsson L. Bantu Tanzanians have a decreased capacity to metabolize omeprazole and mephenytoin in relation to their CYP2C19 genotype. Clin Pharmacol Ther. 1998;64:391–401. doi: 10.1016/S0009-9236(98)90070-4. [DOI] [PubMed] [Google Scholar]
- 24.Persson I, Aklillu E, Rodrigues F, Bertilsson L, Ingelman-Sundberg M. S-mephenytoin hydroxylation phenotype and CYP2C19 genotype among Ethiopians. Pharmacogenetics. 1996;6:521–526. doi: 10.1097/00008571-199612000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Masimirembwa C, Bertilsson L, Johansson I, Hasler JA, Ingelman-Sundberg M. Phenotyping and genotyping of S-mephenytoin hydroxylase (cytochrome P450 2C19) in a Shona population of Zimbabwe. Clin Pharmacol Ther. 1995;57:656–661. doi: 10.1016/0009-9236(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 26.Kobori L, Kohalmy K, Porrogi P, Sárváry E, Gerlei Z, Fazakas J, Nagy P, Járay J, Monostory K. Drug-induced liver graft toxicity caused by cytochrome P450 poor metabolism. Br J Clin Pharmacol. 2008;65:428–436. doi: 10.1111/j.1365-2125.2007.03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Qin S, Xie J, Tang J, Yang L, Shen W, Zhao X, Du J, He G, Feng G. Genetic polymorphism analysis of CYP2C19 in Chinese Han populations from different geographic areas of mainland China. Pharmacogenomics. 2008;9:691–702. doi: 10.2217/14622416.9.6.691. [DOI] [PubMed] [Google Scholar]
- 28.Ota T, Kamada Y, Hayashida M, Iwao-Koizumi K, Murata S, Kinoshita K. Combination analysis in genetic polymorphisms of drug-metabolizing enzymes CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A5 in the Japanese population. Int J Med Sci. 2015;12:78–82. doi: 10.7150/ijms.10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet. 2002;41:913–958. doi: 10.2165/00003088-200241120-00002. [DOI] [PubMed] [Google Scholar]
- 30.Lin R, Zhang L, Zhang P, Zhou L, Liu T, Li Y, Zhang W, Wang W, Zhang J. Influence of CYP2C19 loss-of-function variants on the metabolism of clopidogrel in patients from north-western China. J Clin Pharm Ther. 2015;40:308–314. doi: 10.1111/jcpt.12254. [DOI] [PubMed] [Google Scholar]
- 31.Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, Payot L, Brugier D, Cayla G, Beygui F. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373:309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 32.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]


