Abstract
Background: Dysregulation of long non-coding RNAs (lncRNAs) plays critical roles in tumor progression. The purpose of this study was to investigate the relationship between lncRNA CCAT2 expression and cervical squamous cell cancer susceptibility and prognosis. Methods: Expression levels of lncRNA CCAT2 in 123 cervical squamous cell tumor specimens were determined by quantitative real-time PCR (qRT-PCR), to clarify the clinical significance of lncRNA CCAT2 in cervical squamous cell cancer, we further discussed the relationship between lncRNA CCAT2 expression and overall survival (OS) and progression-free survival (PFS). Results: In the present study, we found that lncRNA CCAT2 was up-regulated in cervical squamous cell cancer tissues compared to the adjacent non-tumor tissues. In addition, the high lncRNA CCAT2 expression was significantly associated with the FIGO stage, lymph node metastasis and depth of cervical invasion (P<0.05). Furthermore, patients with high expression of lncRNA CCAT2 had poor OS (HR=2.813, 95% CI: 1.504-6.172; P=0.017), and PFS rates (HR=3.072, 95% CI: 1.716-8.174; P=0.008). Multivariate Cox proportional hazard model analysis demonstrated that high lncRNA CCAT2 expression was an independent poor prognostic factor for cervical squamous cell cancer patients. Conclusions: Our study suggested that high expression of lncRNA CCAT2 is related to the prognosis of cervical squamous cell cancer; it may be a new prognostic biomarker and potential therapeutic target for cervical squamous cell cancer intervention.
Keywords: Cervical squamous cell cancer, lncRNA CCAT2, overall survival, progression-free survival
Introduction
Cervical cancer is the second leading cause of death among women worldwide, with an estimated 530000 deaths per year [1]. Although it has made a notable progress with treatment developed, including surgical techniques, chemotherapy, and radiotherapy in the past two decades, there are still some early cases appeared invasion and metastasis, which directly affected the prognosis of cervical cancer [2]. In recent years, the incidence of cervical cancer increases every year, most of them are squamous cell carcinoma, and patients tend to be increasingly younger, it had became a serious threat to women’s lives and health [3]. Therefore, an exploration of the molecular pathogenesis of cervical cancer and the identification of potential markers for early detection may play a significant role in treatment and prognosis.
The long non-coding RNAs (lncRNAs) are a class of non-coding RNA over 200 nucleotides with no protein-coding potential [4]. Recently, increasing evidence showed that lncRNAs play crucial roles in the regulation of multiple biological processes, including development, differentiation, and carcinogenesis [5,6]. For example, Jiang et al. showed that lncRNA DEANR1 could facilitate human endoderm differentiation by activating FOXA2 expression [7]. Xie et al. showed that decreased lncRNA SPRY4-IT1 contributed to gastric cancer cell metastasis partly via affected epithelial-mesenchymal transition [8]. Wang et al. showed that up-regulated lncRNA UCA1 contributed to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway [9].
In the present study, we focus on lncRNA CCAT2, a novel long non-coding RNA transcript encompassing the rs6983267 SNP, which was highly over-expressed in microsatellite-stable colorectal cancer and promotes tumor growth, metastasis, and chromosomal instability [10]. Recently, Redis et al. showed that lncRNA CCAT2 represent a valuable predictive marker of clinical outcome (shorter MFS and OS) for breast cancer patients, and high levels of lncRNA CCAT2 indicated that these patients will not benefit from CMF adjuvant chemotherapy [11]. However, little is known about the role of lncRNA CCAT2 in cervical cancer.
The aim of this study is to identify the role of lncRNA CCAT2 in the progression of cervical cancer; we investigated the relationship of lncRNA CCAT2 expression with clinicopathological features, including the survival of patients. Our results indicated that lncRNA CCAT2 expression levels were higher in tumor tissues than those in adjacent non-tumor tissues. Moreover, the relatively higher expression of lncRNA CCAT2 was significantly correlated with malignant status and poor prognosis of cervical cancer patients.
Materials and methods
Tissue specimens
Squamous cell cervical cancer tissue and adjacent non-tumor tissue were obtained from 123 consecutive patients with cervical squamous cell cancer that was confirmed by histo-pathological analysis at the Department of Gynecology and Obstetrics, The Second Affiliated Hospital of Soochow University, between 2006 and 2009. All specimens were immediately frozen in liquid nitrogen and stored at -80°C until RNA extraction. None of enrolled participants were exposed to radiotherapy before the samples were collected. The clinical stage was classified according to the International Federation of Gynecology and Obstetrics criteria. Written informed consent was obtained from all patients prior to participation in the study. The medical ethics committee of The Second Affiliated Hospital of Soochow University approved the study.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from tumor tissue and adjacent non-tumor tissues by Trizol reagent (Invitrogen), according to the manufacturer’s instructions. After purification, cDNA was synthesized from 10 μg total RNA using the Prime Script RT Master Mix (Takara). The primers were designed as follows: for CCAT2, the forward primer was 5’-CCACATCGCTCAGACACCAT-3’ and the reverse primer was 5’-ACCAGGCGCCCAATACG-3’. For human GAPDH, the forward primer was 5’-CGCTCTCTGCTCCTCCTGTTC-3’ and the reverse primer was 5’-ATCCGTTGACTCCGACCTTCAC-3’. The RT-PCR was conducted by SYBR Premix Ex TaqTMII (Takara) on LightCycler (Roche). Relative quantification of RNA expression was calculated by using the 2-ΔΔCt method. Each experiment was performed in triplicate.
Follow-up
All the patients on the study were regularly followed-up for survival analysis until death or until the closing date of study. The median follow-up time among the 123 patients was 48 months, ranging from 6 to 60 months. Clinical records of the patients were obtained from the Second Affiliated Hospital of Soochow University. Examinations conducted during the follow-up period included pelvic MRI, color Doppler ultrasound of the abdominal and urinary tract, chest X-rays for every 3 months for 2 years, at 6 months intervals in years 3 to 5 thereafter, and annually thereafter. Progression-free survival (PFS) was defined as the interval from the date of surgery to confirm local recurrence or distant metastasis, and overall survival (OS) was defined as the interval from the date of surgery to death due to any cause or to the date of last contact.
Statistical analysis
All statistical analyses were performed using SPSS 18.0 software (IBM). Data are expressed as the mean ± SD from at least three separate experiments. The association between the lncRNA CCAT2 and clinicopathologic features was tested using the chi-square test. The relevance between lncRNA CCAT2 expression and the OS/PFS of patients were assessed by the log-rank test with the Kaplan-Meier method. A Cox proportional hazard model was constructed to evaluate the association of lncRNA CCAT2 expression with OS and PFS, respectively. Differences were considered statistically significant when P was less than 0.05.
Results
Up-regulation of lncRNA CCAT2 in cervical cancer
qRT-PCR was performed to detect the expression levels of lncRNA CCAT2 in 123 pairs of cervical cancer and adjacent non-tumor tissues normalized to GAPDH. As shown in Figure 1A, the expression levels of CCAT2 were found to be distinctly increased in cervical cancer tissues compared to adjacent non-tumor tissues (P<0.05). Those data indicated that CCAT2 might play an oncogenic role in cervical cancer progression.
Figure 1.
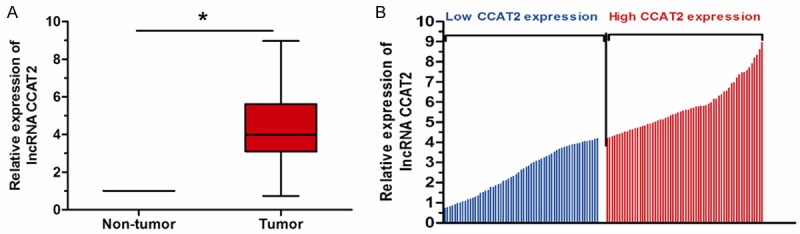
LncRNA CCAT2 expression is up-regulated in cervical cancer tissues. A. lncRNA CCAT2 expression was examined by qRT-PCR in 123 paired cervical cancer tissues and adjacent non-tumor tissues. B. The 123 total cervical patients included in the study were divided into a high CCAT2 expression group and a low CCAT2 expression group according to the median value of relative CCAT2 expression. *P<0.05.
Correlation of lncRNA CCAT2 expression with clinicopathological features
In order to investigate the relationship between lncRNA CCAT2 expression and clinicopathological features in cervical squamous cell cancer. The median expression level of CCAT2 was used as a cutoff point to divide all 123 patients into two groups: cervical cancer patients expressing CCAT2 less than the median expression level were assigned to the low expression group (n=61), and those samples with expression equal or above the median expression level were assigned to the high expression group (n=62) (Figure 1B). The relationships between CCAT2 expression levels and clinicopathological features were shown in Table 1. High CCAT2 expression was observed to be closely associated with FIGO stage, lymph node metastasis and depth of cervical invasion (P<0.05). In contrast, there was no association between CCAT2 expression and other clinical factors, such as age, tumor size, and histology grade (P>0.05).
Table 1.
Clinicopathological features associated with lncRNA CCAT2 expression in 123 cervical squamous cell cancer patients
| Clinicopathological features | Total | LncRNA CCAT2 expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age | ||||
| <45 | 59 | 27 | 32 | 0.415 |
| ≥45 | 64 | 34 | 30 | |
| Tumor size (cm) | ||||
| <4.0 | 52 | 24 | 28 | 0.514 |
| ≥4.0 | 71 | 37 | 34 | |
| Histologic grade | ||||
| G1 + G2 | 64 | 29 | 35 | 0.323 |
| G3 | 59 | 32 | 27 | |
| FIGO stage | ||||
| Ib~IIa | 62 | 39 | 23 | 0.003 |
| IIb~IIIa | 61 | 22 | 39 | |
| Lymph node metastasis | ||||
| No | 78 | 50 | 28 | 0.000 |
| Yes | 45 | 11 | 34 | |
| Depth of cervical invasion | ||||
| <2/3 | 70 | 44 | 26 | 0.000 |
| ≥2/3 | 53 | 17 | 36 | |
Relationship between lncRNA CCAT2 expression and cervical cancer patients’ survival
To further verify the potential clinical utility of the lncRNA CCAT2 high expression, we evaluated the prognostic power of lncRNA CCAT2 on OS and PFS in 123 cervical squamous cell cancer patients. The Kaplan-Meier method and log-rank test was used to determine the relationship between lncRNA CCAT2 and prognosis, and we found the high expression of lncRNA CCAT2 was correlated with a shorter OS or PFS of patients (P<0.05, Figure 2A and 2B). Furthermore, multivariate analyses were utilized to evaluate whether lncRNA CCAT2 expression level and various clinicopathological features were independent prognostic parameters of cervical cancer patient outcomes. Our data revealed that lncRNA CCAT2 expression level was an independent prognostic factor for OS (HR=2.813, 95% CI: 1.504-6.172; P=0.017), as well as PFS (HR=3.072, 95% CI: 1.716-8.174; P=0.008) of cervical cancer patients (Table 2).
Figure 2.
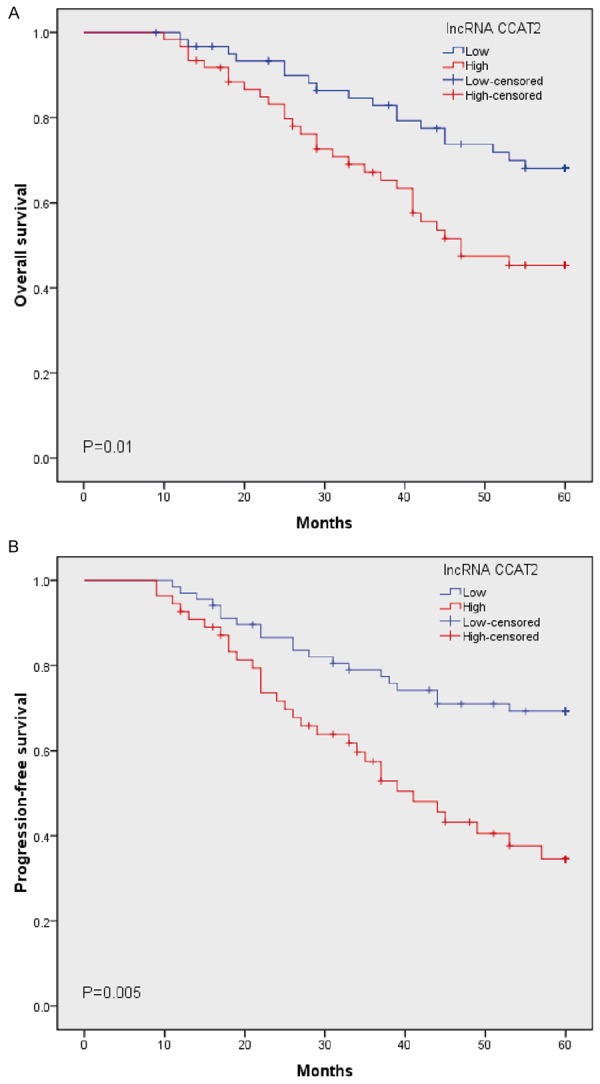
Kaplan-Meier survival curves for cervical cancer patients according to the expression of lncRNA CCAT2. OS and PFS patients with high vs. Low CCAT2 expression levels are shown. A. OS rate of cervical cancer patients with high CCAT2 was significantly poorer compared to those patients with low CCAT2 (P<0.05). B. PFS rate of cervical cancer patients with high CCAT2 was significantly poorer compared to those patients with low CCAT2 (P<0.05).
Table 2.
Multivariate Cox proportional hazard model analysis of overall survival and progression-free survival in cervical cancer patients
| Overall survival | Progression-free survival | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Hazard ratio | 95% CI | P | Hazard ratio | 95% CI | P | |
| Age | 1.083 | 0.517-3.016 | 0.292 | 1.265 | 0.378-2.157 | 0.237 |
| Tumor size | 2.026 | 0.753-6.218 | 0.227 | 1.857 | 0.663-5.829 | 0.135 |
| Histologic grade | 2.784 | 0.508-5.376 | 0.144 | 2.436 | 0.629-5.017 | 0.098 |
| FIGO stage | 3.017 | 1.315-8.683 | 0.013 | 2.789 | 1.571-7.746 | 0.009 |
| Lymph node metastasis | 3.542 | 1.642-9.215 | 0.012 | 3.018 | 1.447-8.726 | 0.007 |
| Depth of cervical invasion | 2.311 | 1.165-5.815 | 0.009 | 2.704 | 1.393-6.741 | 0.015 |
| CCAT2 expression | 2.813 | 1.504-6.172 | 0.017 | 3.072 | 1.716-8.174 | 0.008 |
Discussion
Cervical cancer remains one of the leading causes of cancer death in women worldwide [12]. Even though radiotherapy, chemotherapy and surgery are used as standard treatment modalities for cervical cancer patients, the prognosis of patients is still un-satisfactory. Therefore, characterizations of identifiable molecular markers should be of diagnostic, prognostic and therapeutic value in the management of cervical cancer.
Many studies suggested that lncRNAs play critical roles in various physiological and pathological processes [13]. Recently, dysregulated expression of lncRNA has been found in various types of cancers, including cervical cancer. For example, Cao et al. reported that lncRNA GAS5 was decreased in cervical cancer and associated with poorer overall survival of patients [14]. Yang et al. showed that lncRNA CCHE1 could promote cervical cancer cell proliferation via up-regulating PCNA [15]. Kim et al. showed that lncRNA HOTAIR was up-regulated and associated with poor prognosis of cervical cancer, and in vitro analysis revealed that HOTAIR could promote tumor aggressiveness through the up-regulation of VEGF and MMP-9 and EMT-related genes [16]. However, there were no reports about the clinicopathologic and prognostic significance of lncRNA CCAT2 expression in human cervical squamous cell cancer.
In the present study, based on qRT-PCR data, we explored the association of lncRNA CCAT2 expression with clinicopathological features and prognosis in cervical cancer. Our findings indicated that lncRNA CCAT2 was significantly increased in cervical squamous cell cancer compared with that in adjacent non-tumor tissues. In addition, we found that lncRNA CCAT2 expression was associated with FIGO stage, lymph node metastasis and depth of cervical invasion. More important, we found patient with high expression of CCAT2 was significantly associated with a shorter OS and PFS. These results strongly suggested that lncRNA CCAT2 was involved in the progression and development of cervical cancer. In fact, not only in cervical cancer, CCAT2 over-expression also found to be associated with progression in other cancers. For example, Qiu et al. found that lncRNA CCAT2 was up-regulated in non-small cell lung cancer tissues and correlated with lymph node metastasis. Silencing CCAT2 by siRNA could inhibit the proliferation and invasion ability of lung cancer cells [17]. Zhang et al. showed that lncRNA CCAT2 was elevated in esophageal squamous cell carcinoma and associated with tumor progression [18]. Wang et al. that lncRNA CCAT2 was up-regulated in gastric cancer and correlated with advanced clinical features and shorter overall survival time [19]. Our study expanded the clinical value of lncRNA CCAT2 in cervical cancer progression.
In conclusion, our studies demonstrated that lncRNA CCAT2 was an independent prognostic factor of cervical squamous cell cancer patients. These findings suggested that lncRNA CCAT2 may be a potential prognostic factor and therapeutic target in patients with cervical cancer. However, the molecular mechanisms of lncRNA CCAT2 that involved in cervical cancer need to be further studied.
Acknowledgements
This work was partially supported by Suzhou Municipal Science and Technology Development Plan (BASIC) (No. SYSD2013096).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Hu X, Schwarz JK, Lewis JS Jr, Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70:1441–1448. doi: 10.1158/0008-5472.CAN-09-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waggoner SE. Cervical cancer. Lancet. 2003;361:2217–2225. doi: 10.1016/S0140-6736(03)13778-6. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang W, Liu Y, Liu R, Zhang K, Zhang Y. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 2015;11:137–148. doi: 10.1016/j.celrep.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie M, Nie FQ, Sun M, Xia R, Liu YW, Zhou P, De W, Liu XH. Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J Transl Med. 2015;13:250. doi: 10.1186/s12967-015-0595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, Buiga R, Pop V, Irimie A, Fodde R, Bedrosian I, Martens JW, Foekens JA, Berindan-Neagoe I, Calin GA. CCAT2, a novel long non-coding RNA in breast cancer: Expression study and clinical correlations. Oncotarget. 2013;4:1748–1762. doi: 10.18632/oncotarget.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arbyn M, Castellsague X, de Sanjose S, Bruni L, Saraiya M, Bray F, Ferlay J. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 13.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao S, Liu W, Li F, Zhao W, Qin C. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7:6776–6783. [PMC free article] [PubMed] [Google Scholar]
- 15.Yang M, Zhai X, Xia B, Wang Y, Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol. 2015;36:7615–22. doi: 10.1007/s13277-015-3465-4. [DOI] [PubMed] [Google Scholar]
- 16.Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, Yin R. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35:5375–80. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Xu Y, He C, Guo X, Zhang J, Zhang L, Kong M, Chen B, Zhu C. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2015;111:834–839. doi: 10.1002/jso.23888. [DOI] [PubMed] [Google Scholar]
- 19.Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ, Hu JH. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2015;8:779–785. [PMC free article] [PubMed] [Google Scholar]


