Abstract
Survivin, a member of the inhibitor of apoptosis gene family regulates two critical processes in neoplastic transformation, namely, cell proliferation and apoptosis. This study aimed to detect the effect of survivin on tumor growth of colorectal cancer (CRC) in vivo. We found that inhibition of survivin by interference decreased the sizes and weights of xenografts in nude mice. The number of apoptotic cells of the shRNA survivin group was higher than that of the black group and the shRNA control group. The downregulated expression of survivin decreased the expression levels of bcl2 and ki67. Our results indicated that inhibition of survivin significantly enhanced the inhibition of tumor growth and induced apoptosis. Survivin is an attractive target for CRC treatment.
Keywords: Survivin, colorectal cancer, RNA interference, proliferation, apoptosis
Introduction
Colorectal cancer (CRC) is one of the most common digestive malignant tumors. In the United States, the incidence and mortality of CRC rank third in both men and women [1]. In China, the incidence and mortality of CRC are increasing rather rapidly because of the improvement in people’s living standards, dietary changes, and increase in the aging population. Early symptoms of CRC are usually not detected. When diagnosed, CRC is often in the advanced stage, occasionally with metastatic spread to the liver (15%-25%). The median survival time of metastatic CRC is less than two years. The principle therapeutic method is surgery, assisted by radiotherapy and chemotherapy. Although surgical techniques have rapidly developed during the past few years, the five-year survival rate of CRC has not significantly increased. Recurrence and metastasis are the major causes of death in CRC. Non-surgical treatment, such as chemotherapy and radiotherapy, lack sensitivity and specificity, and result in toxicity and side effects. Therefore, investigations on the molecular mechanisms of CRC, novel biomarkers, and optimized diagnosis and treatment program are essential. Survivin is an epigenetic marker that has been the focus of numerous tumor studies.
Survivin, a member of the inhibitor of apoptosis protein (IAP) family, is on the telomeric position of chromosome 17, and it encodes 16.5 kD protein of 142 amino acids [2]. Its function is related to cell proliferation, cell division, and cell apoptosis [3-5]. Studies indicated that survivin is found during embryonic development, and it is completely downregulated and undetectable in normal adult tissues. However, it becomes prominently re-expressed in several human malignancies, including cancers of the colon, stomach, prostate, and breast [2]. Furthermore, survivin plays an important role in cancer development and it is involved in the resistance of tumor cells to both radiotherapy and chemotherapy [6]. Evidence from cDNA microarray demonstrated that survivin also plays an important role in the pathogenesis of CRC [7]. A recent meta-analysis showed that upregulation of survivin is associated with poor prognosis in patients with CRC [8].
RNA interference (RNAi) technology has been recently used for the efficient and rapid identification of gene function [9,10]. Short hairpin RNA (ShRNA) is a sequence of RNA that makes a tight hairpin turn that can be used to silence target gene expression via RNAi. The expression of shRNA in cells is accomplished by delivery of plasmids or through viral or bacterial vectors [11-13]. Given the ability of shRNA to provide specific and long-term gene silencing, the use of shRNA for gene therapy applications has attracted much interest [14]. In our study, our results indicated that shRNA survivin could actively inhibit tumor growth in vivo. The results of the present study suggested that suppression of survivin expression by RNAi may induce CRC apoptosis and provide a novel approach for anticancer gene therapy.
In this study, after transfection with shRNA, low expression of survivin inhibited tumorigenesis, promoted the apoptosis of CRC in nude mice, and inhibited the expression of downstream molecules, such as ki67 and bcl2. These results confirmed the role of survivin on tumor growth of transplanted CRC.
Materials and methods
Cell culture and transfection
Human CRC cell line (SW1116) was cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 g/ml streptomycin at 37°C in a humidified incubator with 5% CO2. Survivin shRNA plasmids were obtained from Santa Cruz Biotechnology. Cells were transfected using Lipofectamine 2000 (Invitrogen, USA).
Immunoblotting and RT-PCR analysis
Cells were lysed, and immunoblotting assay was conducted according to standard methods. Antibodies against survivin, bcl2, ki67, and β-actin were purchased from Cell Signaling Technology. Total RNAs were isolated from cells using TRIzol (Invitrogen, USA). cDNAs were synthesized using 1 g of total RNAs and Prime ScriptTM RT reagent kit (KeyGen, China) and then used for RT-PCR analysis with PCR MasterMix (Toyobo, Japan). The expression levels of survivin were normalized to those of GAPDH. Primers used in the study are listed in Supplementary Table 1.
In vivo tumor growth assay
Four-week-old nude mice [BALB/cA-nu (nu/nu)] were purchased from Shanghai Experimental Animal Center (Chinese Academy of Sciences, China), maintained in pathogen-free conditions, and sustained with standard diets. All procedures involving animals were approved by the Institutional Committee on Animal Care, Anhui Medical University. Fifteen mice were randomly divided into three groups, with five mice per group. SW1116 cells (1.0 × 106 cells in 100 μl) expressing shRNA survivin was injected subcutaneously into nude mice. SW1116 cells and SW1116 cells with shRNA control were used as the negative control. Bidimensional tumor measurements were measured with calipers once every two days. Tumor volumes were calculated according to the formula: (width2×length)/2. The mice were euthanized after 28 days, and tumors were weighed.
TUNEL assay
Tumor samples were fixed in Bouin’s solution and embedded in paraffin. Tumor sections at 5 mm were cut and deparaffinized. After incubation with PBS for 15 min, the sections were washed with PBS buffer for three times. After incubation with 1% Triton-100 for 15 min and PBS for 5 min, the sections were washed with PBS buffer for three times. Subsequently, 3% hydrogen peroxide blocked endogenous peroxidase activity for 15 min. After washing for three times, the sections were added with 100 µl of TdT enzyme, and the mixture was incubated at 37°C for 1 h. After washing for three times, the protein signals were detected using 3,39-diaminobenzidine (Gene Tech, Shanghai, China). The nuclei of apoptotic cells were brown. The site of intensive apoptosis was selected by two pathologists blinded to the study. They counted 5-10 high-power microscope field of vision to obtain the number of apoptotic cells as a percentage of total cell apoptosis index.
Results
Effect of shRNA survivin on tumor growth
After transfection, SW1116 cells were harvested. Results of Western blot (Figure 1A) showed that the survivin expression levels of the shRNA survivin group decreased compared with those of the blank group and the shRNA control group. We then studied the effects of survivin on anchorage-independent growth. Blank cells, shRNA control cells, and shRNA survivin cells were trypsinized, resuspended, and implanted into the right armpit of nude mice. The lengths and widths of tumors were measured, and the volumes were calculated (Figure 1B). As shown in Figure 1C, the sizes of xenografts from the blank group and the shRNA control group were similar, whereas the sizes of xenografts from the two control groups were much larger than those from the shRNA survivin group. These results indicated that the downregulated expression of survivin in CRC cells reduced tumor growth. Consistent with these results, the downregulated expression of survivin decreased tumor weight by 75% (Figure 1D). TUNEL assay showed that the number of apoptotic cells of the shRNA survivin group was higher than that of the blank group and the shRNA control group (Figure 1E).
Figure 1.
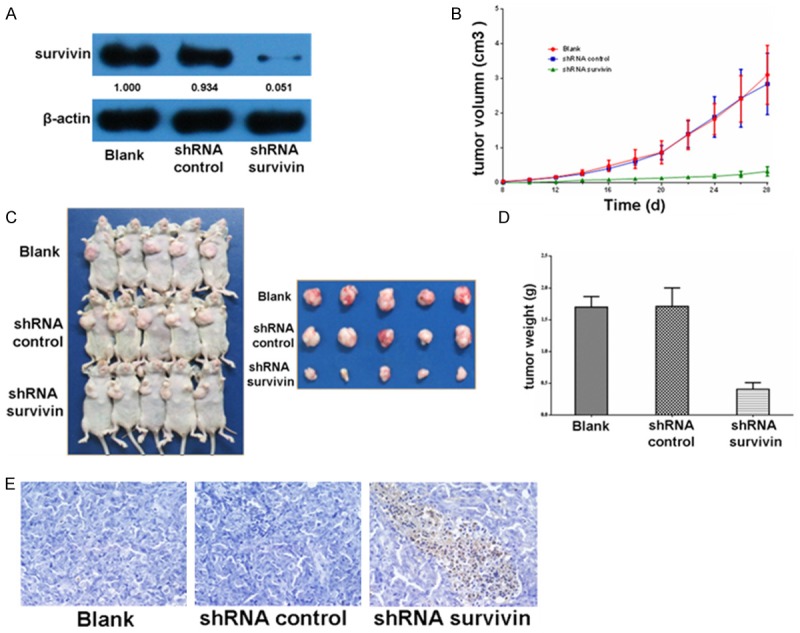
Roles of survivin on tumor growth. A. SW1116 cells were transfected, and immunoblotting was performed to detect the expression levels of survivin in three groups. B. The blank SW1116 cells, shRNA control SW1116 cells, and shRNA survivin SW1116 cells were implanted into nude mice. The xenograft volumes was calculated and presented as the mean ± SD (n = 5). C. Xenografts were obtained and photographed after implantation for 28 days. D. Tumor weight was obtained and presented as the mean ± SD (n = 5). E. TUNEL method was used to detect the apoptosis of tumor tissues.
Effect of shRNA survivin on the expression of downstream molecules (ki67 and bcl2)
To understand the regulation of survivin on its downstream genes, we extracted RNA and proteins of the tumor tissues. RT-PCR and Western blot showed that the RNA (Figure 2A and 2B) and protein (Figure 2C and 2D) expression levels of bcl2 and ki67 decreased in the shRNA survivin group in vivo compared with those in the blank group and the shRNA control group.
Figure 2.
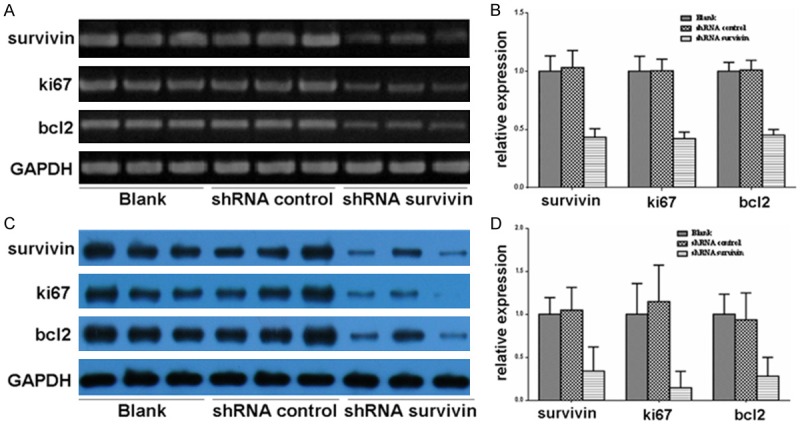
Effects of survivin on the expression of downstream molecules. A and B. The expression levels of survivin, bcl2, ki67, and β-actin were analyzed in the tumor tissues by RT-PCR. C and D. The expression levels of survivin, bcl2, ki67, and β-actin were analyzed in the tumor tissues by Western blot.
Discussion
Apoptosis (programmed cell death) is a highly conserved and strictly regulated biological process that is essential for the maintenance of normal tissue homeostasis [15]. Abnormalities in the control of apoptosis play an important role in tumorigenesis, and inhibition of apoptosis is a common property of cancer cells, increasing their survival and facilitating their escape from immune surveillance and resistance to drug and radiation therapy [3].
Survivin, a member of the IAP family, has been shown to be overexpressed in all human cancer types’ studies, including CRC. Huang YJ. et al. conducted a meta-analysis of 14 studies including 1784 patients with CRC, and their results showed that survivin overexpression in patients with CRC is significantly associated with poor overall survival and disease-free survival [7]. Previous studies indicated that survivin is a regulator of spindle microtubule function during mitosis and an inhibitor of apoptosis [3]. The downregulation of survivin expression by antisense oligonucleotides has been shown to significantly inhibit cell growth and induce apoptosis in melanoma [16] and endometrial cancer [17]. In our study, to reveal the role of survivin in the development of CRC, we inhibited survivin expression in human CRC cells by RNAi. Transfection with survivin shRNA resulted in a decrease of 50%-60% in the expression of both survivin mRNA and survivin protein, whereas transfection with the black group and the shRNA control group failed to reduce the expression of survivin mRNA or protein. Meanwhile, in animal experiments, the sizes and weights of xenografts from the two control groups were much bigger than those from the shRNA survivin group. TUNEL staining showed that the number of apoptotic cells of the shRNA survivin group was greater than that of the two control groups. These data indicated that inhibition of survivin resulted in a rapid induction of apoptosis and reduced cell proliferation in CRC cells.
We found compelling evidence that the presence of survivin in CRC was strongly associated with the expression of bcl2 and ki67. In our study, RT-PCR and Western blot analysis showed decreased bcl2 and ki67 levels in cells transfected with shRNA survivin compared with cells transfected with the black and shRNA survivin control. These results indicated that low expression of survivin inhibited the expression of bcl2 and ki67. Tanaka et al. used immunohistochemical analysis and Cox proportional hazard model to demonstrate that the expression of survivin is significantly associated with bcl2 expression and reduced apoptotic indices, which are strongly associated with poor prognosis in breast cancer [18]. Bcl2 was the first protein shown to lead to tumor initiation and progression by preventing apoptosis. The localization of bcl2 in normal colonic epithelium is crypt bases, suggesting that bcl2 may be physiologically important for the viability of regenerating stem cells [19]. Hague A. et al. observed overexpression of bcl2 in colorectal adenomas and carcinomas, implying a role for this oncoprotein in apoptotic inhibition in colorectal neoplasia [20]. Both survivin and bcl2 genes are regulated by TATA-less, GC-rich promoter sequence in similar manners, and both are markedly transcribed in actively proliferating cell types [21]. However, our results suggested that bcl2 may be a downstream gene of survivin. Regardless of the pathway of co-expression, survivin and bcl2 proteins may mediate non-overlapping antiapoptotic mechanisms. In CRC and in many of the other human cancers, the expression of survivin plus other antiapoptotic genes, such as bcl2, may cause more distinguished antiapoptotic effects. Recently, several in vitro studies showed that simultaneous gene silencing of bcl2, survivin, and other antiapoptotic genes results in a significantly higher induction of apoptosis and a decrease in tumor growth in pancreatic cancer cells and bladder cancer cells; these findings implied that survivin and bcl2 are interesting candidates for target-directed molecular-based antitumor therapy [22,23].
The nuclear protein ki67 is an established prognostic and predictive indicator for human cancers. Clinically, ki67 has been shown to correlate with metastasis and clinical stage of tumors. Marian et al. found a significant positive correlation between immunoexpression of survivin and the ki67 index [24]. Similarly, our results demonstrated that survivin expression was related to CRC proliferation. No study has documented such a relationship using RT-PCR and Western blot analysis for ki67 expression. In addition, Thomas W. et al. found that nuclear staining of ki67, P21, and survivin is significantly associated with disease-specific survival and increased predictive ability in a multivariate model of clear cell renal cell carcinoma [25]. Assessment of these markers, such as survivin, bcl2, and ki67, in combination with other conventional prognostic factors could help identify the prognostic group more accurately for patients with CRC.
Acknowledgements
This study was supported by the National Science Foundation of China (No. 81201536).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 3.LaCasse EC, Baird S, Korneluk RG, MacKenzie AE. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene. 1998;17:3247–3259. doi: 10.1038/sj.onc.1202569. [DOI] [PubMed] [Google Scholar]
- 4.Sah NK, Khan Z, Khan GJ, Bisen PS. Structural, functional and therapeutic biology of survivin. Cancer Lett. 2006;244:164–171. doi: 10.1016/j.canlet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Reed JC, Bischoff JR. BIRinging chromosomes through cell division and survivin the experience. Cell. 2000;102:545–548. doi: 10.1016/s0092-8674(00)00076-3. [DOI] [PubMed] [Google Scholar]
- 6.Waligorska-Stachura J, Jankowska A, Wasko R, Liebert W, Biczysko M, Czarnywojtek A, Baszko-Błaszyk D, Shimek V, Ruchała M. Survivin-prognostic tumor biomarker in human neoplasms-review. Ginekol Pol. 2012;83:537–540. [PubMed] [Google Scholar]
- 7.Williams NS, Gaynor RB, Scoggin S, Verma U, Gokaslan T, Simmang C, Fleming J, Tavana D, Frenkel E, Becerra CI. Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res. 2003;9:931–946. [PubMed] [Google Scholar]
- 8.Huang YJ, Qi WX, He AN, Sun YJ, Shen Z, Yao Y. The prognostic value of survivin expression in patients with colorectal carcinoma: a meta-analysis. Jpn J Clin Oncol. 2013;43:988–995. doi: 10.1093/jjco/hyt103. [DOI] [PubMed] [Google Scholar]
- 9.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 10.Klaus A, Birchmeier W. Wet signaling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Rao DD, Senzer N, Nemunaitis J. RNA interference and cancer therapy. Pharm Res. 2011;28:2983–2995. doi: 10.1007/s11095-011-0604-5. [DOI] [PubMed] [Google Scholar]
- 12.Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat Biotechnol. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 13.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 14.Senzer N, Barve M, Kuhn J, Melnyk A, Beitsch P, Lazar M, Lifshitz S, Magee M, Oh J, Mill SW, Bedell C, Higgs C, Kumar P, Yu Y, Norvell F, Phalon C, Taquet N, Rao DD, Wang Z, Jay CM, Pappen BO, Wallraven G, Brunicardi FC, Shanahan DM, Maples PB, Nemunaitis J. Phase I trial of “bi-shRNAi(furin)/GMCSF DNA/autologous tumor cell” vaccine (FANG) in advanced cancer. Mol Ther. 2012;20:679–686. doi: 10.1038/mt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 16.Grossman D, Kim PJ, Schechner JS, Altieri DC. Inhibition of melanoma tumor growth in vivo by survivin targeting. Proc Natl Acad Sci U S A. 2001;98:635–640. doi: 10.1073/pnas.230450097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ai Z, Yin L, Zhou X, Zhu Y, Zhu D, Yu Y, Feng Y. Inhibition of survivin reduces cell proliferation and induces apoptosis in human endometrial cancer. Cancer. 2006;107:746–756. doi: 10.1002/cncr.22044. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res. 2000;6:127–134. [PubMed] [Google Scholar]
- 19.Hockenbery DM, Zutter M, Hickey W, Nahm M, Korsmeyer SJ. BCL2 protein is topographically restricted in tissues characterized by apoptotic cell death. Proc Natl Acad Sci U S A. 1991;88:6961–6965. doi: 10.1073/pnas.88.16.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hague A, Moorghen M, Hicks D, Chapman M, Paraskeva C. BCL-2 expression in human colorectal adenomas and carcinomas. Oncogene. 1994;9:3367–3370. [PubMed] [Google Scholar]
- 21.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 22.Rückert F, Samm N, Lehner AK, Saeger HD, Grützmann R, Pilarsky C. Simultaneous gene silencing of Bcl-2, XIAP and Survivin re-sensitizes pancreatic cancer cells towards apoptosis. BMC Cancer. 2010;10:379. doi: 10.1186/1471-2407-10-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunze D, Kraemer K, Erdmann K, Froehner M, Wirth MP, Fuessel S. Simultaneous siRNA-mediated knockdown of antiapoptotic BCL2, Bcl-xL, XIAP and survivin in bladder cancer cells. Int J Oncol. 2012;41:1271–1277. doi: 10.3892/ijo.2012.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danilewicz M, Stasikowska-Kanicka O, Wągrowska-Danilewicz M. Augmented immunoexpression of survivin correlates with parameters of aggressiveness in prostate cancer. Pol J Pathol. 2015;66:44–48. doi: 10.5114/pjp.2015.51152. [DOI] [PubMed] [Google Scholar]
- 25.Weber T, Meinhardt M, Zastrow S, Wienke A, Fuessel S, Wirth MP. Immunohistochemical analysis of prognostic protein markers for primary localized clear cell renal cell carcinoma. Cancer Invest. 2013;31:51–59. doi: 10.3109/07357907.2012.749267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


