Abstract
Background: It is well-established that differences among ethnic groups in drug responses are primarily due to the genetic diversity of pharmacogenes. A number of genes or variants that play a crucial role in drug responses have been designated Very Important Pharmacogenes (VIP) by the PharmGKB database. Clarifying the polymorphic distribution of VIPs in different ethnic groups will aid in personalized medicine for specific populations. Methods: We sequenced 85 VIP variants in the Lhoba population based on the PharmGKB database. The polymorphic distribution of the 85 VIP variants in 100 Lhoba subjects was determined and compared with that of 11 major HapMap populations, including ASW, CEU, CHB, CHD, GIH, JPT, LWK, MEX, MKK, TSI, and YRI. We used χ2 tests to identify significantly different loci between these populations. We downloaded SNP allele frequencies from the ALlele FREquency Database to observe the global genetic variation distribution for these specific loci. And then we used Structure software to perform the genetic structure analysis of 12 populations. Results: Based on comparisons of selected available loci, we found that 23, 28, 16, 10, 20, 16, 24, 19, 22, 21 and 36 of the selected VIP variant genotype frequencies in the Lhoba population differed from those of the ASW, CEU, CHB, CHD, GIH, JPT, LWK, MEX, MKK, TSI, and YRI populations, respectively. In addition, Pairwise FST values and clustering analyses also showed the VIP variants in Lhoba exhibited a close genetic affinity with CHD, CHB and JPT populations. Conclusion: Our results complement pharmacogenomic data on the Lhoba ethnic group and may be helpful in the diagnosis of certain diseases in minorities.
Keywords: Genetic polymorphism, pharmacogenomics, VIP variants, Lhoba
Introduction
The goals of personalized medicine are to maximize therapeutic efficacy and minimize the risk of drug toxicity for an individual patient. Pharmacogenomics plays a major role in personalized medicine. Pharmacogenetics is the study of how genetic variation influences the response to drugs. Race, ethnicity, and ancestry have a strong influence on pharmacogenetics and on our understanding of population-level differences in drug response [1,2]. The primary goal of pharmacogenetics, however, is to identify the individual genetic determinants of drug activity so that therapy can be tailored to the individual patient. The Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB: http://www.pharmgkb.org) was created in order to aggregate data from the primary research literature and distill and curate that information in multiple forms [3,4].
A number of studies have highlighted the importance of ethnicity in influencing genetic variability. China is a multinational country, with 55 minority ethnicities, including the Lhoba ethnic minority. The 2,965 people of the Lhoba ethnic minority reside primarily in Mainling, Medog, Lhunze, and Nangxian counties in southeastern Tibet. Additionally, a small number live in Luoyu in southern Tibet. Because of differences in genetics, physiology, pathology, eating habits, living environment, and nutritional status, the same drug dosage regimen for every ethnic group may be inappropriate [5]. Genetic variations, such as single nucleotide polymorphisms (SNPs), play an important role clinically [6]. SNPs have been shown to alter enzyme activity, resulting in abnormally increased or decreased metabolism of drugs [7]. For instance, it has been estimated that CYP2C9 is responsible for the metabolic clearance of up to 15-20% of all drugs undergoing phase I metabolism [8]. Significant differences are found in CYP2C9*2 allele frequencies in different populations. For example, the CYP2C9*2 allele frequency in Spain is 26.7% but zero in east Asia [9]. Genotyping all patients for relevant genetic variants before beginning a drug treatment is not feasible. As an alternative, population allele frequency data may be able to substitute for individual genotyping. However, minimal relevant data are available for minorities. The goal of this study was to address the lack of data regarding the allele frequencies of clinically relevant SNPs within the Lhoba population. Our results complement current pharmacogenomic data on the Lhoba ethnic group. Further genotyping studies of specific populations are necessary to provide the best medical care to all individuals.
Materials and methods
Study participants
We recruited a random sample of 100 unrelated Lhoba adults from the Xinjiang Region of China. In general, all control subjects were healthy, without chronic diseases or disorders related to the vital organs. To ensure that controls were cancer-free, they were tested for the presence of plasma carcinoembryonic antigen and alpha-fetoprotein. In addition, we selected individuals who had exclusively Lhoba ancestry for at least the previous three generations. All participants signed informed consent forms, after which 5 ml of peripheral blood was drawn from each subject. The study protocol was approved by the Clinical Research Ethics Committees of Northwest University.
Variant selection and genotyping
We selected genetic variants from published polymorphisms associated with VIP variants from the PharmGKB database and excluded loci that could not be designed. A total of 86 genetic variant loci were selected. Genomic DNA was extracted from whole blood using a GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag Co. Ltd., Xi’an City, China). The DNA concentration was measured using a NanoDrop 2000 (Thermo Scientific, Waltham, Massachusetts, USA). Sequenom MassARRAY Assay Design 3.0 software (Sequenom, San Diego, California, USA) was used to design a Multiplexed SNP MassEXTEND assay (Sequenom) [10]. A Sequenom Mass ARRAY RS1000 was used to genotype SNPs using the standard protocol recommended by the manufacturer [10]. Finally, Sequenom Typer 4.0 software was used for data management and analysis [10,11]. Laboratory personnel were blinded to genotyping results.
Statistical analyses
Microsoft Excel and SPSS 19.0 statistical software (SPSS, Chicago, IL) were used to perform Hardy-Weinberg equilibrium (HWE) and chi-square tests. All p values in this study were two-sided and a p value of ≤ 0.05 after Bonferroni correction was considered the threshold for statistical significance. The Bonferroni correction was applied to determine the p value threshold of significance: 0.05/(74*4). Validation of the frequency of each variant in the Lhoba population was tested for departure from the HWE using an exact test. We calculated and compared the genotype frequencies in the Lhoba population with those of 11 other populations (African ancestry in the southwestern USA (ASW); a northwestern European population (CEU); the Han Chinese in Beijing, China (CHB); a Chinese population of metropolitan Denver, Colorado, USA (CHD); the Gujarati Indians in Houston, Texas, USA (GIH); the Japanese population in Tokyo, Japan (JPT); the Luhya people in Webuye, Kenya (LWK); people of Mexican ancestry living in Los Angeles, California, USA (MEX); the Maasai people in Kinyawa, Kenya (MKK); the Tuscan people of Italy (TSI); and the Yoruba in Ibadan, Nigeria (YRI). (data from HapMap: http://hapmap.ncbi.nlm.nih.gov) separately using a chi-square test. The program Arlequin v3.5 was used to calculate global FST together with pairwise FST values among all of the populations using loci that were polymorphic. The Structure (version 2.3.4) software were used to analysis the genetic structure within a hypothetical K number in 12 populations.
Results
Basic information about the selected VIP loci of the Lhoba population listed in Table 1. Twelve loci were disregarded in our study because genotype data were lacking or the call rate was < 90%. In addition, 27 loci were removed from the analyses of VIP variants for lack of genotype results in the HapMap database, and therefore, data for comparisons were unavailable.
Table 1.
Basic characteristics of selected variants in the Lhoba
| SNP ID | Genes | Allele | Categories | Lhoba | HWE | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| A | B | Family | Phase | MAF | AA-Count | AB-Count | BB-Count | |||
| rs10264272 | CYP3A5 | C | T | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs1042713 | ADRB2 | G | A | Adrenergic receptors family | Others | 0.485 | 22 | 53 | 25 | 0.542 |
| rs1042714 | ADRB2 | G | C | Adrenergic receptors family | Others | 0.145 | 2 | 25 | 73 | 0.934 |
| rs1045642 | ABCB1 | A | G | ATP-binding cassette (ABC) transporters superfamiy | Others | 0.475 | 20 | 55 | 25 | 0.304 |
| rs1051266 | SLC19A1 | T | C | Solute carrier family | Others | 0.332 | 44 | 43 | 11 | 0.919 |
| rs1065776 | P2RY1 | T | C | G-protein coupled receptor family | Others | 0.01 | 0 | 2 | 98 | 0.920 |
| rs10929302 | UGT1A10 | G | A | UDP-glucuronosyltransferase family | Phase II | 0.071 | 86 | 12 | 1 | 0.440 |
| rs1128503 | ABCB1 | A | G | ATP-binding cassette (ABC) transporters superfamiy | Others | 0.28 | 50 | 44 | 6 | 0.361 |
| rs1142345 | TPMT | T | C | Methyltransferase superfamily | Phase II | 0.015 | 97 | 3 | 0 | 0.879 |
| rs1229984 | ADH1B | T | C | Alcohol dehydrogenase family | Phase I | 0.06 | 1 | 10 | 89 | 0.256 |
| rs12720441 | KCNH2 | G | A | Eag family | Others | 0 | 100 | 0 | 0 | / |
| rs12721634 | CYP3A4 | T | C | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs1540339 | VDR | C | T | Nuclear receptor famiy | Others | 0.285 | 11 | 35 | 54 | 0.158 |
| rs1544410 | VDR | C | T | Nuclear receptor famiy | Others | 0.07 | 87 | 12 | 1 | 0.433 |
| rs1695 | GSTP1 | A | G | Glutathione S-transferase family | Phase II | 0.105 | 80 | 19 | 1 | 0.913 |
| rs17238540 | HMGCR | T | C | HMGCR | Phase I | 0 | 100 | 0 | 0 | / |
| rs17244841 | HMGCR | A | T | HMGCR | Phase I | 0 | 100 | 0 | 0 | / |
| rs1799853 | CYP2C9 | C | T | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs1800460 | TPMT | C | G | Methyltransferase superfamily | Phase II | 0 | 0 | 0 | 100 | / |
| rs1800497 | DRD2 | G | A | G-protein coupled receptor family | Others | 0.24 | 57 | 38 | 5 | 0.677 |
| rs1800888 | ADRB2 | C | T | Adrenergic receptors family | Others | 0 | 100 | 0 | 0 | / |
| rs1801030 | SULT1A2 | C | T | Sulfotransferase family | Phase II | 0 | 100 | 0 | 0 | / |
| rs1801131 | MTHFR | T | G | Methylenetetrahydrofolate reductase family | Phase I | 0.301 | 47 | 43 | 8 | 0.673 |
| rs1801133 | MTHFR | G | A | Methylenetetrahydrofolate reductase family | Phase I | 0.227 | 58 | 37 | 4 | 0.524 |
| rs1801252 | ADRB1 | A | G | Adrenergic receptors family | Others | 0.135 | 75 | 23 | 2 | 0.879 |
| rs1801253 | ADRB1 | G | C | Adrenergic receptors family | Others | 0.211 | 6 | 29 | 62 | 0.310 |
| rs1801272 | CYP2A6 | A | T | Cytochrome P450 superfamily | Phase I | 0.015 | 0 | 3 | 97 | 0.879 |
| rs1805124 | SCN5A | T | C | Sodium channel gene family | Others | 0.115 | 77 | 23 | 0 | 0.194 |
| rs2032582 | ABCB1 | A | C | ATP-binding cassette (ABC) transporters superfamiy | Others | 0.459 | 23 | 47 | 16 | 0.352 |
| rs20417 | PTGS2 | C | G | PTGS2 | Phase I | 0.025 | 2 | 1 | 97 | 0.000 |
| rs2046934 | P2RY12 | G | A | G-protein coupled receptor family | Others | 0.21 | 4 | 34 | 62 | 0.805 |
| rs2066702 | ADH1B | G | A | Alcohol dehydrogenase family | Phase I | 0 | 100 | 0 | 0 | / |
| rs2066853 | AHR | G | A | AHR | Others | 0.235 | 57 | 39 | 4 | 0.397 |
| rs2228570 | VDR | T | C | Nuclear receptor famiy | Others | 0.43 | 14 | 58 | 28 | 0.067 |
| rs2239179 | VDR | T | C | Nuclear receptor famiy | Others | 0.18 | 69 | 26 | 5 | 0.233 |
| rs2239185 | VDR | G | A | Nuclear receptor famiy | Others | 0.371 | 41 | 40 | 16 | 0.251 |
| rs2740574 | CYP3A4 | A | T | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs28371706 | CYP2D6 | C | G | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs28371725 | CYP2D6 | G | A | Cytochrome P450 superfamily | Phase I | 0.02 | 96 | 4 | 0 | 0.838 |
| rs28399433 | CYP2A6 | G | T | Cytochrome P450 superfamily | Phase I | 0.145 | 1 | 27 | 72 | 0.374 |
| rs28399444 | CYP2A6 | A | T | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs28399454 | CYP2A6 | C | T | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs28399499 | CYP2B6 | T | C | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs3211371 | CYP2B6 | C | T | Cytochrome P450 superfamily | Phase I | 0.5 | 0 | 100 | 0 | 0.000 |
| rs3745274 | CYP2B6 | G | T | Cytochrome P450 superfamily | Phase I | 0.153 | 70 | 26 | 2 | 0.818 |
| rs3782905 | VDR | G | C | Nuclear receptor famiy | Others | 0.1 | 1 | 18 | 81 | 1.000 |
| rs3807375 | KCNH2 | C | T | Eag family | Others | 0.275 | 6 | 43 | 51 | 0.433 |
| rs3814055 | NR1I2 | C | T | Nuclear receptor famiy | Others | 0.101 | 79 | 20 | 0 | 0.264 |
| rs3815459 | KCNH2 | C | T | Eag family | Others | 0.474 | 27 | 46 | 22 | 0.778 |
| rs3846662 | HMGCR | A | G | HMGCR | Phase I | 0.485 | 22 | 53 | 25 | 0.542 |
| rs3918290 | DPYD | C | T | DPYD | Phase I | 0 | 100 | 0 | 0 | / |
| rs4124874 | UGT1A10 | T | G | UDP-glucuronosyltransferase family | Phase II | 0.165 | 69 | 29 | 2 | 0.600 |
| rs4148323 | UGT1A10 | G | A | UDP-glucuronosyltransferase family | Phase II | 0.39 | 41 | 40 | 19 | 0.111 |
| rs4149056 | SLCO1B1 | T | C | Solute carrier family | Others | 0.045 | 91 | 9 | 0 | 0.637 |
| rs4244285 | CYP2C19 | G | A | Cytochrome P450 superfamily | Phase I | 0.28 | 51 | 42 | 7 | 0.677 |
| rs4986893 | CYP2C19 | G | A | Cytochrome P450 superfamily | Phase I | 0.02 | 96 | 4 | 0 | 0.838 |
| rs4986910 | CYP3A4 | A | G | Cytochrome P450 superfamily | Phase I | 0 | 99 | 0 | 0 | / |
| rs4986913 | CYP3A4 | G | A | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs5030656 | CYP2D6 | AAG | / | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs5219 | KCNJ11 | C | T | Inward-rectifier potassium channel family | Others | 0.434 | 28 | 55 | 15 | 0.158 |
| rs59421388 | CYP2D6 | C | G | Cytochrome P450 superfamily | Phase I | 0.01 | 98 | 2 | 0 | 0.920 |
| rs6025 | F5 | T | C | F5 | Others | 0 | 0 | 0 | 100 | / |
| rs61736512 | CYP2D6 | C | G | Cytochrome P450 superfamily | Phase I | 0 | 100 | 0 | 0 | / |
| rs6277 | DRD2 | G | A | G-protein coupled receptor family | Others | 0 | 100 | 0 | 0 | / |
| rs6791924 | SCN5A | G | A | Sodium channel gene family | Others | 0.005 | 99 | 1 | 0 | 0.960 |
| rs689466 | PTGS2 | T | C | PTGS2 | Phase I | 0.44 | 31 | 50 | 19 | 0.884 |
| rs698 | ADH1C | T | C | Alcohol dehydrogenase family | Phase I | 0.116 | 78 | 19 | 2 | 0.516 |
| rs701265 | P2RY1 | A | G | G-protein coupled receptor family | Others | 0.135 | 74 | 25 | 1 | 0.481 |
| rs7294 | VKORC1 | C | T | VKORC1 | Phase I | 0.055 | 90 | 9 | 1 | 0.180 |
| rs7626962 | SCN5A | G | T | Sodium channel gene family | Others | 0 | 100 | 0 | 0 | / |
| rs776746 | CYP3A5 | C | T | Cytochrome P450 superfamily | Phase I | 0.19 | 66 | 30 | 4 | 0.800 |
| rs7975232 | VDR | C | A | Nuclear receptor famiy | Others | 0.38 | 40 | 44 | 16 | 0.508 |
| rs975833 | ADH1A | G | C | Alcohol dehydrogenase family | Phase I | 0.42 | 32 | 45 | 17 | 0.865 |
| rs9934438 | VKORC1 | G | A | VKORC1 | Phase I | 0.055 | 1 | 9 | 90 | 0.180 |
Table 2 lists the genotypic frequencies of tested variants in the Lhoba population and illustrates the variants with frequencies in the Lhoba population that were significantly different from the 11 HapMap populations. Based on comparisons of selected available loci, we found that 23, 28, 16, 10, 20, 16, 24, 19, 22, 21 and 36 of the selected VIP variant genotype frequencies in the Lhoba population differed from those of the ASW, CEU, CHB, CHD, GIH, JPT, LWK, MEX, MKK, TSI, and YRI populations, respectively (P < 0.05). However, after validation using the Bonferroni correction (P <0.05/ (74×11)), the differences in frequency distribution of the variants in the Lhoba population versus the 11 populations were 14, 21, 7, 0, 13, 8, 16, 8, 18, 13 and 26 respectively. The rs3814055 (NR1I2), rs4124874 (UGT1A1), and rs2066853 (AHR) locus showed the greatest number of significant differences between Lhoba and 11 HapMap populations.
Table 2.
Lhoba compared with the11 HapMap populations after Bonferroni’s multiple adjustment
| SNP | ASW | CEU | CHB | CHD | GIH | JPT | LWK | MEX | MKK | TSI | YRI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10264272 | / | / | / | / | / | / | 1.445E-12 | / | 8.823E-08 | / | 8.95E-09 |
| rs1042713 | 0.6380937 | 0.0044716 | 0.5270236 | 0.444125 | 0.1707646 | 0.2674132 | 0.772293 | 0.5414963 | 0.811307 | 0.0016608 | 0.3529788 |
| rs1042714 | / | 6.805E-08 | 0.6277193 | / | / | 0.1785214 | / | / | / | / | 0.5692257 |
| rs1045642 | 1.001E-05 | 0.1065589 | 0.0577659 | 0.0802092 | 0.0280032 | 0.9051566 | / | 0.9568834 | 5.145E-13 | 0.6065329 | 5.413E-15 |
| rs1051266 | 0.0043283 | 6.895E-06 | 0.0063808 | 0.0003241 | 7.061E-07 | 0.1000146 | 0.2811009 | 8.062E-07 | 0.042643 | 0.000215 | 0.4522463 |
| rs1065776 | / | / | / | / | / | / | / | / | / | / | / |
| rs10929302 | / | 6.513E-06 | 0.5364754 | / | / | 0.5128563 | / | / | / | / | 3.074E-08 |
| rs1128503 | 6.812E-16 | 6.961E-08 | 0.6064446 | 0.6460303 | 0.0325144 | 0.0187387 | 3.476E-25 | 4.099E-05 | 6.934E-28 | 1.293E-07 | 3.563E-27 |
| rs1142345 | / | / | / | / | / | / | 0.0004143 | 0.0312395 | / | / | 0.2549644 |
| rs1229984 | / | 0.0289084 | 7.297E-23 | / | / | 2.618E-22 | / | / | / | / | 0.0306175 |
| rs12720441 | / | / | / | / | / | / | / | / | / | / | / |
| rs12721634 | / | / | / | / | / | / | / | / | / | / | / |
| rs1540339 | 1.703E-11 | 4.77E-10 | 0.6820561 | 0.4519886 | 8.313E-10 | 0.8053552 | 3.919E-21 | 4.041E-06 | 3.041E-23 | 1.172E-09 | 3.378E-19 |
| rs1544410 | 5.43E-05 | 2.932E-14 | 0.4616421 | 0.4491027 | 2.135E-14 | 0.060301 | 8.076E-06 | 0.0001279 | 4.388E-12 | 6.733E-13 | 1.573E-07 |
| rs1695 | 4.163E-10 | 7.76E-12 | 0.0563114 | 0.0184164 | 6.755E-07 | 0.886209 | 3.546E-16 | 1.97E-12 | 5.743E-09 | 6.537E-06 | 3.281E-10 |
| rs17238540 | / | / | / | / | / | / | / | / | / | / | / |
| rs17244841 | / | / | / | / | / | / | / | / | / | / | / |
| rs1799853 | / | / | / | / | / | / | / | / | / | / | / |
| rs1800460 | / | / | / | / | / | / | / | / | / | / | / |
| rs1800497 | 0.0082188 | 0.3206538 | 0.0010863 | 0.0002928 | 0.3810381 | 0.002664 | 0.0272732 | 0.0074558 | 0.0215364 | 0.6832066 | 0.0009302 |
| rs1800888 | / | / | / | / | / | / | / | / | / | / | / |
| rs1801030 | / | / | / | / | / | / | / | / | / | / | / |
| rs1801131 | 0.0910937 | 0.6534614 | 0.1965392 | 0.0670376 | 0.1433041 | 0.0333442 | 0.0275723 | 0.2363873 | 0.4899573 | 0.3715165 | 1.416E-05 |
| rs1801133 | 0.0010973 | 0.1472415 | 6.751E-06 | 0.0451049 | 0.3713918 | 0.0159539 | 0.0009765 | 0.002124 | 4.855E-05 | 2.086E-05 | 0.0004851 |
| rs1801252 | / | / | 0.0011173 | / | / | 0.001287 | / | / | / | / | 0.0006274 |
| rs1801253 | / | 0.1442617 | 0.4732605 | / | / | 0.4736514 | / | / | / | / | 0.0008045 |
| rs1801272 | / | 9.854E-34 | 5.38E-32 | / | / | 9.095E-31 | / | / | / | / | 1.805E-35 |
| rs1805124 | 0.0032287 | 0.1268706 | / | / | 0.0849444 | / | 0.0002433 | 0.3269952 | 1.05E-07 | 0.0057815 | 9.724E-06 |
| rs2032582 | / | / | / | / | / | / | / | / | / | / | / |
| rs20417 | / | 1.167E-29 | 2.058E-29 | / | / | 3.392E-29 | / | / | / | / | 3.207E-24 |
| rs2046934 | / | 0.8499027 | 0.9614884 | / | / | 0.8712248 | / | / | / | / | 0.6454923 |
| rs2066702 | 1.065E-10 | / | / | / | / | / | 4.081E-07 | / | / | / | 5.646E-15 |
| rs2066853 | 0.0141483 | 0.0004519 | 0.0011956 | 0.0048595 | 0.0023018 | 2.2E-05 | 1.45E-06 | 0.0472198 | 0.0013065 | 0.0003466 | 3.642E-05 |
| rs2228570 | / | / | / | / | / | / | / | / | / | / | / |
| rs2239179 | 0.0019673 | 3.597E-07 | 0.2087307 | 0.6354083 | 6.432E-09 | 0.6468278 | 0.0010861 | 0.0869567 | 9.07E-08 | 3.708E-06 | 0.0159804 |
| rs2239185 | / | / | 0.4685249 | / | / | 0.2766129 | / | / | / | / | 0.0172396 |
| rs2740574 | / | / | / | / | / | / | / | / | / | / | / |
| rs28371706 | / | / | / | / | / | / | / | / | / | / | / |
| rs28371725 | / | / | / | / | / | / | / | / | / | / | / |
| rs28399433 | / | / | / | / | / | / | / | / | / | / | / |
| rs28399444 | / | / | / | / | / | / | / | / | / | / | / |
| rs28399454 | / | / | / | / | / | / | / | / | / | / | / |
| rs28399499 | 0.0003479 | / | / | / | / | / | / | / | 0.1678005 | / | 2.037E-06 |
| rs3211371 | / | / | / | / | / | / | / | / | / | / | / |
| rs3745274 | 0.0210484 | 0.0147724 | 0.8048011 | 0.9542118 | 7.743E-07 | 0.603502 | 0.0012926 | 0.0102078 | 1.156E-06 | 0.0094913 | 6.105E-08 |
| rs3782905 | / | 2.278E-19 | 2.913E-21 | / | / | 7.343E-25 | / | / | / | / | 1.07E-22 |
| rs3807375 | 0.7813935 | 1.863E-11 | 0.7706129 | 0.9413499 | 4.484E-10 | 0.081591 | 0.0162642 | 0.0206186 | 0.9907614 | 1.658E-11 | 0.2726303 |
| rs3814055 | 2.4E-05 | 1.382E-07 | 5.46E-05 | 0.0006412 | 3.023E-11 | 0.0001352 | 2.845E-05 | 6.538E-05 | 0.0015888 | 2.053E-09 | 2.662E-05 |
| rs3815459 | / | / | 0.0007393 | / | / | 1.5E-06 | / | / | / | / | 0.1291254 |
| rs3846662 | 5.804E-09 | 0.1248809 | 0.825542 | 0.6531327 | 0.0221528 | 0.8459821 | 2.521E-20 | 0.0480714 | 5.189E-13 | 0.3144964 | 4.179E-22 |
| rs3918290 | / | / | / | / | / | / | / | / | / | / | / |
| rs4124874 | 3.298E-19 | 1.126E-08 | 0.0017157 | 0.0003381 | 2.688E-16 | 0.0010809 | 5.565E-30 | 2.673E-08 | 5.62E-33 | 2.929E-07 | 7.398E-35 |
| rs4148323 | / | 6.646E-13 | 0.0003685 | 0.0001079 | 2.382E-14 | 1.467E-06 | / | 7.694E-09 | / | / | 6.646E-13 |
| rs4149056 | 0.3727746 | 0.000974 | 0.002254 | 0.0263308 | / | 0.0638509 | / | / | 0.0257473 | 3.261E-06 | / |
| rs4244285 | / | 0.0151862 | 0.1531883 | / | / | 0.9912602 | / | / | / | / | 0.0136854 |
| rs4986893 | / | / | / | / | / | / | / | / | / | / | / |
| rs4986910 | / | / | / | / | / | / | / | / | / | / | / |
| rs4986913 | / | / | / | / | / | / | / | / | / | / | / |
| rs5030656 | / | / | / | / | / | / | / | / | / | / | / |
| rs5219 | / | / | / | / | / | / | / | / | / | / | / |
| rs59421388 | / | / | / | / | / | / | / | / | / | / | / |
| rs6025 | / | 0.1646743 | / | / | / | / | / | / | / | / | / |
| rs61736512 | / | / | / | / | / | / | / | / | / | / | / |
| rs6277 | / | 1.54E-26 | / | / | / | / | / | / | / | / | / |
| rs6791924 | / | / | / | / | / | / | / | / | / | / | / |
| rs689466 | 3.389E-08 | 1.522E-08 | 0.534223 | 0.6954382 | 1.963E-08 | 0.214661 | 1.783E-17 | 0.005605 | 4.43E-24 | 4.283E-07 | 7.182E-14 |
| rs698 | 0.2472551 | 3.218E-13 | 0.1091256 | 0.3145579 | 0.0027601 | 0.2195671 | 0.4639062 | 0.0115411 | 0.0084764 | 0.0001312 | 0.1139771 |
| rs701265 | 6.476E-16 | 0.301935 | 0.0005385 | 0.0151595 | 0.0445677 | 0.0055455 | 6.36E-27 | 0.34792 | 1.494E-30 | 0.3207243 | 1.784E-29 |
| rs7294 | 1.998E-16 | 2.767E-13 | 0.612706 | 0.3582965 | 1.542E-31 | 0.0691227 | 5.411E-17 | 6.888E-09 | 5.207E-21 | 8.073E-12 | 2.356E-21 |
| rs7626962 | / | / | / | / | / | / | / | / | / | / | 0.0012409 |
| rs776746 | 3.516E-12 | 5.209E-06 | 0.0524418 | 0.5040015 | 0.4270217 | 0.1157965 | 7.21E-27 | 0.2287979 | 3.231E-11 | 0.0003761 | 4.227E-29 |
| rs7975232 | 0.0002258 | 0.0009613 | 0.3768935 | 0.2806556 | 0.0085925 | 0.6861881 | 1.387E-09 | 0.0949649 | 3.44E-09 | 0.0003739 | 4.224E-06 |
| rs975833 | / | 0.0756969 | 1.535E-09 | / | / | 6.723E-08 | / | / | / | / | 0.0474908 |
| rs9934438 | 8.943E-27 | 3.766E-24 | 0.612706 | 0.4507347 | 1.051E-33 | 0.0738706 | 8.196E-37 | 2.08E-16 | 2.051E-43 | 6.33E-20 | 2.593E-43 |
p value <0.05/(74×11) indicates statistical significance.
In Table 3 we listed the Pairwise FST values between Lhoba and other 11 populations. FST value is less than 0.15 represent there is no genetic differentiation between the two populations. populations. Comparing other populations, the lower level of differentiation were found between Lhoba and CHD (FST = 0.022), JPT (FST = 0.027) and CHB (FST = 0.03) populations, while the greater divergence were observed in YRI (FST = 0.331), MKK (FST = 0.264), and GIH (FST = 0.192).
Table 3.
Distribution of pairwise FST distances among the among the 12 populations
| Lhoba | ASW | CEU | CHB | CHD | GIH | JPT | LWK | MEX | MKK | TSI | YRI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lhoba | 0 | |||||||||||
| ASW | 0.26016 | 0 | ||||||||||
| CEU | 0.1452 | 0.14514 | 0 | |||||||||
| CHB | 0.03002 | 0.21775 | 0.14398 | 0 | ||||||||
| CHD | 0.02218 | 0.21406 | 0.13922 | -0.00152 | 0 | |||||||
| GIH | 0.19204 | 0.09749 | 0.03622 | 0.17875 | 0.17232 | 0 | ||||||
| JPT | 0.02671 | 0.20065 | 0.1332 | 0.00419 | 0.00331 | 0.16642 | 0 | |||||
| LWK | 0.33588 | 0.02041 | 0.223 | 0.29502 | 0.29455 | 0.17418 | 0.27481 | 0 | ||||
| MEX | 0.11532 | 0.10756 | 0.02912 | 0.08894 | 0.08553 | 0.05365 | 0.08526 | 0.18942 | 0 | |||
| MKK | 0.26376 | 0.02066 | 0.143 | 0.23816 | 0.23644 | 0.10624 | 0.21391 | 0.02616 | 0.1355 | 0 | ||
| TSI | 0.13181 | 0.14758 | 0.00249 | 0.1233 | 0.12296 | 0.04433 | 0.11399 | 0.22728 | 0.0279 | 0.14895 | 0 | |
| YRI | 0.33185 | 0.01407 | 0.22394 | 0.29007 | 0.2897 | 0.16559 | 0.26752 | 0.00487 | 0.1878 | 0.02375 | 0.22898 | 0 |
STRUCTURE analysis provided complementary methods for visualizing patterns of genetic similarity and differentiation between Lhoba and other eleven populations. Best model at K = 4, where the proportion of each ancestral component in a single individual is represented by a vertical bar divided into four colors. In Figure 1, the bar plot revealed that the individuals sampled in Lhoba were near to cluster of CHD population.
Figure 1.
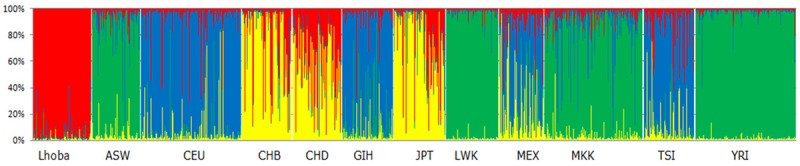
Results of STRUCTURE clustering analysis (K = 4) for Lhoba and Hapmap populations. ASW: African ancestry in southwest USA; CEU: Utah residents with Northern and Western European ancestry from the CEPH collection; CHB: Han Chinese in Beijing, China; CHD: Chinese in Metropolitan Denver, Colorado; GIH, Gujarati Indians in Houston, Texas; JPT: Japanese in Tokyo, Japan; LWK: Luhya in Webuye, Kenya; MEX: Mexican ancestry in Los Angeles, California; MKK: Maasai in Kinyawa, Kenya; TSI: Toscans in Italy; YRI: Yoruba in Ibadan, Niger.
Discussion
This study assessed the polymorphic distribution of 86 pharmacogenomic VIP variants from the PharmGKB database in the Lhoba population as compared with four major HapMap populations. We found that the allelic frequencies in the Lhoba population are generally similar to those in the CHD, CHB and JPT populations but differ significantly from those in the MKK and YRI populations. The differences are particularly pronounced in the YRI population. Pairwise FST values and clustering analyses also showed the VIP variants in Lhoba exhibited a close genetic affinity with most similar to CHD population. These results suggest that genotype data from one group or subgroup (i.e., nation or ethnicity) should not be overly generalized and applied to genetically distinct groups.
In our study, we found that the genotype frequency of rs3814055 in NR1I2 differed significantly among the 12 selected populations. The pregnane X receptor (PXR), also known as nuclear receptor subfamily 1, group I, member 2 (NR1I2), pregnane-activated receptor (PAR), and steroid and xenobiotic receptor (SXR), is a transcription factor belonging to the nuclear hormone receptor superfamily [12,13]. PXR/NR1I2 is a key regulator of the expression of genes involved in all stages of drug metabolism and transport [14,15]. Phase I drug-metabolizing enzymes regulated by PXR/NR1I2 include several CYPs (e.g., CYP3A4, CYP3A5, CYP2B6, and CYP2C8), carboxylesterases, and dehydrogenases [16-19]. The most common clinical reason for the activation of PXR/NR1I2 is the occurrence of drug-drug interactions mediated by up-regulated CYP3A4 expression. Surprisingly, the level of CYP3A4 expression can vary by up to 100-fold between individuals [20,21]. It is well known that ethnicity is an important factor contributing to the large inter-individual variability in drug metabolism, therapeutic response, and toxicity [22,23]. The frequency of rs3814055 in the NR1I2 gene varies in different ethnic groups. In Han Chinese, the frequency of rs3814055 (-25385T), a variant that leads to enhanced CYP3A4 metabolic activity [24], is lower than in Caucasians, Europeans, and African Americans and similar to that of Asians and African Americans. The variant occurs at a frequency of 0.218 in Han Chinese [25], 0.21 in Asians [26], 0.39 in Caucasians [24], 0.50 in Europeans [26], 0.36 in the Dutch [27], and 0.34 in African Americans [26]. In our study, the frequency of the rs3814055 SNP variant was 0.101 in the Lhoba population.
The allelic frequencies in the Lhoba population were generally similar to those in the CHD population. However, the frequency of rs1801133 in the MTHFR gene differed in the two populations. The allelic frequency was 0.227 in the Lhoba population and 0.341 in the CHB population. The enzyme 5,10-methylenetetrahydrofolate reductase (MTHFR) catalyzes the conversion of 5,10-methylenetetrahydrofolate (5,10-methylene-THF) into 5-methyltetrahydrofolate (5-methyl-THF), a major circulating form of folate that provides a methyl group for homocysteine methylation [28,29]. MTHFR plays a key role in folate metabolism by channeling one-carbon units between nucleotide synthesis and methylation reactions. Reduced 5-methyl-THF levels may decrease methylation of homocysteine to methionine, resulting in hyperhomocysteinemia and DNA hypomethylation [30]. On the other hand, reduced levels of 5,10-methylene-THF, which is required for thymidylate synthesis, could lead to misincorporation of uracil into DNA, increasing the frequency of chromosome damage [30], an effect that facilitates the action of certain chemotherapeutics, including MTX. MTHFR deficiency is the most common inherited folate metabolism disorder. More attention should also be paid to capecitabine, cisplatin, pemetrexed, cyanocobalamin, and related agents in the Lhoba population.
Conclusions
Our results demonstrate that national populations in similar geographic regions, such as the Lhoba, may have widely varying genetic allele frequencies for clinically relevant SNPs. The genotype frequencies of VIP variants significantly affect a population’s response to a given drug. Determination of the genotype distribution and frequencies of VIP variants in different populations would provide a theoretical foundation for safer drug administration and enhanced curative effects. Our results complement current data contained in the pharmacogenomics database as it pertains to the Lhoba ethnic group. Additional genotyping studies of specific populations are necessary to provide the best medical care to all individuals. However, the sample size of the Lhoba population in our study was relative small, and further investigation involving a larger cohort of Lhoba individuals is necessary to determine the generalizability of our results to these and other conditions in the Lhoba population.
Acknowledgements
This work was supported by the Key Program of Natural Science Foundation of Xizang (Tibet) Autonomous Region (201122, 20152R-13-11), the Major Training Program of Tibet University for Nationalities (No. 13myZP06), the National Natural Science Foundations (No. 81560516), and the Major science and technology research projects of Xizang (Tibet) Autonomous Region (2015).
Disclosure of conflict of interest
None.
References
- 1.Pilotto A, Panza F, Seripa D. Pharmacogenetics in geriatric medicine: challenges and opportunities for clinical practice. Curr Drug Metab. 2011;12:621–634. doi: 10.2174/138920011796504545. [DOI] [PubMed] [Google Scholar]
- 2.Roses AD. Pharmacogenetics and the practice of medicine. Nature. 2000;405:857–865. doi: 10.1038/35015728. [DOI] [PubMed] [Google Scholar]
- 3.Altman RB. PharmGKB: a logical home for knowledge relating genotype to drug response phenotype. Nat Genet. 2007;39:426. doi: 10.1038/ng0407-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sangkuhl K, Berlin DS, Altman RB, Klein TE. PharmGKB: understanding the effects of individual genetic variants. Drug Metab Rev. 2008;40:539–551. doi: 10.1080/03602530802413338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balant LP, Balant-Gorgia EA. Cultural differences: implications on drug therapy and global drug development. Int J Clin Pharmacol Ther. 2000;38:47–52. doi: 10.5414/cpp38047. [DOI] [PubMed] [Google Scholar]
- 6.Codd V, Mangino M, van der Harst P, Braund PS, Kaiser M, Beveridge AJ, Rafelt S, Moore J, Nelson C, Soranzo N, Zhai G, Valdes AM, Blackburn H, Mateo Leach I, de Boer RA, Kimura M, Aviv A Wellcome Trust Case Control Consortium. Goodall AH, Ouwehand W, van Veldhuisen DJ, van Gilst WH, Navis G, Burton PR, Tobin MD, Hall AS, Thompson JR, Spector T, Samani NJ. Common variants near TERC are associated with mean telomere length. Nat Genet. 2010;42:197–199. doi: 10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engen RM, Marsh S, Van Booven DJ, McLeod HL. Ethnic differences in pharmacogenetically relevant genes. Curr Drug Targets. 2006;7:1641–1648. doi: 10.2174/138945006779025446. [DOI] [PubMed] [Google Scholar]
- 8.Van Booven D, Marsh S, McLeod H, Carrillo MW, Sangkuhl K, Klein TE, Altman RB. Cytochrome P450 2C9-CYP2C9. Pharmacogenet Genomics. 2010;20:277–281. doi: 10.1097/FPC.0b013e3283349e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong X, Zhang S, Mao G, Jiang S, Zhang Y, Yu Y, Tang G, Xing H, Xu X. CYP2C9*3 allelic variant is associated with metabolism of irbesartan in Chinese population. Eur J Clin Pharmacol. 2005;61:627–634. doi: 10.1007/s00228-005-0976-8. [DOI] [PubMed] [Google Scholar]
- 10.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;Chapter 2:Unit 2.12. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 11.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, Lee JC, Nicoletti R, Hatton C, Goyette M, Girard L, Majmudar K, Ziaugra L, Wong KK, Gabriel S, Beroukhim R, Peyton M, Barretina J, Dutt A, Emery C, Greulich H, Shah K, Sasaki H, Gazdar A, Minna J, Armstrong SA, Mellinghoff IK, Hodi FS, Dranoff G, Mischel PS, Cloughesy TF, Nelson SF, Liau LM, Mertz K, Rubin MA, Moch H, Loda M, Catalona W, Fletcher J, Signoretti S, Kaye F, Anderson KC, Demetri GD, Dummer R, Wagner S, Herlyn M, Sellers WR, Meyerson M, Garraway LA. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 12.Blumberg B, Sabbagh W Jr, Juguilon H, Bolado J Jr, van Meter CM, Ong ES, Evans RM. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 14.Dussault I, Lin M, Hollister K, Wang EH, Synold TW, Forman BM. Peptide mimetic HIV protease inhibitors are ligands for the orphan receptor SXR. J Biol Chem. 2001;276:33309–33312. doi: 10.1074/jbc.C100375200. [DOI] [PubMed] [Google Scholar]
- 15.Matic M, Mahns A, Tsoli M, Corradin A, Polly P, Robertson GR. Pregnane X receptor: promiscuous regulator of detoxification pathways. Int J Biochem Cell Biol. 2007;39:478–483. doi: 10.1016/j.biocel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin B, Moore LB, Stoltz CM, McKee DD, Kliewer SA. Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol. 2001;60:427–431. [PubMed] [Google Scholar]
- 18.Scheer N, Ross J, Rode A, Zevnik B, Niehaves S, Faust N, Wolf CR. A novel panel of mouse models to evaluate the role of human pregnane X receptor and constitutive androstane receptor in drug response. J Clin Invest. 2008;118:3228–3239. doi: 10.1172/JCI35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerbal-Chaloin S, Pascussi JM, Pichard-Garcia L, Daujat M, Waechter F, Fabre JM, Carrere N, Maurel P. Induction of CYP2C genes in human hepatocytes in primary culture. Drug Metab Dispos. 2001;29:242–251. [PubMed] [Google Scholar]
- 20.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- 21.Lown KS, Mayo RR, Leichtman AB, Hsiao HL, Turgeon DK, Schmiedlin-Ren P, Brown MB, Guo W, Rossi SJ, Benet LZ, Watkins PB. Role of intestinal P-glycoprotein (mdr1) in interpatient variation in the oral bioavailability of cyclosporine. Clin Pharmacol Ther. 1997;62:248–260. doi: 10.1016/S0009-9236(97)90027-8. [DOI] [PubMed] [Google Scholar]
- 22.Dishy V, Sofowora GG, Xie HG, Kim RB, Byrne DW, Stein CM, Wood AJ. The effect of common polymorphisms of the beta2-adrenergic receptor on agonist-mediated vascular desensitization. N Engl J Med. 2001;345:1030–1035. doi: 10.1056/NEJMoa010819. [DOI] [PubMed] [Google Scholar]
- 23.Chowbay B, Zhou S, Lee EJ. An interethnic comparison of polymorphisms of the genes encoding drug-metabolizing enzymes and drug transporters: experience in Singapore. Drug Metab Rev. 2005;37:327–378. doi: 10.1081/dmr-28805. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Kuehl P, Green ED, Touchman JW, Watkins PB, Daly A, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Wrighton SA, Hancock M, Kim RB, Strom S, Thummel K, Russell CG, Hudson JR Jr, Schuetz EG, Boguski MS. The human pregnane X receptor: genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics. 2001;11:555–572. doi: 10.1097/00008571-200110000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Wang XD, Li JL, Su QB, Deng XY, Lu Y, Chen J, Zhang JX, Zhao LZ, Zuo Z, Chan E, Chen X, Chowbay B, Xue CC, Huang M, Zhou SF. A pharmacogenetic study of pregnane X receptor (NR1I2) in Han Chinese. Curr Drug Metab. 2007;8:778–786. doi: 10.2174/138920007782798199. [DOI] [PubMed] [Google Scholar]
- 26.King CR, Xiao M, Yu J, Minton MR, Addleman NJ, Van Booven DJ, Kwok PY, McLeod HL, Marsh S. Identification of NR1I2 genetic variation using resequencing. Eur J Clin Pharmacol. 2007;63:547–554. doi: 10.1007/s00228-007-0295-3. [DOI] [PubMed] [Google Scholar]
- 27.Bosch TM, Deenen M, Pruntel R, Smits PH, Schellens JH, Beijnen JH, Meijerman I. Screening for polymorphisms in the PXR gene in a Dutch population. Eur J Clin Pharmacol. 2006;62:395–399. doi: 10.1007/s00228-006-0108-0. [DOI] [PubMed] [Google Scholar]
- 28.Shane B. Folylpolyglutamate synthesis and role in the regulation of one-carbon metabolism. Vitam Horm. 1989;45:263–335. doi: 10.1016/s0083-6729(08)60397-0. [DOI] [PubMed] [Google Scholar]
- 29.Schwahn B, Rozen R. Polymorphisms in the methylenetetrahydrofolate reductase gene: clinical consequences. Am J Pharmacogenomics. 2001;1:189–201. doi: 10.2165/00129785-200101030-00004. [DOI] [PubMed] [Google Scholar]
- 30.Kim YI. Folate and cancer prevention: a new medical application of folate beyond hyperhomocysteinemia and neural tube defects. Nutr Rev. 1999;57:314–321. doi: 10.1111/j.1753-4887.1999.tb06905.x. [DOI] [PubMed] [Google Scholar]


