Abstract
Background/Aims: Gemcitabine (GEM) is the first-line chemotherapy in patients with unresectable pancreatic cancer. However, the clinical outcomes of this regimen are still unsatisfactory in prolonging survival. Resistant to GEM is one of the reasons for poor prognosis. Therefore, looking for molecular biomarkers to predict chemosensitivity to GEM is important for treatment in unresectable pancreatic cancer patients. The aim of this study was to analyze S100A4 mRNA in tissues of unresectable pancreatic cancer obtained by endoscopic ultrasound-guided fine-needle aspiration biopsy (EUS-FNA), and to determine the relation between S100A4 mRNA level and chemosensitivity to GEM. Methods: The analysis was performed on samples from 36 patients with unresectable pancreatic cancer who were treated with gemcitabine alone. The patients were assigned to receive GEM at 1,000 mg/m2/wk for weeks 1 to 6, followed by 1 week rest, then for 4 weeks. mRNA was extracted for S100A4 mRNA assay from patients above by EUS-FNA before GEM-treatment. The 36 patients were divided into the following two groups. Patients with partial response and those with stable disease whose tumor markers decreased by 50% or more were classified as the effective group. The rest of patients were classified as the non effective group. The relationship between GEM efficacy and S100A4 mRNA expression was then examined by chi-squared test. Results: S100A4 mRNA showed a significant correlation with GEM efficacy. Patients in the effective group had low S100A4 mRNA expression, whereas patients in non-effective group had high S100A4 mRNA expressions (P = 0.0059). Conclusion: S100A4 mRNA level analyzed in EUS-FNA samples is an important molecular biomarker for prediction of chemosensitivity to GEM in unresectable pancreatic cancer.
Keywords: Unresectable pancreatic cancer, gemcitabine, S100A4
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of death resulting from cancer [1]. It has the exceedingly high rates of distant metastatic recurrence even after successful surgical resection of early-stage tumors. Since 1997, gemcitabine (GEM) has been the standard treatment for unresectable pancreatic cancer. GEM results in a tumor response rate of 5-27% and offers a median survival time of 5-8 months [2]. Unfortunately, this means that the best current treatment offers very modest benefits.
PDAC patients receive little benefit from GEM largely because most PDAC cells are resistant to chemotherapeutic agent GEM. Endogenous gene in PDAC cells might confer resistance to GEM chemotherapy. Therefore, it is important for us to find a useful marker for predicting response to GEM in PDAC, especially in unresectable pancreatic cancer.
S100A4 was cloned in the 1980s and early 1990s from various cell systems [3,4]. It has been identified as a cytoplasmic protein in normal cells, which is naturally expressed in various cell types including both cancer and normal cells, and its elevation is usually associated with cell invasion and metastasis [5-8], cell death and apoptosis [9,10], chemotherapy resistance and poor clinical outcome [11,12].
Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) is widely used for diagnosis of pancreatic lesions. Many studies have explored using endoscopic ultrasound-guided fine needle aspiration of pancreatic cancer tissue to identify molecular targets. Grading of PNETs by the highest Ki-67 index in EUS-FNA specimens with adequate cellularity has a high concordance with grading of resected specimens, and can predict long term patient survival with high accuracy [13]. EUS-FNA is capable of non-operative detection of ZIP4, thus offering the potential to direct pre-operative detection and targeted therapy of PDAC [14]. Camus et al. has found that Combined EUS-FNA and biliary and/or duodenal stenting is feasible in almost all patients with suspected pancreatic cancer, with no additional hazard and a high histological yield [15].
In pancreatic cancer, S100A4 could be a marker for malignancy in pancreatic tumors and for poor prognosis in patients with pancreatic cancer [16]. Kozono S et al. has reported S100A4 mRNA expression may predict radioresistance of pancreatic cancer cells and may play an important role in the poor response of pancreatic cancer cells to radiation therapy [17]. In the present study, we used the endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) technique to acquire samples of unresectable pancreatic cancer, and to identify possible predictors of chemosensitivity to GEM by S100A4 mRNA analysis using Q-PCR.
Materials and methods
Patients
36 patients with unresectable adenocarcinoma of the pancreas were selected. All the patients were not amenable for complete surgical resection based on clinical or radiogrephic evaluation (laparascopy was not required). Patients with small-cell carcinoma, mucinous cystadenocarcinoma, or islet cell or papillary cystic neoplasm were not eligible. Patients must also have been at least 18 years of age, and received no prior chemotherapy or radiotherapy.
The 36 patients with unresectable pancreatic cancer, who were admitted to Oncology, the affiliated hospital of Qingdao University from June 1999 to June 2010, and were planned to treat with GEM mono therapy, were consecutively entered into this study. GEM mono therapy was performed for all patients by administering intravenous GEM at 1,000 mg/m2/wk for weeks 1 to 6, followed by 1 week rest, then for 3 of 4 weeks. The patients were assessed for definitive GEM efficacy, and were thus investigated for correlations between GEM sensitivity and S100A4 mRNA levels. Clinicopathologic data for the 36 patients are shown in Table 1. Evaluation of response to GEM by imaging study was based on the Response Evaluation Criteria in Solid Tumors (RECIST). The GEM-effective patients were defined as having a partial response (PR) by imaging studies or as having stable disease (SD) by imaging studies and a 50% or more decrease in both of abnormal CA 19-9 and CEA titers in sera, as compared to pretreatment values. Informed consent to use the specimens for this study according to the institutional rules of the hospital was obtained from all subjects.
Table 1.
Clinical characteristics of patients receiving GEM monotherapy
| No. of patients | 36 | |
| Age (Y) | Mean ± SD (Range) | 64.6 ± 9.3 (42-71) |
| Gender | Male:Female | 16:20 |
| Location | Head:Body/tail | 9:27 |
| Follow-up time from commencement of GEM (month) | ||
| Median (Range) | 8.1 (3.7-22.5) | |
| Number of courses of GEM monotherapy | ||
| Mean ± SD (Range) | 6.1 ± 4.0 (2-17) | |
| GEM efficacy | Effective*:Non-effective | 7:29 |
GEM, gemcitabine;
Effective, partial response by imaging study or stable disease by imaging study with 50% or more decrease in tumor markers compared to pretreatment values.
Endoscopic ultrasound-guided fine-needle aspiration biopsy (EUS-FNA) procedure
36 patients with un-resectable pancreatic ductal cancers treated with GEM were studied. EUS-FNA was performed as the procedures described before [18]: Briefly, the lesion was recognized on B-mode imaging. No vessels in the target area were confirmed with the color Doppler mode. After determination of the adequate angle to the tumor, an aspiration needle was introduced into the lesion. While the catheter connected to the needle was sucked by a 20 ml syringe, the needle was moved back and forth 20-30 times within the tumor. The negative pressure was released before the needle was removed from the lesion. To obtain sufficient tissue for RNA extraction and pathological diagnosis, several biopsy specimens were collected from each tumor by EUS-FNA using 19 or 22-gauge aspiration needles. A 19-gauge needle can take more amount of specimen than a 22-gauge needle. However, a 22-gauge needle gives less damage to tissue than a 19-gauge needle and can take enough specimens for the diagnosis and the analysis. We used 19-gauge needles for the first nine cases. For the following 26 cases, the tissues were obtained by 22-gauge needles. A cytopathologist immediately examined the specimens for cancer cells using part of the obtained tissue.
RNA extraction and quantitative PCR
To ensure RNA quality, the obtained tissue was instantly immersed in 1 ml of RNAlater (Ambion, Austin, TX, USA) and incubated overnight in reagent at 4°C. RNA was isolated from tissue samples by using standard RNAzol procedures. For reverse transcription-PCR, complementary DNA (cDNA) was synthesized in a 20-uL reaction mixture with 1 μg of RNA, as described in the protocol (AB Gene Reverse Transcription System; ABGene, Surrey, UK). The Q-PCR system used was the Amplofluor Uniprimer system (Intergen Company, Oxford, UK) and Thermo-Start (ABgene, Epsom, Surrey, UK). Specific primer pairs for S100A4 were designed by the authors by using Beacon Designer software and were manufactured by Shenggong, Shanghai, China. Q-PCR conditions was as follows: enzyme activation at 95°C for 12 min for 1 cycle, followed by 60 cycles of denaturing at 94°C for 15 s, annealing at 55°C for 35 s, and extension at 72°C for 30 s. The levels of molecule cDNA (copies per 50 ng of RNA) in the samples were calculated. Q-PCR for β-actin was also performed on the same samples to correct for any residual differences in the initial level of RNA in the specimens (in addition to spectrophotometry). The products of Q-PCR were verified on agarose gels.
Statistical analysis
The relationship between mRNA expression and GEM efficacy was examined by chi squared test (Fisher’s exact test). Survival data were estimated by the Kaplan-Meier method and were examined by log-rank test. For all tests, one-sided P values <0.05 were defined as statistically significant. The SPSS software program was used for the analysis.
Results
Clinical outcome
When GEM efficacy was named as PR or SD with a 50% or more decrease in tumor markers compared to baseline, 7 patients were classified into the effective group, and 29 patients were classified into the uneffective group (Table 1). There was a significant difference between the survival periods of the effective and the non-effective groups (Median survival time, 15.4 months vs. 8.3 months, respectively; P = 0.016) (Figure 1).
Figure 1.
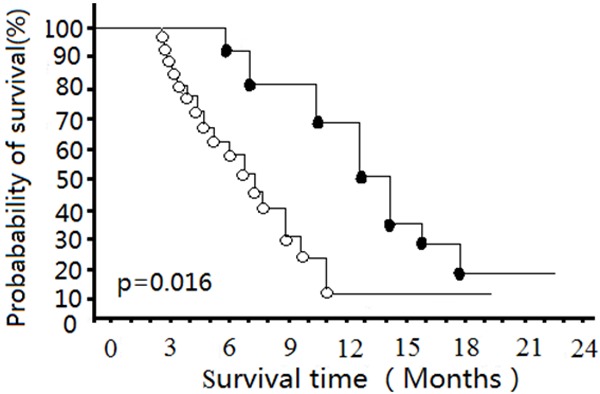
Probability of survival for patients with unresectable pancreatic ductal cancer stratified by GEM efficacy. Closed circles, GEM-effective group. Open circles, GEM-non-effective group. There is a significant difference between survivals in the two groups.
S100A4 mRNA show increased expression in GEM-non-effective group
In GEM-non-effective group, the average RNA value of the 29 samples (expressed as transcript copy number per 50 μg of messenger RNA and standardized with β-actin) was 16.29 ± 1.84; In GEM-effective group, the average RNA value of the 7 samples (expressed as transcript copy number per 50 ng of messenger RNA and standardized with β-actin) was 2.18 ± 0.47 (Figure 2A, P = 0.0059). Agarose gel indicated that low S100A4 mRNA was expressed in GEM-effective group, and high S100A4 mRNA was expressed in GEM-uneffective group (Figure 2B).
Figure 2.

Levels of transcripts of S100A4 in tumor samples of GEM-non-effective group and GEM-effective group (expressed as transcript copy number per 50 ng of messenger RNA and standardized with β-actin). *P = 0.0059.
Discussion
Endoscopic ultrasound-guided fine-needle aspiration biopsy (EUS-FNA) is widely performed as a histological and cytological diagnostic method for pancreatic carcinoma (PC) [19,20]. However, few reports on gene analysis of PC using EUS-FNA samples have been used. Furthermore, much few reports on assessment of chemosensitivity to GEM from patients with unresectable PC using EUS-FNA samples have been used [21].
S100A4 expression could be a marker for malignancy and prediction radioresistance of PC [22,23], but it is unclear whether S100A4 mRNA using EUS-FNA samples in patients with unresectable PC could be a marker for assessment of chemosensitivity to GEM.
The objective response rate of GEM monotherapy for PC has been reported to be 5-26% [2]. In this study, PR was observed in 10 of 36 (25.8%) patients treated with GEM monotherapy, which was correlated with the response rates reported previously [2]. There was a significant difference between the survival periods of the effective and the non-effective groups (Median survival time, 15.4 months vs. 8.3 months, respectively; P = 0.016). CA 19-9 has been demonstrated to be correlated with clinical efficacy of GEM in pancreatic cancer. In this study, the GEM-effective group had a significantly better prognosis than the non-effective group, indicating that the grouping based on GEM efficacy was appropriate.
In the GEM-effective group, the average RNA value was 2.18 ± 0.47, which was significantly lower compared to the GEM-non-effective group (16.29 ± 1.84, P = 0.0059). RNA extraction and quantitative PCR of S100A4 mRNA analysis may therefore be suitable to predict GEM efficacy by using a samples taken by EUS-FNA from unresectable pancreatic cancer. Our results suggested that high S100A4 mRNA expression is a predictor of GEM resistance. However, the contamination of normal tissue into tumor tissue obtained by EUS-FNA may be a major obstacle to an accurate analysis.
In conclusion, S100A4 mRNA expression in EUS-FNA biopsy specimens may be a predictor for response to GEM in patients with unresectable pancreatic cancer. These data may be helpful for future cancer treatments that target specific molecules.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Ward E. Cancer Statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.O’Reilly EM, Abou-Alfa GK. Cytotoxic therapy for advanced pancreatic adenocarcinoma. Semin Oncol. 2007;34:347–353. doi: 10.1053/j.seminoncol.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Garrett SC, Varney KM, Weber DJ, Bresnick AR. S100A4, a mediator of metastasis. J Biol Chem. 2006;281:677–680. doi: 10.1074/jbc.R500017200. [DOI] [PubMed] [Google Scholar]
- 4.Mazzucchelli L. Protein S100A4: too long overlooked by pathologists? Am J Pathol. 2002;160:7–13. doi: 10.1016/S0002-9440(10)64342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natarajan J, Hunter K, Mutalik VS, Radhakrishnan R. Overexpression of S100A4 as a biomarker of metastasis and recurrence in oral squamous cell carcinoma. J Appl Oral Sci. 2014;22:426–33. doi: 10.1590/1678-775720140133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D, Zheng XF, Yang ZY, Liu DX, Zhang GY, Jiao XL, Zhao H. S100A4 silencing blocks invasive ability of esophageal squamous cell carcinoma cells. World J Gastroenterol. 2012;18:915–22. doi: 10.3748/wjg.v18.i9.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukamoto N, Egawa S, Akada M, Abe K, Saiki Y, Kaneko N, Yokoyama S, Shima K, Yamamura A, Motoi F, Abe H, Hayashi H, Ishida K, Moriya T, Tabata T, Kondo E, Kanai N, Gu Z, Sunamura M, Unno M, Horii A. The expression of S100A4 in human pancreatic cancer is associated with invasion. Pancreas. 2013;42:1027–33. doi: 10.1097/MPA.0b013e31828804e7. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Fu S, Xu Y, Zheng Z. RNA interference targeting inhibition of S100A4 suppresses cell growth and promotes apoptosis in human laryngeal carcinoma Hep-2 cells. Mol Med Rep. 2014;10:1389–94. doi: 10.3892/mmr.2014.2345. [DOI] [PubMed] [Google Scholar]
- 9.Mahon PC, Baril P, Bhakta V, Chelala C, Caulee K, Harada T, Lemoine NR. S100A4 contributes to the suppression of BNIP3 expression, chemoresistance, and inhibition of apoptosis in pancreatic cancer. Cancer Res. 2007;67:6786–95. doi: 10.1158/0008-5472.CAN-07-0440. [DOI] [PubMed] [Google Scholar]
- 10.Roh J, Knight S, Chung JY, Eo SH, Goggins M, Kim J, Cho H, Yu E, Hong SM. S100A4 expression is a prognostic indicator in small intestine adenocarcinoma. J Clin Pathol. 2014;67:216–21. doi: 10.1136/jclinpath-2013-201883. [DOI] [PubMed] [Google Scholar]
- 11.Niu Y, Wang L, Cheng C, Du C, Lu X, Wang G, Liu J. Increased expressions of SATB1 and S100A4 are associated with poor prognosis in human colorectal carcinoma. APMIS. 2015;123:93–101. doi: 10.1111/apm.12310. [DOI] [PubMed] [Google Scholar]
- 12.Cao CM, Yang FX, Wang PL, Yang QX, Sun XR. Clinicopathologic significance of S100A4 expression in osteosarcoma. Eur Rev Med Pharmacol Sci. 2014;18:833–839. [PubMed] [Google Scholar]
- 13.Lee SH, Kim H, Hwang JH, Shin E, Lee HS, Hwang DW, Cho JY, Yoon YS, Han HS, Cha BH. CD24 and S100A4 expression in resectable pancreatic cancers with earlier disease recurrence and poor survival. Pancreas. 2014;43:380–8. doi: 10.1097/MPA.0000000000000097. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Liu H, Pan H, Du Q, Liang J. Clinicopathological significance of S100A4 expression in human hepatocellular carcinoma. J Int Med Res. 2013;41:457–62. doi: 10.1177/0300060513478086. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa T, Yamao K, Hijioka S, Bhatia V, Mizuno N, Hara K, Imaoka H, Niwa Y, Tajika M, Kondo S, Tanaka T, Shimizu Y, Kinoshita T, Kohsaki T, Nishimori I, Iwasaki S, Saibara T, Hosoda W, Yatabe Y. Evaluation of Ki-67 index in EUS-FNA specimens for the assessment of malignancy risk in pancreatic neuroendocrine tumors. Endoscopy. 2014;46:32–8. doi: 10.1055/s-0033-1344958. [DOI] [PubMed] [Google Scholar]
- 16.Xu C, Wallace MB, Yang J, Jiang L, Zhai Q, Zhang Y, Hong C, Chen Y, Frank TS, Stauffer JA, Asbun HJ, Raimondo M, Woodward TA, Li Z, Guha S, Zheng L, Li M. ZIP4 is a novel diagnostic and prognostic marker in human pancreatic cancer: a systemic comparison between EUS-FNA and surgical specimens. Diagn Cytopathol. 2014;42:751–8. doi: 10.2174/1566524013666131217112921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camus M, Trouilloud I, Villacis AL, Mangialavori L, Duchmann JC, Gaudric M, Roseau G, Terris B, Mitry E, Chaussade S, Prat F. Effectiveness of combined endoscopic ultrasound-guided fine-needle aspiration biopsy and stenting in patients with suspected pancreatic cancer. Eur J Gastroenterol Hepatol. 2012;24:1281–7. doi: 10.1097/MEG.0b013e328357cdfd. [DOI] [PubMed] [Google Scholar]
- 18.Ikenaga N, Ohuchida K, Mizumoto K, Yu J, Fujita H, Nakata K, Ueda J, Sato N, Nagai E, Tanaka M. S100A4 mRNA is a diagnostic and prognostic marker in pancreatic carcinoma. J Gastrointest Surg. 2009;13:1852–8. doi: 10.1007/s11605-009-0978-4. [DOI] [PubMed] [Google Scholar]
- 19.Kozono S, Ohuchida K, Ohtsuka T, Cui L, Eguchi D, Fujiwara K, Zhao M, Mizumoto K, Tanaka M. S100A4 mRNA expression level is a predictor of radioresistance of pancreatic cancer cells. Oncol Rep. 2013;30:1601–8. doi: 10.3892/or.2013.2636. [DOI] [PubMed] [Google Scholar]
- 20.Wiersema MJ, Kochman ML, Cramer HM, Tao LC, Wiersema LM. Endosonograpy-guided real-time fine-needle aspiration biopsy. Gastrointest Endosc. 1994;40:700–707. [PubMed] [Google Scholar]
- 21.Khalid A, Nodit L, Zahid M, Bauer K, Brody D, Finkelstein SD, McGrath KM. Endoscopic ultrasound fine needle aspiration DNA analysis to differentiate malignant and benign pancreatic masses. Am J Gastroenterol. 2006;101:2493–2500. doi: 10.1111/j.1572-0241.2006.00740.x. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K, Yamao K, Okubo K, Sawaki A, Mizuno N, Ashida R, Koshikawa T, Ueyama Y, Kasugai K, Hase S, Kakumu S. Differential diagnosis of pancreatic cancer and focal pancreatitis by using EUS-guided FNA. Gastrointest Endosc. 2005;61:76–79. doi: 10.1016/s0016-5107(04)02224-2. [DOI] [PubMed] [Google Scholar]
- 23.Tada M, Komatsu Y, Kawabe T, Sasahira N, Isayama H, Toda N, Shiratori Y, Omata M. Quantitative analysis of K-ras gene mutation in pancreatic tissue obtained by endoscopic ultrasonography-guided fine needle aspiration: Clinical utility for diagnosis of pancreatic tumor. Am J Gastroenterol. 2002;97:2263–2270. doi: 10.1111/j.1572-0241.2002.05980.x. [DOI] [PubMed] [Google Scholar]


