Abstract
The genetic or abnormal activation of PI3K/PTEN signaling pathway play an important role with regard to disease progression in variety of human malignancies. Experimental and epidemiologic studies indicated that the genetic polymorphisms in the PTEN, PI3K genes are associated with cancer risk, yet little evidence exists for those 2 genes and colorectal cancer (CRC) risk. To address this, we evaluated whether PTEN rs701848, PIK3CA rs2699887 variants are associated with CRC susceptibility, clinicopathological parameters and clinical outcomes in CRC patients treated with FOLFOX (Oxaliplatin, Leucovorin, 5-Fluorouracil) regimen. A case-control study was performed in 780 CRC patients and 764 healthy controls using the TaqMan assay method. A significant increased risk of CRC was observed in patients carrying PTEN rs701848 TC or CC genotype (adjusted OR=1.306, 95% CI=1.030-1.655, P=0.027; adjusted OR=1.543, 95% CI=1.148-2.075, P=0.004, respectively), TC/CC genotype (adjusted OR=1.367, 95% CI=1.090-1.714, P=0.043) in the dominant model, and C allele (adjusted OR=1.229, 95% CI=1.067-1.416, P=0.004). However, no association was detected between rs2699887 in the PIK3CA gene and CRC risk. A significant association was found between pathological grade (Dukes A and B vs. Dukes C and D) and PIK3CA rs2699887 genotypes. Furthermore, Kaplan-Meier analysis revealed that PTEN rs701848 genotypes were significantly associated with the overall survival (OS) of CRC patients treated with FOLFOX regimen (n=780). Individuals carrying PTEN rs701848 TC or TC/CC genotypes showed significantly longer median survival time (MST) than TT genotype and significant hazard ratio (TC: adjusted HR=0.523, 95% CI=0.325-0.840, P=0.007; TC/CC: adjusted HR=0.545, 95% CI=0.351-0.845, P=0.007). Therefore, rs701848 polymorphism in the PTEN gene is associated with susceptibility to CRC, and C allele of rs701848 showed significant independent better prognosis of CRC patients treated with FOLFOX regimen. These results indicate that rs701848 in the PTEN gene might be a candidate pharmacogenomic factor to assess the susceptibility and prognosis in CRC patients.
Keywords: PTEN, PIK3CA, polymorphisms, susceptibility, colorectal cancer, prognosis
Introduction
In China, Colorectal cancer (CRC) is the most common gastrointestinal tract malignancy with almost 400,000 new cases diagnosed per annum posing a significant public health burden [1,2]. Since the change to a western dietary pattern in China, the incidence and mortality rates of CRC have increased markedly [3]. Observational studies lend support to environmental factors and genetic factors such as germline mutations on colorectal carcinogenesis are involved in the pathogenesis of CRC [4,5]. Accumulating significant advances have been made in understanding the biology of CRC carcinogenesis in particular polymorphisms of tumor susceptibility candidate genes [6,7]. Moreover, previous studies demonstrated that the prognosis of CRC varies considerably even among patients with the same stage and receiving the similar treatment, partially because of genetic variants in individuals affecting the effectiveness of chemotherapy and clinical outcomes [8-10]. Hence, a better understanding of the genetic risk factors that underlie CRC could lead to improved strategies for therapy screening and prognosis prediction of CRC patients.
The phosphoinositide-3-kinase (PI3K)/Akt signaling pathway is one of the most important kinase cascades and mediates a wide range of cellular functions such as survival, proliferation, migration, differentiation and angiogenesis [11]. PI3Ks are family of lipid kinases capable of phosphorylating the 3’OH of the inositol ring of phosphoinositides, which are activated by receptor tyrosine kinases such as epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor (VEGFR) and insulin-like growth factor receptor (IGFR). Phosphatase and tensin homolog (PTEN) gene, a tumor suppressor gene located on chromosome 10q23, encodes a dual specificity protein phosphatase, which negatively regulates PI3K/AKT signaling [12]. Genetic alterations of the genes related to this pathway, including mutations of PI3K and PTEN, facilitate tumorigenesis and are common in human cancers [13-17]. In the present study, we expanded the exploration to essential components of the AKT signaling pathway comprising PI3K and PTEN.
More evidences showed that single nucleotide polymorphisms (SNPs) of many genes involved in influencing individuals’ susceptibility to CRC [18,19]. In light of the critical role of the AKT pathway in CRC, it is possible that SNPs in this pathway may play an important role in CRC development. However, no published study has yet addressed the genetic effect of PTEN, PIK3CA polymorphisms on the susceptibility and prognosis of CRC. Accordingly, in this large prospective cohort, we analyzed the functional genetic polymorphisms of PTEN rs701848, PIK3CA rs2699887 in CRC patients, attempting to analyze between 2 potentially functional SNPs and their impacting on the occurrence of CRC and clinical outcomes after FOLFOX (Oxaliplatin, Leucovorin, 5-Fluorouracil) regimen in a Chinese population.
Materials and methods
Study subjects
Overall, in this case-control study 780 patients with CRC and a group of 764 age- and gender- matched cancer-free controls were recruited at Cancer Hospital (Institute) of Peking Union Medical College, Peking, China between September 2007 and October 2013. Briefly, this cohort was newly diagnosed incident CRC patients based on histopathologically confirmed. They were consecutively recruited without the restriction of age and gender and were without prior history of other cancers or previous chemotherapy or radiotherapy. The principal demographic data were obtained from interviewer-administered health risk questionnaires or medical records. Tumor differentiation and pathological grade of the CRC patients was classified by the World Health Organization criteria and staged according to Duke’s criteria. All included patients received FOLFOX regimen. The FOLFOX regimen consists of oxaliplatin (100 mg/m2, iv gtt (2 h), day 1, every 4 weeks), leucovorin (200 mg/m2, iv gtt, day 1-day 5, every 4 weeks), 5-fluorouracil (500 mg/m2, day 1-day 5, every 4 weeks). The treatment was given until disease progression, or patient’s refusal to continue treatment.
In this study, 764 unrelated age- and gender- matched healthy controls were recruited from individuals who visited the same hospital for a physical examination. The cancer-free controls had no known medical illness or hereditary disorders, and were not taking any medications. Before recruitment, this study was approved by the Research Ethics Committee of Peking Union Medical College. A standard questionnaire was administered through face-to-face interviews by trained interviewers to collect demographic data and related factors. Each patient donated 5 mL of venous blood after providing a written informed consent.
Genotyping
Genomic DNA was extracted from a leukocyte cell pellet of each blood sample (5 mL) using the QIAamp DNA Blood Mini Kit (Dynal Biotech (Beijing) Ltd, Beijing, China), and stored at -80°C. The genotyping of these 2 SNPs was performed using predesigned TaqMan SNP Genotyping Assays on the ABI 7500 Real-Time PCR platform (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA) following the manufacturer’s instructions. The TaqMan probes were synthesized by Life Technology (Shanghai, China). The sequences of the primers and probes are listed in Table S1. The reaction mixture of 5 mL contained 10 ng genomic DNA, 2.5 mL of TaqMan Genotyping Master Mix, 1.25 mL of the primers and probes mix and 1.25 mL of double distilled water. The amplification was performed under the following conditions: 95°C for 10 min, 45 cycles of 95°C for 15 s, 60°C for 60 s and 60°C for 30 s. The genotyping rates of these SNPs were all above 97%. For quality control, 6 negative controls were included in each plate and 10% of the samples were randomly selected for repeated genotyping for confirmation; and the results were 100% concordant.
Statistical analysis
Analysis was performed with SPSS 16.0 for Windows (SPSS Inc. Chicago, Illinois, USA). All tests were two-sided and P<0.05 was considered statistical significant. Two-sided χ2 test was used to assess differences in distributions of demographic, epidemiologic, and clinical variables between CRC patients and controls, as well as between alleles and genotypes. Hardy-Weinberg equilibrium test (HWE) was determined using a goodness-of-fit χ2 test to compare expected genotype frequencies with the observed genotype frequency (p 2+2pq+q 2=1). Unconditional logistic regression models were used to analyze the association between the genotypes and CRC susceptibility, and clinical variables. Disease-free survival (DFS) and overall survival (OS) were compared with the Kaplan-Meier method and the significance was determined by the log-rank test. Multivariate Cox proportional hazards regression models were applied to obtain the adjusted hazard ratio (HR) and 95% CI for evaluating the effects of clinical variables, genotypes on DFS and OS in CRC patients.
Results
Characteristics of CRC patients and controls
Selected baseline characteristics and clinical variables of the CRC patients are presented in Table 1. In total, 780 patients with pathologically confirmed CRC and a group of 764 age- and gender- matched cancer-free healthy controls were included in this study. There were no significant differences in the distributions of gender and age between CRC patients and controls (P=0.544 and P=0.637, respectively). The age was matched between CRC patients (range: 26-75 years old; mean: 58 years old) and controls (range: 28-73 years; mean: 58 years old). Among the CRC patients, 56.4% of CRC patients were male, and 84.6% of them were in the status of ever smoking. Tumor differentiations of the majority patients were in grade 2 (G2, moderate, 80.3%), while only 9.5% of them were in grade 1 (G1, well). In this included cohort, all patients underwent oxaliplatin-based chemotherapy (FOLFOX regimen). The clinical variables of age, gender, smoking status, and first-degree family history of cancer were adjusted for any residual confounding effects in later logistic regression analyses.
Table 1.
Baseline characteristics of CRC patients
| Variables | Patients, N (%) |
|---|---|
| Age at diagnosis, yrs | |
| ≤58 | 361 (46.3) |
| >58 | 419 (53.7) |
| Gender | |
| Male | 440 (56.4) |
| Female | 340 (43.6) |
| First-degree family history of cancer | |
| No | 678 (86.9) |
| Yes | 102 (13.1) |
| Smoking status | |
| Ever | 660 (84.6) |
| Never† | 120 (15.4) |
| Tumor size (cm) | |
| ≤3.5 | 181 (23.2) |
| >3.5 | 599 (76.8) |
| Tumor differentiation | |
| Grade 1 (G1, Well) | 74 (9.5) |
| Grade 2 (G2, moderate) | 626 (80.3) |
| Grade 3 (G3, poor) | 80 (10.3) |
| Pathological grade | |
| Dukes A | 154 (19.7) |
| Dukes B | 286 (36.7) |
| Dukes C | 270 (34.6) |
| Dukes D | 70 (9.0) |
| Lymph node metastases | |
| No | 293 (37.6) |
| Yes | 487 (62.4) |
| Therapeutic regimens | |
| FOLFOX regimen | 780 (100) |
Defined as ≤100 cigarettes in lifetime.
Genotype and allele frequencies of PTEN, PIK3CA polymorphisms and CRC risk
The frequencies of allelic and genotype distribution for PTEN rs701848, PIK3CA rs2699887 in both CRC patients and controls are presented in Table 2. Genotype frequencies of rs701848 in the PTEN gene, rs2699887 in the PIK3CA gene in controls all conformed well to Hardy-Weinberg equilibrium (P=0.134, P=0.171, respectively). We observed that the alleles and genotypes from PTEN rs701848 genetic variant were statistically associated with the risk of CRC. There was statistically increased risk of CRC in the genotypes comparison (TC vs. TT: adjusted OR=1.306, 95% CI=1.030-1.655, P=0.027; CC vs. TT: adjusted OR=1.543, 95% CI=1.148-2.075, P=0.004). Furthermore, a significant increased CRC risk was observed in the dominant model (TC/CC vs. TT: adjusted OR=1.367, 95% CI=1.090-1.714, P=0.007), meanwhile decreased risk in the recessive model (TC/TT vs. CC: adjusted OR=0.773, 95% CI=0.602-0.993, P=0.044). Moreover, compared with the T allele, the C allele had a significant increased risk of developing CRC (T vs. C: adjusted OR=1.229, 95% CI=1.067-1.416, P=0.004). However, no significant difference was detected between rs2699887 of the PIK3CA gene and CRC risk, as shown in Table 2.
Table 2.
Frequency distribution of genotypes and their associations with the risk of developing CRC
| Genotypes | Patients, N (%) | Controls†, N (%) | P ‡ | Adjusted OR§ | 95% CI§ |
|---|---|---|---|---|---|
| PTEN rs701848 | |||||
| TT | 186 (23.8) | 229 (30.0) | 1.000 | ||
| TC | 421 (54.0) | 397 (51.9) | 0.027 | 1.306 | 1.030-1.655 |
| CC | 173 (22.2) | 138 (18.1) | 0.004 | 1.543 | 1.148-2.075 |
| Dominant model | |||||
| TT | 186 (23.8) | 229 (30.0) | 1.000 | ||
| TC/CC | 594 (76.2) | 535 (70.0) | 0.007 | 1.367 | 1.090-1.714 |
| Recessive model | |||||
| CC | 173 (22.2) | 138 (18.1) | 1.000 | ||
| TT/TC | 607 (77.8) | 626 (81.9) | 0.044 | 0.773 | 0.602-0.993 |
| Allelic frequency (%) | |||||
| T allele | 50.8 | 56.0 | 1.000 | ||
| C allele | 49.2 | 44.0 | 0.004 | 1.229 | 1.067-1.416 |
| PIK3CA rs2699887 | |||||
| GG | 596 (76.4) | 579 (75.8) | 1.000 | ||
| GA | 173 (22.2) | 167 (21.8) | 0.959 | 1.006 | 0.791-1.281 |
| AA | 11 (1.4) | 18 (2.4) | 0.173 | 0.594 | 0.278-1.268 |
| Dominant model | |||||
| GG | 596 (76.4) | 579 (75.8) | 1.000 | ||
| GA/AA | 184 (23.6) | 185 (24.2) | 0.773 | 0.966 | 0.765-1.221 |
| Recessive model | |||||
| AA | 11 (1.4) | 18 (2.4) | 1.000 | ||
| GA/GG | 769 (98.6) | 746 (97.6) | 0.171 | 1.687 | 0.791-3.595 |
| Allelic frequency (%) | |||||
| G allele | 87.5 | 86.7 | 1.000 | ||
| A allele | 12.5 | 13.3 | 0.515 | 0.932 | 0.755-1.151 |
OR indicates odds ratio; CI, confidence interval; the significance levels are P<0.05 for all the bold values.
The observed genotype frequency among individuals in the control group was in agreement with Hardy-Weinberg equilibrium (p2+2pq+q2=1: P=0.134 for PTEN rs701848, P=0.171 for PIK3CA rs2699887).
P values were calculated by unconditional logistic regression adjusted for age, gender, smoking status, and first-degree family history of cancer.
OR and 95% CI values were calculated by unconditional logistic regression adjusted for age, gender, smoking status, and first-degree family history of cancer.
Association of PTEN, PIK3CA polymorphisms and clinical parameters in CRC patients
In the case-only analysis (n=780), we further investigated the association between rs701848 in the PTEN gene and rs2699887 in the PIK3CA gene and clinical variables and environmental risk factors using two-sided χ2 test and adjusted unconditional logistic regression adjusted by age, gender, smoking status, and first-degree family history of cancer, outlined in Table 3. We found that the frequency (27.1%) of the PIK3CA rs2699887 GA/AA genotypes in CRC patients with pathological grade (Dukes C and D) was significantly higher than that (20.9%) in those with Dukes A and B (adjusted OR=1.420, 95% CI=1.017-1.984, P=0.040), as shown in Table 3. Furthermore, a tendency towards higher frequency of the PIK3CA rs2699887 GA/AA genotypes were observed in CRC patients with older ages (>58 years old) (26.0%) in comparison with those ≤58 years old (20.8%) (adjusted OR=1.357, 95% CI=0.968-1.902, P=0.077) (Table 3). However, there was no significant association detected between the PTEN rs701848 and clinical characteristics.
Table 3.
Association between genotypes and clinicopathological features in CRC patients
| Variables | PTEN rs701848 | PIK3CA rs2699887 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| TT N (%) | TC/CC N (%) | P †,‡ | Adjusted OR§ | 95% CI§ | GG N (%) | GA/AA N (%) | P †,‡ | Adjusted OR§ | 95% CI§ | |
| Age, years | ||||||||||
| ≤58 | 81 (22.4) | 280 (77.6) | 0.392† | 1.000 | 286 (79.2) | 75 (20.8) | 0.086† | 1.000 | ||
| >58 | 105 (25.1) | 314 (74.9) | 0.516‡ | 0.895 | 0.640-1.251 | 310 (74.0) | 109 (26.0) | 0.077‡ | 1.357 | 0.968-1.902 |
| Gender | ||||||||||
| Male | 107 (24.3) | 333 (75.7) | 0.725† | 1.000 | 334 (75.9) | 106 (24.1) | 0.708† | 1.000 | ||
| Female | 79 (23.2) | 261 (76.8) | 0.676‡ | 1.074 | 0.769-1.500 | 262 (77.1) | 78 (22.9) | 0.682‡ | 0.932 | 0.666-1.304 |
| First-degree family history of cancer | ||||||||||
| No | 167 (24.6) | 511 (75.4) | 0.185† | 1.000 | 512 (75.5) | 166 (24.5) | 0.129† | 1.000 | ||
| Yes | 19 (18.6) | 83 (81.4) | 0.130‡ | 1.515 | 0.885-2.593 | 84 (82.4) | 18 (17.6) | 0.224‡ | 0.713 | 0.413-1.229 |
| Smoking status | ||||||||||
| Ever | 151 (22.9) | 509 (77.1) | 0.137† | 1.000 | 498 (75.5) | 162 (24.5) | 0.140† | 1.000 | ||
| Never† | 35 (29.2) | 85 (70.8) | 0.100‡ | 0.690 | 0.444-1.074 | 98 (81.7) | 22 (18.3) | 0.161‡ | 0.698 | 0.423-1.154 |
| Tumor size (cm) | ||||||||||
| ≤3.5 | 38 (21.0) | 143 (79.0) | 0.304† | 1.000 | 132 (72.9) | 49 (27.1) | 0.208† | 1.000 | ||
| >3.5 | 148 (24.7) | 451 (75.3) | 0.226‡ | 0.778 | 0.519-1.168 | 464 (77.5) | 135 (22.5) | 0.265‡ | 0.804 | 0.548-1.179 |
| Tumor differentiation | ||||||||||
| Grade 1 | 13 (17.6) | 61 (82.4) | 0.183† | 1.000 | 60 (81.1) | 14 (18.9) | 0.320† | 1.000 | ||
| Grade 2/Grade 3 | 173 (24.5) | 533 (75.5) | 0.176‡ | 0.649 | 0.347-1.214 | 536 (75.9) | 170 (24.1) | 0.320‡ | 1.363 | 0.740-2.511 |
| Pathological grade | ||||||||||
| Dukes A and B | 108 (24.5) | 332 (75.5) | 0.602† | 1.000 | 348 (79.1) | 92 (20.9) | 0.045† | 1.000 | ||
| Dukes C and D | 78 (22.9) | 262 (77.1) | 0.565‡ | 1.104 | 0.789-1.543 | 248 (72.9) | 92 (27.1) | 0.040‡ | 1.420 | 1.017-1.984 |
| Lymph node metastases | ||||||||||
| No | 73 (24.9) | 220 (75.1) | 0.587† | 1.000 | 220 (75.1) | 73 (24.9) | 0.499† | 1.000 | ||
| Yes | 113 (23.2) | 374 (76.8) | 0.700‡ | 1.069 | 0.760-1.504 | 376 (77.2) | 111 (22.8) | 0.529‡ | 0.896 | 0.636-1.261 |
P values were calculated from 2-sided chi-square tests or Fisher’s Exact Test.
P values were calculated by unconditional logistic regression adjusted for age, gender, smoking status, and first-degree family history of cancer.
OR and 95% CI values were calculated by unconditional logistic regression adjusted for age, gender, smoking status, and first-degree family history of cancer.
Effects of PTEN, PIK3CA polymorphisms on CRC survival
Multivariate Cox regression analysis and Kaplan-Meier analysis were performed to further evaluate the correlations between genetic polymorphisms of rs701848 in the PTEN gene, rs2699887 in the PIK3CA gene and the prognosis of CRC patients after treated with FOLFOX regimen chemotherapy (n=780).
Kaplan-Meier analysis revealed that rs701848 genotypes in the PTEN gene were significantly associated with the OS of CRC patients (log-rank test: P=0.036 in the genotypes and P=0.010 in the dominant model, respectively). CRC patients carrying PTEN rs701848 TC or CC genotype had a significantly longer OS time (TC genotype: median survival time, MST=126 months, 95% CI=113-140 months, CC genotype: MST=82 months, 95% CI=75-90 months, respectively) in comparison to the carriers who had TT genotype (MST=75 months, 95% CI=57-94 months), as illustrated in Figure 1A. Furthermore, multivariate Cox regression analysis also established that rs701848 TC genotypes acted as prognostic factors (TC genotype: adjusted HR=0.523, 95% CI=0.325-0.840, P=0.007), meanwhile CC genotype showed a tendency toward prolonged OS (adjusted HR=0.767, 95% CI=0.568-1.035, P=0.083) adjusted by age, gender, smoking status, and first-degree family history of cancer, outlined in Table 4. Additionally, in the dominant model, rs701848 TC/CC genotype carriers (MST=126 months, 95% CI=117-137 months; Figure 1B) showed significantly prolonged OS time, and verified in multivariate Cox regression model analysis (adjusted HR=0.545, 95% CI=0.351-0.845, P=0.007), as illustrated in Table 4. However, in this study we did not found significant association between the PIK3CK polymorphisms and clinical outcomes in FOLFOX treated CRC patients.
Figure 1.
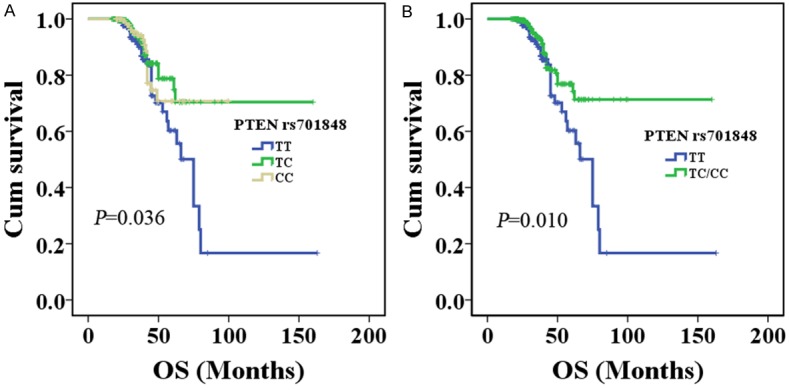
The relationship between the PTEN rs701848 polymorphism and CRC prognosis according to Kaplan-Meier analysis. A. PTEN rs701848 TC, CC genotype had longer overall survival in CRC patients treated with FOLFOX regimen (log-rank test: P=0.036); B. PTEN rs701848 TC/CC genotypes had longer overall survival in CRC patients treated with FOLFOX regimen (log-rank test: P=0.010).
Table 4.
Multivariate COX regression analysis of influencing prognosis factors in the CRC patients with postoperative chemotherapy
| Variables | RFS | OS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Total N | Events N (%) | Adjusted HR† | 95% CI† | P ‡ | Total N | Events N (%) | Adjusted HR† | 95% CI† | P ‡ | |
| PTEN rs701848 | ||||||||||
| TT | 186 | 35 (18.8) | 1.000 | 186 | 34 (18.3) | 1.000 | ||||
| TC | 421 | 84 (20.0) | 1.240 | 0.826-1.861 | 0.300 | 421 | 39 (9.3) | 0.523 | 0.325-0.840 | 0.007 |
| CC | 173 | 27 (15.6) | 0.989 | 0.765-1.279 | 0.932 | 173 | 17 (9.8) | 0.767 | 0.568-1.035 | 0.083 |
| TC/CC | 594 | 111 (18.7) | 1.126 | 0.764-1.660 | 0.549 | 594 | 56 (9.4) | 0.545 | 0.351-0.845 | 0.007 |
| PIK3CA rs2699887 | ||||||||||
| GG | 596 | 113 (19.0) | 1.000 | 596 | 71 (11.9) | 1.000 | ||||
| GA | 173 | 29 (16.8) | 0.880 | 0.584-1.326 | 0.542 | 173 | 17 (9.8) | 0.886 | 0.520-1.510 | 0.657 |
| AA | 11 | 4 (36.4) | 1.354 | 0.820-2.234 | 0.236 | 11 | 2 (18.2) | 1.077 | 0.531-2.186 | 0.837 |
| GA/AA | 184 | 33 (17.9) | 1.375 | 0.930-0.629 | 0.717 | 184 | 19 (10.3) | 0.958 | 0.575-1.597 | 0.869 |
| Age, years | ||||||||||
| ≤58 | 361 | 69 (19.1) | 1.000 | 361 | 41 (11.4) | 1.000 | ||||
| >58 | 419 | 77 (18.4) | 1.113 | 0.798-1.552 | 0.527 | 419 | 49 (11.7) | 1.144 | 0.748-1.748 | 0.535 |
| Gender | ||||||||||
| Male | 440 | 83 (18.9) | 1.000 | 440 | 52 (11.8) | 1.000 | ||||
| Female | 340 | 63 (18.5) | 1.097 | 0.788-1.528 | 0.583 | 340 | 38 (11.2) | 1.069 | 0.698-1.639 | 0.758 |
| First-degree family history of cancer | ||||||||||
| No | 678 | 126 (18.6) | 1.000 | 678 | 74 (10.9) | 1.000 | ||||
| Yes | 102 | 20 (19.6) | 0.956 | 0.591-1.546 | 0.854 | 102 | 16 (15.7) | 1.486 | 0.860-2.568 | 0.156 |
| Smoking status | ||||||||||
| Ever | 660 | 126 (19.1) | 1.000 | 660 | 80 (12.1) | 1.000 | ||||
| Never | 120 | 20 (16.7) | 0.855 | 0.527-1.388 | 0.527 | 120 | 10 (8.3) | 0.531 | 0.271-1.041 | 0.065 |
HR, Hazard Ratio; 95% CI, 95% confidence interval.
Adjusted HR and 95% CI values were assessed using multivariate Cox regression analysis adjusted for age, gender, smoking status, and first-degree family history of cancer.
P values were calculated by multivariate Cox regression analysis adjusted for age, gender, smoking status, and first-degree family history of cancer.
Discussion
Mutations or dysregulation of genes involved in PI3K/PTEN/AKT pathways have been associated with invasion, metastasis, and prognosis of a variety of cancers, including CRC [20,21]. PTEN, phosphatase and tensin homolog, an important regulator of cell cycle progression and cellular survival, exerts its tumor suppressor function by acting as a negative regulator via the PI3K signaling pathway [22]. Inactivating mutations in PTEN gene and activating mutations in PIK3CA gene have been reported to occur about 25 and 30% in CRC patients, respectively [23,24]. The role of genetic mutations in these pathways is complex and dependent on interactions with environmental factors; however evaluation of the role of germline mutations in initiation and progression of CRC is clearly warranted. Therefore, we for the first time performed a case-control study to explore systematically the correlation between PTEN rs701848, PIK3CA rs2699887 polymorphisms and the susceptibility, clinical variables, and clinical outcomes of CRC patients after FOLFOX chemotherapy.
In this study, we found that rs701848 variants in the PTEN gene were associated with increased susceptibility to CRC. Individuals with heterozygotes, homozygotes, or variant alleles of rs701848 in the PTEN gene showed 1.2-1.5-fold increased CRC risk. Similarly, Cao Q et al. reported that patients with rs701848 in the 3’UTR region of PTEN was associated with increased renal cell cancer risk (CC vs.TT, P=0.014, OR=1.45, 95% CI=1.08-1.96) [25]. Another study reconstructed the PTEN haplotypes according to genotyping data and linkage disequilibrium status of polymorphisms (rs10490920, rs532678, rs701848, and insertion/deletion polymorphism rs3442166 0), and found that the T-C-C-del haplotype was associated with decreased hepatocellular carcinoma risk (n=134) [13]. SNP rs701848 of the PTEN gene is located at 3’-UTR region, not able to change the encoded amino acids; however it might influence genetic splicing, protein expression, regulation of cell cycle, etc. Furthermore, the 3’-UTR region targeted by microRNAs might alter the strength of microRNAs binding site near the SNP rs701848, with consequence on regulation of target genes, thereby influencing gene regulation and protein expression. However, the function of the SNP still needs to be further investigated in future studies. In addition, no significant difference was determined between rs2699887 in the PIK3CA gene and CRC risk in this Chinese population.
We further analyzed the relationship between polymorphisms of rs701848 in the PTEN gene and rs2699887 in the PIK3CA gene with clinical characteristics and environmental risk factors (smoking status, first-degree family history of cancer). We found that the frequency of the PIK3CA rs2699887 GA/AA genotypes in CRC patients with pathological grade (Dukes C and D) was significantly higher, suggesting that rs2699887 polymorphism may be involved in the development of pathological grade-associated CRC. This polymorphism may be used as a potential marker for identification of the malignant and invasive pathological grade-associated CRC patients.
Little is known regarding the polymorphisms of rs701848 in the PTEN gene and rs2699887 in the PIK3CA gene in terms of the potential impact on the CRC clinical outcomes. We further investigated the association of these polymorphisms with the survival time of CRC patients treated with FOLFOX regimen. It is worth to noting that the PTEN rs701848 TC genotype and TC/CC genotype were related to a significantly prolonged OS time. Moreover, multivariate analysis further confirmed the independent prognostic value of rs701848 polymorphism in CRC patients treated with FOLFOX regimen. On the contrary, Wang X et al. found that CC allele of PTEN rs701848 was associated with the increased risk of recurrence (HR=2.06, 95% CI=1.19-3.58) and patient death (HR=2.01, 95% CI=1.15-3.53) in gastric cancer patients (n=221) [8]. Since the difference between this study and Wang X et al. may be due to different tumors, thus further investigations are warranted to understand the precise mechanism of this polymorphism involved in different tumors.
To our best knowledge, this is the first investigation that we developed the association between PTEN rs701848, PIK3CA rs2699887polymorphisms and susceptibility, clinical characteristics, and chemotherapeutic outcomes in a large sample population of CRC patients. We provided evidence that PTEN rs701848 polymorphisms are associated with susceptibility and therapeutic outcome in CRC patients treated with FOLFOX therapy in a large and well characterized cohort. The data suggested that PTEN rs701848 polymorphisms may play an important role in the development of CRC, and therefore, may be a vital prognostic indicator for CRC, and employed as candidate biomarker for the prediction of susceptibility and potential-adjuvant in CRC patients for FOLFOX therapy in the future.
Acknowledgements
The authors gratefully acknowledge the efforts and contributions of doctors, nurses and technical staff at the Cancer Hospital (Institute), Peking Union Medical College.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Li Q, Gan L, Liang L, Li X, Cai S. The influence of marital status on stage at diagnosis and survival of patients with colorectal cancer. Oncotarget. 2015;6:739–747. doi: 10.18632/oncotarget.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai MH, Xirasagar S, Li YJ, de Groen PC. Colonoscopy Screening Among US Adults Aged 40 or Older With a Family History of Colorectal Cancer. Prev Chronic Dis. 2015;12:E80. doi: 10.5888/pcd12.140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zauber AG. The impact of screening on colorectal cancer mortality and incidence: has it really made a difference? Dig Dis Sci. 2015;60:681–691. doi: 10.1007/s10620-015-3600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karoui M, Tresallet C, Julie C, Zimmermann U, Staroz F, Brams A, Muti C, Boulard C, Robreau AM, Puy H, Malafosse R, Penna C, Pruvot FR, Thiery JP, Boileau C, Rougier P, Nordlinger B, Radvanyi F, Franc B, Hofmann-Radvanyi H. Loss of heterozygosity on 10q and mutational status of PTEN and BMPR1A in colorectal primary tumours and metastases. Br J Cancer. 2004;90:1230–1234. doi: 10.1038/sj.bjc.6601687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, Symmans WF, Pusztai L, Nolden LK, Horlings H, Berns K, Hung MC, van de Vijver MJ, Valero V, Gray JW, Bernards R, Mills GB, Hennessy BT. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Board RE, Thelwell NJ, Ravetto PF, Little S, Ranson M, Dive C, Hughes A, Whitcombe D. Multiplexed assays for detection of mutations in PIK3CA. Clin Chem. 2008;54:757–760. doi: 10.1373/clinchem.2007.098376. [DOI] [PubMed] [Google Scholar]
- 7.Maehama T, Taylor GS, Dixon JE. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem. 2001;70:247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Lin Y, Lan F, Yu Y, Ouyang X, Wang X, Huang Q, Wang L, Tan J, Zheng F. A GG allele of 3’-side AKT1 SNP is associated with decreased AKT1 activation and better prognosis of gastric cancer. J Cancer Res Clin Oncol. 2014;140:1399–1411. doi: 10.1007/s00432-014-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, Hortobagyi GN, Hung MC, Yu D. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 10.Burris HA 3rd. Overcoming acquired resistance to anticancer therapy: focus on the PI3K/AKT/mTOR pathway. Cancer Chemother Pharmacol. 2013;71:829–842. doi: 10.1007/s00280-012-2043-3. [DOI] [PubMed] [Google Scholar]
- 11.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumor suppressor gene, MMAC1, at chromosome 10q23. 3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–364. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 13.Ding J, Gao Y, Liu R, Xu F, Liu H. Association of PTEN polymorphisms with susceptibility to hepatocellular carcinoma in a Han Chinese population. DNA Cell Biol. 2011;30:229–234. doi: 10.1089/dna.2010.1126. [DOI] [PubMed] [Google Scholar]
- 14.Ma J, Zhang J, Ning T, Chen Z, Xu C. Association of genetic polymorphisms in MDM2, PTEN and P53 with risk of esophageal squamous cell carcinoma. J Hum Genet. 2012;57:261–264. doi: 10.1038/jhg.2012.15. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Yang J, Yu Q, Wu H, Liu B, Xiong H, Hu G, Zhao J, Yuan X, Liao Z. Associations between Single-Nucleotide Polymorphisms in the PI3K-PTEN-AKT-mTOR Pathway and Increased Risk of Brain Metastasis in Patients with Non-Small Cell Lung Cancer. Clin Cancer Res. 2013;19:6252–6260. doi: 10.1158/1078-0432.CCR-13-1093. [DOI] [PubMed] [Google Scholar]
- 16.Wang LE, Ma H, Hale KS, Yin M, Meyer LA, Liu H, Li J, Lu KH, Hennessy BT, Li X, Spitz MR, Wei Q, Mills GB. Roles of genetic variants in the PI3K and RAS/RAF pathways in susceptibility to endometrial cancer and clinical outcomes. J Cancer Res Clin Oncol. 2012;138:377–385. doi: 10.1007/s00432-011-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu JL, Wang ZW, Hu LM, Yin ZQ, Huang MD, Hu ZB, Shen HB, Shu YQ. Genetic variants in the PI3K/PTEN/AKT/mTOR pathway predict platinum-based chemotherapy response of advanced non-small cell lung cancers in a Chinese Population. Asian Pac J Cancer Prev. 2012;13:2157–2162. doi: 10.7314/apjcp.2012.13.5.2157. [DOI] [PubMed] [Google Scholar]
- 18.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 19.Coughlin CM, Johnston DS, Strahs A, Burczynski ME, Bacus S, Hill J, Feingold JM, Zacharchuk C, Berkenblit A. Approaches and limitations of phosphatidylinositol-3-kinase pathway activation status as a predictive biomarker in the clinical development of targeted therapy. Breast Cancer Res Treat. 2010;124:1–11. doi: 10.1007/s10549-010-1108-4. [DOI] [PubMed] [Google Scholar]
- 20.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA, Malmström P, Memeo L, Isola J, Bendahl PO, Rosen N, Hibshoosh H, Ringnér M, Borg A, Parsons R. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oda K, Okada J, Timmerman L, Rodriguez-Viciana P, Stokoe D, Shoji K, Taketani Y, Kuramoto H, Knight ZA, Shokat KM, McCormick F. PIK3CA cooperates with other phosphatidy-linositol 3’-kinase pathway mutations to effect oncogenic transformation. Cancer Res. 2008;68:8127–8136. doi: 10.1158/0008-5472.CAN-08-0755. [DOI] [PubMed] [Google Scholar]
- 22.Colakoglu T, Yildirim S, Kayaselcuk F, Nursal TZ, Ezer A, Noyan T, Karakayali H, Haberal M. Clinicopathological significance of PTEN loss and the phosphoinositi de 3 kinase/Aktp athway in sporadic colorectal neoplasms: is PTEN loss predictor of local recurrence? Am J Surg. 2008;195:719–725. doi: 10.1016/j.amjsurg.2007.05.061. [DOI] [PubMed] [Google Scholar]
- 23.Lin PC, Lin JK, Lin HH, Lan YT, Lin CC, Yang SH, Chen WS, Liang WY, Jiang JK, Chang SC. A comprehensive analysis of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) loss in colorectal cancer. World J Surg Oncol. 2015;13:186. doi: 10.1186/s12957-015-0601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sood A, McClain D, Maitra R, Basu-Mallick A, Seetharam R, Kaubisch A, Rajdev L, Mariadason JM, Tanaka K, Goel S. PTEN gene expression and mutations in the PIK3CA gene as predictors of clinical benefit to anti-epidermal growth factor receptor antibody therapy in patients with KRAS wild-type metastatic colorectal cancer. Clin Colorectal Cancer. 2012;11:143–150. doi: 10.1016/j.clcc.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Q, Ju X, Li P, Meng X, Shao P, Cai H, Wang M, Zhang Z, Qin C, Yin C. A functional variant in the mTOR promoter modulates its expression and is associated with renal cell cancer risk. PLoS One. 2012;7:e50302. doi: 10.1371/journal.pone.0050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


