Abstract
Gastric cancer (GC) is one of the most common malignancies and one of the major causes of cancer-related deaths worldwide. In the present study, we investigated the association between miR-449a rs112310158 SNP and GC risk. Our findings revealed that a variant GG genotype increased the risk of occurrence of GC compared to a wild type AA genotype (OR = 2.542, 95% CI: 1.304-4.954, P = 0.005). Specifically, the G allele reduced the risk of occurrence of cervical cancer in women compared to the A allele (OR = 1.279, 95% CI: 1.012-1.617, P = 0.043). In conclusion, our findings suggest that miR-449a rs112310158 is a genetic risk factor for GC.
Keywords: Gastric cancer, miR-449a, polymorphism
Introduction
Gastric cancer (GC) is one of the most common malignancies and one of the major causes of cancer-related deaths worldwide [1]. Studies found that gastric carcinogenesis is a complex, long-lasting multistep and multifactorial process [2]. Epidemiological studies have shown that environmental factors, including high salt intake, consumption of alcoholic and tobacco, and Helicobacter pylori (H. pylori) infection, contribute to the risk of gastric cancer [3]. However, not all the people exposed to the same environmental risk factors eventually developed gastric carcinoma, suggesting that host or genetic factors may also play a role in the occurrence of gastric cancer [4].
In recent years, many studies explored the potential association between single nucleotide polymorphisms (SNPs) and risk of gastric cancer [5]. Among the reported SNPs, MiRNA-related single nucleotide polymorphisms (miR-SNPs), which are defined as single-nucleotide polymorphisms (SNPs) in miRNA genes or the miRNA binding site are of great interest [6,7]. MiRNAs are a class of conservative, small, single-strand, non-coding RNA molecules composed of around 22 nucleotides. MiRNAs regulate gene expression through degradation of target mRNAs or inhibition of translation by binding to complementary sequences in 3’-untranslated regions of messenger RNAs [8-10]. SNPs in miRNA genes or the miRNA binding site reportedly influence miRNAs expression, functions and individual susceptibility to cancers [11]. SNPs in miRNA may also play important roles in gastric cancer susceptibility through altering the expression or function of miRNAs, subsequently leading to aberrant expression of their target genes [12]. However, so far only very few studies have attempted to identify single nucleotide polymorphisms (SNPs) in miRNA genes which are associated with the occurrence of gastric cancer.
In the present study, we genotyped a mir-449a single nucleotide polymorphisms (rs112310158) in GC patients to evaluate the relationship of this genetic variant with risk for development of GC.
Materials and methods
Sample collection and DNA extraction
Blood samples were collected from GC patients who underwent GC resection at the Department of General Surgery at the Affiliated Hospital of Nantong University from 2012-2014. Blood samples were also collected from healthy volunteers without a history of any cancer. Genomic DNA was immediately extracted using QIAamp® Blood Mini Kit (QIAGEN Inc., Valencia, CA, USA) according to the manufacturers instruction and was stored at -20°C. This study was supervised and approved by the Human Tissue Research Committee of the Affiliated Hospital of Nantong University. Written consent was obtained from all the participants enrolled in this study.
Genotyping
The genotypes of rs112310158 were detected by polymerase-chain reaction (PCR)-direct sequencing assay. The 178-bp DNA fragment containing the polymorphic site was amplified using two primers (forward 5’-AGTGGCTTGCTTCATAGCAGAA-3’, and reverse 5’-GTGCTCTGGATACCTGTGTGT-3’). Polymerase chain reaction was performed in a total volume of 25 μl reaction mixture containing 50 ng of genomic DNA, 0.2 μl Taq DNA Polymerase (Takara, Dalian, China), 0.5 μl dNTP, 0.5 μl each primer, 2 μl 10 × Buffer and ddH2O. The polymerase chain reaction profile consisted of an initial melting step at 94°C for 3 minutes followed by 40 cycles of 94°C for 45 seconds, 60°C for 45 seconds and 72°C for 30 seconds, and an additional extension 72°C for 7 minutes in a thermal cycler (CFX-96, Bio-Rad). Purified products were sequenced on an ABI Prism 3730 × l sequencer (Applied Biosystems, Foster City, CA, USA) using BigDye Terminator Sequencing Standards.
Real-time quantitative polymerase chain reaction (RT-qPCR) of miR-449a
Total RNA was extracted from human tissues using Trizol reagent (Invitrogen, NY, USA) according to the manufacturer’s instructions. miR-449a quantification was performed using TaqMan MicroRNA Assays (Applied Biosystems, Foster City, USA). RT primer and TaqMan probe of miR-449a (Applied Biosystems, Foster City, USA) were used for PCR analysis on a Bio-Rad CFX-96 (Bio-Rad, Hercules, CA, USA) in accordance with the manufacturer’s protocol. RNU6B, as an internal control, was used as endogenous controls to normalize miR-449a.
Immunohistochemical analysis
Immunohistochemistry (IHC) was performed as described previously [13].
Statistical analyses
Statistical analyses were performed using the statistical package SPSS, version 12.0 (Chicago, IL, USA). All values are presented as mean ± standard deviation. Genotype and allelic frequencies were compared between the two groups by the Chi-Square test. The associations of genotypes of rs112310158 with risk of gastric cancer were estimated by calculating the odds ratios (OR) and their 95% confidence intervals (CIs) from multivariate logistic regression models. A P value less than 0.05 was considered the cutoff for statistical significance.
Results
Participant information
Characteristics of GC patients and healthy volunteers are summarized in Table 1. This case-control study enrolled 448 GC patients and 452 healthy volunteers. There was no statistically significant in the age and sex distributions between GC patients and healthy volunteers (P > 0.05). When comparing Clinical characteristics between GC cases and volunteers groups, GC cases were more likely to be cigarette smokers (OR = 1.345, 95% CI = 1.028-1.761) and alcohol drinkers (OR = 1.910, 95% CI = 1.465-2.490), have cancer history in the first relatives (OR = 4.178, 95% CI = 1.554-11.231), and have higher infection rate of Helicobacter pylori (OR = 1.428, 95% CI = 1.098-1.857).
Table 1.
Clinical characteristics of the gastric cancer cases and control subjects
| Cases | Controls | OR (95% CI) | P value | |
|---|---|---|---|---|
| N = 448 | N = 452 | |||
| Age, years | ||||
| < 55 | 179 (40.0%) | 190 (42.0%) | ||
| ≥ 55 | 269 (60.0%) | 262 (58.0) | 1.090 (0.835-1.422) | 0.542 |
| Sex | ||||
| Female | 148 (33.0%) | 159 (35.2%) | ||
| Male | 300 (67.0%) | 293 (64.8%) | 1.100 (0.835-1.449) | 0.527 |
| Cancer history in the first relatives | ||||
| No | 428 (95.5%) | 447 (98.9%) | ||
| Yes | 20 (4.5%) | 5 (1.1%) | 4.178 (1.554-11.231) | 0.002 |
| Alcohol drinkers | ||||
| Never | 202 (45.1%) | 276 (61.6%) | ||
| Ever | 246 (54.9%) | 176 (38.9%) | 1.910 (1.465-2.490) | 0.000 |
| Cigarette smokers | ||||
| Never | 260 (58.0%) | 294 (65.0%) | ||
| Ever | 188 (42.0%) | 158 (35.0%) | 1.345 (1.028-1.761) | 0.034 |
| Helicobacter pylori | ||||
| Negative | 207 (46.2%) | 249 | ||
| Positive | 241 (53.8%) | 203 | 1.428 (1.098-1.857) | 0.009 |
| Histologic type | ||||
| Squamous cell carcinoma | 382 (85.3%) | |||
| Adenocarcinomas | 36 (8.0%) | |||
| Adenosquamous carcinoma | 30 (6.7%) |
Association between miR-449a rs112310158 SNP and GC risk
Genotyping at miR-449a rs112310158 in control patients revealed the following distribution: 72.6% CC, 24.5% CG, and 9.2% GG (Table 2). A chi-squared test showed that, in healthy women, the distribution of genotypes at the miR-449a rs112310158 locus was in accordance with Hardy-Weinberg equilibrium (χ2 = 0.069, P = 0.792), indicating that these subjects are representative of the general population.
Table 2.
Genotype frequencies of the miR-449a rs112310158 polymorphism among GC cases and controls and their association with GC risk
| Genotypes | Cases No. (%) | Controls No. (%) | OR (95% CI) | P value |
|---|---|---|---|---|
| rs112310158 | ||||
| N = 448 | N = 452 | |||
| AA | 288 (64.3%) | 307 (67.9%) | ||
| AG | 129 (28.8%) | 132 (29.2%) | 1.042 (0.779-1.394) | 0.824 |
| GG | 31 (6.9%) | 13 (2.9%) | 2.542 (1.304-4.954) | 0.005 |
| A Allele | 705 (78.7%) | 746 (82.5%) | ||
| G Allele | 191 (21.3%) | 158 (17.5%) | 1.279 (1.012-1.617) | 0.043 |
Unconditional logistic regression analysis showed that a variant GG genotype increased the risk of occurrence of GC compared to a wild type AA genotype (OR = 2.542, 95% CI: 1.304-4.954, P = 0.005); an AG genotype did not confer a statistically significant change in risk. Specifically, the G allele reduced the risk of occurrence of cervical cancer in women compared to the A allele (OR = 1.279, 95% CI: 1.012-1.617, P = 0.043) (Table 2). Further, Stratified analysis of the age showed that subjects over 55 years of age, the G allele further increased the risk of GC compared to the A allele (OR = 3.461, 95% CI: 1.447-8.276, P = 0.004) (Table 3).
Table 3.
Increased genetic susceptibility to GC in subjects over 55 years of age at the miR-449a rs112310158 polymorphism locus
| Genotypes | Cases No. (%) | Controls No. (%) | OR (95% CI) | P value |
|---|---|---|---|---|
| rs112310158 | ||||
| N = 269 | N = 262 | |||
| AA | 169 (62.8%) | 178 (67.9%) | ||
| AG | 77 (28.6%) | 77 (29.4%) | 1.053 (0.721-1.540) | 0.847 |
| GG | 23 (8.6%) | 7 (2.7%) | 3.461 (1.447-8.276) | 0.004 |
| A Allele | 415 (77.1%) | 433 (82.6%) | ||
| G Allele | 123 (22.9%) | 91 (17.4%) | 1.410 (1.042-1.909) | 0.027 |
Correlation between miR-449a rs112310158 polymorphism and risk of being infected with Helicobacter pylori
Unconditional logistic regression analysis showed that the GG genotype reduced the risk of occurrence of Helicobacter pylori infection in women compared to the AA genotype (OR = 2.683, 95% CI: 1.376-5.229); The G allele was associated with reduced risk of occurrence of Helicobacter pylori infection compared to the A allele (OR = 1.385, 95% CI: 1.095-1.752) (Table 4).
Table 4.
The miR-449a rs112310158 polymorphism genotype is correlated with Helicobacter pylori infection
| Helicobacter pylori positive | Helicobacter pylori negative | OR (95% CI) | P value | |
|---|---|---|---|---|
| rs112310158 | ||||
| N = 444 | N = 456 | |||
| AA | 280 | 315 | ||
| AG | 133 | 128 | 1.169 (0.874-1.564) | 0.653 |
| GG | 31 | 13 | 2.683 (1.376-5.229) | 0.003 |
| A Allele | 693 | 758 | ||
| G Allele | 195 | 154 | 1.385 (1.095-1.752) | 0.007 |
Correlation between rs112310158 and the expression of miR-449a and its target gene
Using fluorescence quantitative PCR assay, we detected the expression of miR-449a in GC tissues from various genotypes. We found the miR-449a levels were significantly lower in patients with GG genotype than that in patients with AA genotype (Figure 1A). Furthermore, miR-449a target gene was searched using the public database TargetScan (http://www.targetscan.org), and protein kinase C, epsilon (PRKCE) that possessed a critically conserved binding site for miR-449a was selected for further study (Figure 1B). We further evaluated the clinical significance of PRKCE expression in patient various genotypes. Immunohistochemical staining (Figure 1C) demonstrated that PRKCE expression was higher in GC patients with GG genotype compared with that in patients with AA genotype.
Figure 1.
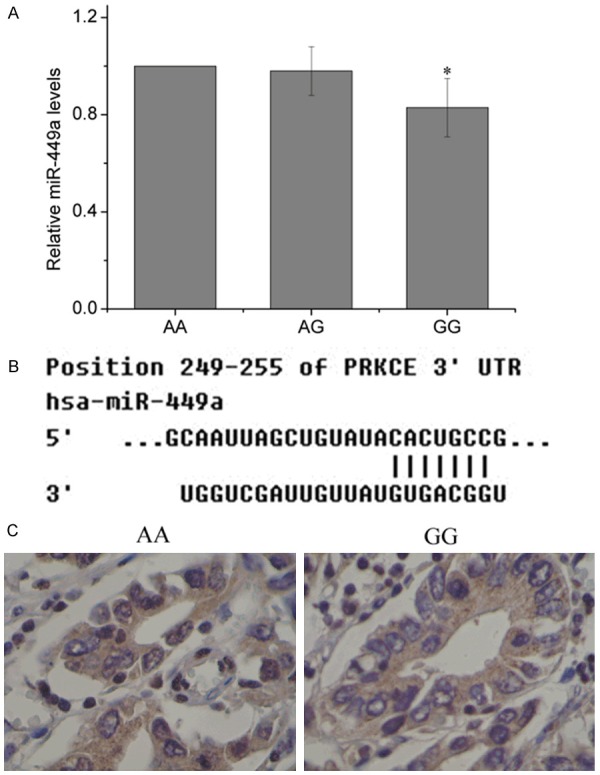
Correlation between rs112310158 and the expression of miR-449a and its target gene. A: The expression of miR-449a was detected by QRT-PCR. B: miR-449a target gene was searched using the public database TargetScan (http://www.targetscan.org). C: PRKCE expression was detected by immunohistochemical staining. *P < 0.05.
Discussion
In the present study, we investigated the association between miR-449a rs112310158 SNP and GC risk. Our findings revealed that miR-449a rs112310158 GG genotype was significantly related to increased risk of GC compared with those carrying the AA genotypes. Furthermore, we found miR-449a rs112310158 polymorphism was associated with the expression of miR-449a and its target gene (PRKCE).
microRNA-449a (miR-449a) is a miRNA that has been previously identified in several types of cancers. miR-449a, which functions as a cancer suppressor gene, is closely associated with a great variety of cancer, including lung [14], liver [15] and gastric [16] cancer; prostatic carcinoma [17,18]; ovarian [19]; and breast [20] and bladder [21] carcinoma. In normal conditions, miR-449a was found to be strongly expressed in testis, lung and trachea tissue [22]. Down-regulation of miR-449a has been detected in several cancers including GC. Li et al reported that miR-449a was downregulated in gastric cancer cell lines and gastric cancer tissues [23]. They found miR-449a/E2F3 axis plays an important role in proliferation and apoptosis in gastric cancer. Similarly, Hu et al. provided evidence that miR-449a could modulate cell cycle and apoptosis through regulating cyclin D1 and BCL2 expression in SGC7901 cells [24]. However, the exact role of miR-449a in gastric cancer remains unknown. In the present study, we found miR-449a rs112310158 SNP was associated with GC risk in a Chinese population. A variant GG genotype of rs112310158 increased the risk of occurrence of GC compared to a wild type AA genotype (OR = 2.542, 95% CI: 1.304-4.954, P = 0.005).
Kinases are central mediators of signal transduction and an important class among them is protein kinase C (PKC) which constitutes 2% of the human kinome [25]. PKC is a series of structurally related serine/threonine kinases that are classified as conventional PKC, novel PKC and atypical PKC based on their structural properties and responsiveness to second messengers [26]. Since their discovery as receptors for tumor-promoting phorbol esters, PKCs have been intensively studied for their contribution to cancer [27]. Common processes regulated by PKCs include cell survival, proliferation, apoptosis, migration and invasion [28]. PKCε is the first isozyme that was shown to possess oncogenic functions and is emerging as an undisputed tumor promoter. PKCε is overexpressed in various tumor types and is associated with different processes related to cancer development namely, cell transformation, cell survival, cell proliferation, EMT, cytoskeletal reorganization, extracellular matrix (ECM) rearrangement, disruption of cell-cell contacts, cell motility, stem cell properties and therapy resistance [29]. In this study, we found protein kinase C epsilon possessed a critically conserved binding site for miR-449a and may be a direct target gene of miR-449a. We found the miR-449a levels were significantly lower in patients with GG genotype than that in patients with AA genotype. Furthermore, we found PRKCE expression was higher in GC patients with GG genotype compared with that in patients with AA genotype. These findings indicated that miR-449a rs112310158 may influence the risk of GC through modulating PRKCE expression.
In conclusion, our findings suggest that miR-449a rs112310158 is a genetic risk factor for GC. Further larger-scale and well-designed clinical studies are warranted to validate this finding.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 3.Kelley JR, Duggan JM. Gastric cancer epidemiology and risk factors. J Clin Epidemiol. 2003;56:1–9. doi: 10.1016/s0895-4356(02)00534-6. [DOI] [PubMed] [Google Scholar]
- 4.Milne AN, Carneiro F, O’Morain C, Offerhaus GJ. Nature meets nurture: molecular genetics of gastric cancer. Hum Genet. 2009;126:615–628. doi: 10.1007/s00439-009-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condorelli G, Latronico MV, Cavarretta E. microRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol. 2014;63:2177–2187. doi: 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 6.Link A, Kupcinskas J, Wex T, Malfertheiner P. Macro-role of microRNA in gastric cancer. Dig Dis. 2012;30:255–267. doi: 10.1159/000336919. [DOI] [PubMed] [Google Scholar]
- 7.Pan HW, Li SC, Tsai KW. MicroRNA dysregulation in gastric cancer. Curr Pharm Des. 2013;19:1273–1284. doi: 10.2174/138161213804805621. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 11.Ni Q, Ji A, Yin J, Wang X, Liu X. Effects of Two Common Polymorphisms rs2910164 in miR-146a and rs11614913 in miR-196a2 on Gastric Cancer Susceptibility. Gastroenterol Res Pract. 2015;2015:764163. doi: 10.1155/2015/764163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua HB, Yan TT, Sun QM. miRNA polymorphisms and risk of gastric cancer in Asian population. World J Gastroenterol. 2014;20:5700–5707. doi: 10.3748/wjg.v20.i19.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen WH, Xin PL, Pan QX, Chen YY, Wang CR, Zhang ZS, Chen YF, Zhang CY, Cai WJ. ERCC1 single nucleotide polymorphism C8092A, but not its expression is associated with survival of esophageal squamous cell carcinoma patients from Fujian province, China. PLoS One. 2014;9:e106600. doi: 10.1371/journal.pone.0106600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeon HS, Lee SY, Lee EJ, Yun SC, Cha EJ, Choi E, Na MJ, Park JY, Kang J, Son JW. Combining microRNA-449a/b with a HDAC inhibitor has a synergistic effect on growth arrest in lung cancer. Lung Cancer. 2012;76:171–176. doi: 10.1016/j.lungcan.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Buurman R, Gurlevik E, Schaffer V, Eilers M, Sandbothe M, Kreipe H, Wilkens L, Schlegelberger B, Kuhnel F, Skawran B. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. 2012;143:811–820. e1–15. doi: 10.1053/j.gastro.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 16.Bou Kheir T, Futoma-Kazmierczak E, Jacobsen A, Krogh A, Bardram L, Hother C, Gronbaek K, Federspiel B, Lund AH, Friis-Hansen L. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 18.Noonan EJ, Place RF, Basak S, Pookot D, Li LC. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget. 2010;1:349–58. doi: 10.18632/oncotarget.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, He XJ, Ma LP, Li N, Yang J, Cheng YX, Cui H. [Expression and significance of microRNAs in the p53 pathway in ovarian cancer cells and serous ovarian cancer tissues] . Zhonghua Zhong Liu Za Zhi. 2011;33:885–890. [PubMed] [Google Scholar]
- 20.Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M, Yu Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388–2393. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Lin YW, Mao YQ, Wu J, Liu YF, Zheng XY, Xie LP. MicroRNA-449a acts as a tumor suppressor in human bladder cancer through the regulation of pocket proteins. Cancer Lett. 2012;320:40–47. doi: 10.1016/j.canlet.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 22.Lize M, Pilarski S, Dobbelstein M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 2010;17:452–458. doi: 10.1038/cdd.2009.188. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Li H, Zhang R, Liu J. MicroRNA-449a inhibits proliferation and induces apoptosis by directly repressing E2F3 in gastric cancer. Cell Physiol Biochem. 2015;35:2033–2042. doi: 10.1159/000374010. [DOI] [PubMed] [Google Scholar]
- 24.Hu J, Fang Y, Cao Y, Qin R, Chen Q. miR-449a Regulates proliferation and chemosensitivity to cisplatin by targeting cyclin D1 and BCL2 in SGC7901 cells. Dig Dis Sci. 2014;59:336–345. doi: 10.1007/s10620-013-2923-3. [DOI] [PubMed] [Google Scholar]
- 25.Cameron AJ, Escribano C, Saurin AT, Kostelecky B, Parker PJ. PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat Struct Mol Biol. 2009;16:624–630. doi: 10.1038/nsmb.1606. [DOI] [PubMed] [Google Scholar]
- 26.Jain K, Basu A. The Multifunctional Protein Kinase C-epsilon in Cancer Development and Progression. Cancers (Basel) 2014;6:860–878. doi: 10.3390/cancers6020860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu A. The potential of protein kinase C as a target for anticancer treatment. Pharmacol Ther. 1993;59:257–280. doi: 10.1016/0163-7258(93)90070-t. [DOI] [PubMed] [Google Scholar]
- 28.Steinberg SF. Structural basis of protein kinase C isoform function. Physiol Rev. 2008;88:1341–1378. doi: 10.1152/physrev.00034.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorin MA, Pan Q. Protein kinase C epsilon: an oncogene and emerging tumor biomarker. Mol Cancer. 2009;8:9. doi: 10.1186/1476-4598-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


