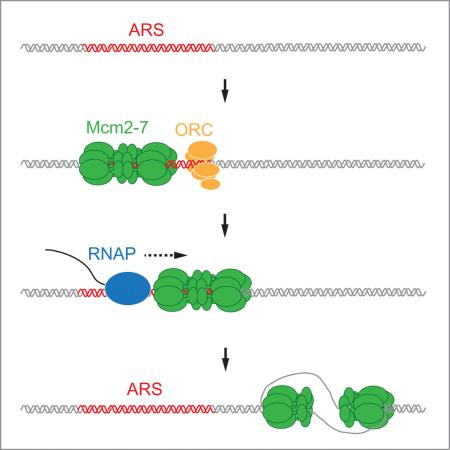
Summary
Eukaryotic genomes are replicated from many origin sites that are licensed by the loading of the replicative DNA helicase, Mcm2-7. How eukaryotic origin positions are specified remains elusive. Here we show that, contrary to the bacterial paradigm, eukaryotic replication origins are not irrevocably defined by selection of the helicase loading site, but can shift in position after helicase loading. Using purified proteins we show that DNA translocases, including RNA polymerase, can push budding yeast Mcm2-7 double hexamers along DNA. Displaced Mcm2-7 double hexamers support DNA replication initiation distal to the loading site in vitro. Similarly, in yeast cells that are defective for transcription termination, collisions with RNA polymerase induce a redistribution of Mcm2-7 complexes along the chromosomes, resulting in a corresponding shift in DNA replication initiation sites. These results reveal a eukaryotic origin specification mechanism that departs from the classical replicon model, helping eukaryotic cells to negotiate transcription-replication conflict.
Graphical Abstract
Introduction
Eukaryotic genomes are replicated from many origin sites that are distributed along each chromosome. How these sites are selected is not well understood. The replicon model proposed an origin specification mechanism that is based on the interaction of an initiator protein with a specific DNA sequence element, termed ‘replicator’, at the chromosomal origin (Jacob, 1964). This model proved sufficient to account for origin site selection in bacteria. For example, in Escherichia coli, specific recognition sequences target the initiator protein, DnaA, to the unique chromosomal origin (Costa et al., 2013). Upon binding to the origin, DnaA locally melts the DNA and directs the loading of a molecule of the replicative DNA helicase, DnaB, around each of the unwound DNA strands. Further origin unwinding by DnaB facilitates subsequent assembly of the replisome. Since DnaB is loaded in active form around the DNA, selection of the helicase loading site effectively determines origin position in E. coli.
The isolation of budding yeast, Saccharomyces cerevisiae, replication origins as autonomously replicating sequence (ARS) elements suggested that eukaryotic origins of replication are similarly defined by specific DNA sequences (Stinchcomb et al., 1979). However, studies in Xenopus demonstrated that specific origin DNA sequences are not principally required for regulated DNA replication in eukaryotes (Harland and Laskey, 1980), a feature that has since been also documented in other eukaryotic systems ranging from yeast to human cells (Bogenschutz et al., 2014; Dershowitz et al., 2007; Gros et al., 2014; Kipling and Kearsey, 1990; Krysan and Calos, 1991; On et al., 2014; Takeda et al., 2005). This plasticity may allow for the very rapid replication of chromosomes from a large number of non-specific origin sites, which occurs, for example, during the cleavage cycles of early Xenopus and Drosophila embryos (Blumenthal et al., 1974; Hyrien et al., 1995). Nonetheless, DNA replication initiates from distinct chromosomal regions in yeast and in higher eukaryotes later in development, raising the questing of how these sites are specified (Gilbert, 2004; Hyrien, 2015).
The lack of specific replicator sequences in eukaryotes suggests that epigenetic and developmentally controlled mechanisms play a more prominent role in defining eukaryotic origin positions. Eukaryotic origins are licensed in late M / G1 phase by pre-replicative complexes (pre-RCs) that load the replicative DNA helicase, Mcm2-7, in inactive form onto chromatin; activation of the Mcm2-7 helicase occurs exclusively in the subsequent S phase, when replisomes assemble around the Mcm2-7 core (Siddiqui et al., 2013). Principally, eukaryotic origin specification mechanisms either actively recruit initiation factors to direct pre-RC assembly at specific chromosomal regions, or conversely exclude pre-RCs from potential origin sites. Transcription, for example, which is known to interfere with pre-RC assembly and thereby inhibit replication origin activity (Donato et al., 2006; Looke et al., 2010; Mori and Shirahige, 2007; Nieduszynski et al., 2005; Snyder et al., 1988), may shape chromosomal replication profiles by restricting pre-RC assembly to non-transcribed regions. Consistent with this notion, onset of zygotic transcription at the midblastula transition promotes specific origin usage in Xenopus and Drosophila embryos by confining replication initiation sites to intergenic regions (Hyrien et al., 1995; Sasaki et al., 1999). Similarly, transcription appears to define the boundaries of the initiation zone located between the convergent dihydrofolate reductase (DHFR) and 2BE2121 genes in chinese hamster ovary (CHO) cells: Deletion of the DHFR promoter extends the initiation zone into the inactive DHFR gene (Saha et al., 2004), whereas defective termination of transcription of the DHFR gene restricts the initiation zone to the remaining non-transcribed intergenic space further downstream (Mesner and Hamlin, 2005).
The strict separation of Mcm2-7 loading and Mcm2-7 activation in the cell cycle is essential to prevent re-replication (Siddiqui et al., 2013). At the same time, this two-step mechanism imposes a requirement for replication origins to maintain competence from the time of licensing until the time of activation. Activation times vary significantly, as origins do not fire synchronously upon entry into S phase, but fire in a predetermined pattern throughout S phase, while some origins are only activated during replication stress (Blow et al., 2011; Rhind and Gilbert, 2013). During pre-RC assembly, Mcm2-7 complexes are loaded as inactive double-hexamers (DHs) around double-stranded DNA, in a reaction that requires the coordinated activities of the initiation factors ORC, Cdc6, and Cdt1 (Yardimci and Walter, 2014). Mcm2-7 DHs form topologically bound complexes on DNA that even resist high-salt washes (Gros et al., 2014; Remus et al., 2009). Nonetheless, in budding yeast cells arrested in G1 phase with α-factor continuous re-loading of Mcm2-7 proteins by ORC and Cdc6 is required to maintain Mcm2-7 at the origin (Aparicio et al., 1997; Chen et al., 2007; Cocker et al., 1996; Semple et al., 2006). Such re-loading, however, cannot occur during S phase when re-replication control mechanisms prevent Mcm2-7 loading (Siddiqui et al., 2013), raising the question how origin function is maintained during normal S phase or during prolonged S phase arrest after checkpoint activation.
It was previously found that reconstituted Mcm2-7 DHs can slide on DNA if exposed to high-salt conditions (Evrin et al., 2009; Remus et al., 2009). The physiological significance of this Mcm2-7 DH mobility, and whether Mcm2-7 DHs remain functional after sliding along DNA, is not known. We reasoned that the ability to slide may help maintain Mcm2-7 DHs on DNA after collision with other proteins. For example, transcriptional activity has been detected at a large fraction of budding yeast origins (Looke et al., 2010), indicating that collisions between RNA polymerase and pre-RCs may occur on a frequent basis. We, therefore, set out to investigate Mcm2-7 DH stability after collision with other proteins on DNA. We find that although ARS elements are preferred loading sites for Mcm2-7 both in vitro and in vivo, DNA replication can initiate distal to ARS elements upon re-localization of Mcm2-7 DHs away from the ARS. Our results uncover facilitated Mcm2-7 DH sliding as a mechanism by which non-replicative DNA transactions, such as transcription, can influence eukaryotic origin positions, and suggest a eukaryote-specific origin maintenance mechanism that is not dependent on Mcm2-7 re-loading.
Results
Isolation of pre-RCs free in solution
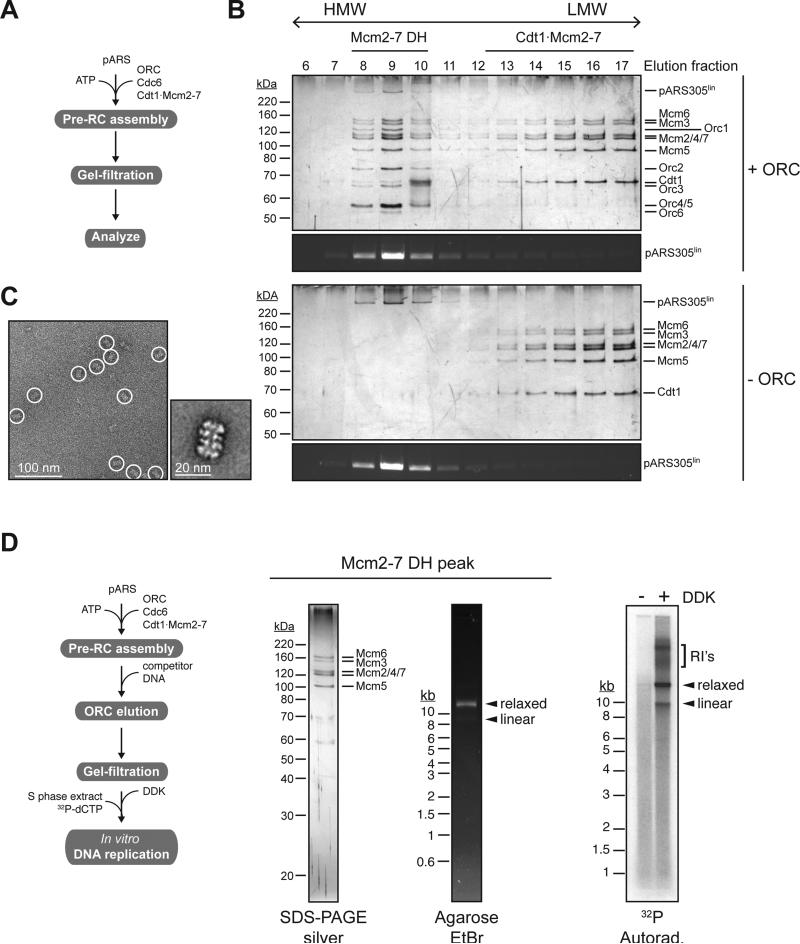
To model collisions between DNA translocases and Mcm2-7 DHs in vitro we developed a gel-filtration-based approach to analyze and isolate reconstituted budding yeast pre-RCs (Remus et al., 2009) bound to DNA free in solution (Figure 1A). Mcm2-7 complexes separate into two distinct populations during gel-filtration of a pre-RC assembly reaction (Figure 1B): A low molecular weight (LMW) fraction corresponding to Cdt1·Mcm2-7 complexes that were not converted into pre-RCs in the course of the reaction; and an ORC-dependent high molecular weight (HMW) fraction that co-fractionates with ORC and template DNA, but is largely devoid of Cdt1, corresponding to pre-RCs. Electron-microscopic analysis of the HMW fraction confirms the double hexameric structure of Mcm2-7 complexes eluting in this fraction (Figure 1C).
Figure 1. Isolation of functional pre-RCs free in solution.
(A) Reaction scheme. (B) Gel-filtration analysis of reconstituted pre-RC assembly reaction using linearized pARS305 (9.8 kb) as a template. Proteins were analyzed by SDS-PAGE and silver stain, DNA was analyzed by agarose gel-electrophoresis and ethidium-bromide staining. Pre-RCs were assembled in the presence (top two panels) or absence (bottom two panels) of ORC. (C) Representative electron micrograph (left) and 2D class average (right) of Mcm2-7 complexes eluting in fractions 8-10 in presence of ORC in B. (D) In vitro DNA replication assay using purified Mcm2-7 DHs isolated on relaxed pARS305 (9.8 kb); reaction scheme on left. Input protein was analyzed by SDS-PAGE and silver stain, input DNA by agarose gel-electrophoresis and ethidium bromide stain. Replication products were visualized by autoradiography after native agarose gel-electrophoresis. RI's = replication intermediates.
Although ORC, unlike Cdc6 and Cdt1, remains bound to the DNA after Mcm2-7 loading, it does not maintain contact with the Mcm2-7 DH (Remus et al., 2009; Sun et al., 2014) and is dispensable for DNA replication after Mcm2-7 loading (Gros et al., 2014; Hua and Newport, 1998; Rowles et al., 1999). We find that ORC is selectively removed from the template DNA after Mcm2-7 loading by competition with an excess of ARS-containing DNA prior to gel-filtration (Figure 1D). Using an in vitro replication approach (Gros et al., 2014) we tested the activity of Mcm2-7 DHs isolated in this manner. Purified Mcm2-7 DHs bound to plasmid DNA were phosphorylated with Cdc7·Dbf4 kinase (DDK), and DNA replication initiated by addition of S phase yeast extract supplemented with 32P-dCTP. The data confirm that isolated Mcm2-7 DHs are functional and support regulated DNA replication in vitro (Figure 1D).
T7 RNAP can push Mcm2-7 DHs along DNA
To model collisions between Mcm2-7 DHs and transcription complexes we analyzed the effect of T7 RNA polymerase (T7 RNAP) on Mcm2-7 DH stability. Under the conditions used for pre-RC assembly, T7 RNAP transcription of pARS1 and pARS305 is dependent on the presence of the T7 promoter (Figure S1A). Transcript lengths greatly exceed the unit length of the plasmids, demonstrating that T7 RNAP transcribes the entire length of the templates, yet T7 RNAP progression is efficiently controlled by a tandem array of T7 terminator sequences (Figure S1B).
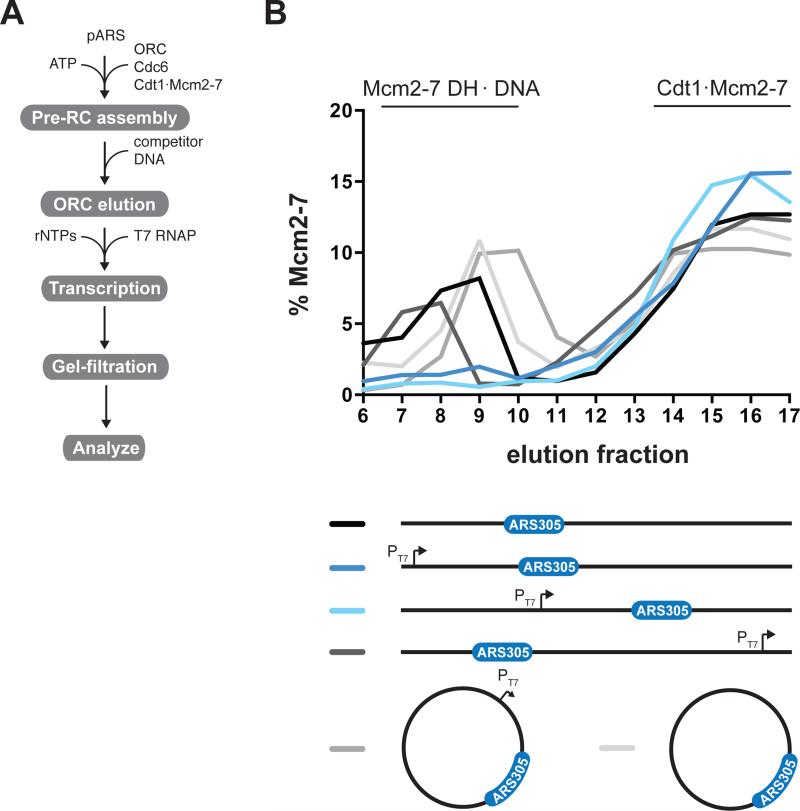
We determined whether Mcm2-7 DHs remain bound to DNA in the presence of T7 transcription using gel-filtration. As templates we used variants of pARS305, which we have previously shown to assemble pre-RCs preferentially at the ARS305 sequence (Gros et al., 2014). Following pre-RC assembly we supplemented the reaction with a ribonucleotide-triphosphate (rNTP) mix and T7 RNAP to initiate transcription (Figure 2A). To prevent reloading of Mcm2-7 in the course of the experiment, ORC was removed from the template DNA by addition of ARS1 competitor DNA prior to transcription initiation. Mcm2-7 DHs were retained on linear pARS305 when T7 RNAP transcription was prevented in the absence of a T7 promoter, but were lost from the DNA if transcription was initiated from a T7 promoter placed upstream of ARS305 (Figure 2B and Figure S2B). Mcm2-7 DHs were lost from the template DNA if the T7 promoter was oriented towards ARS305, but not if the promoter was oriented away from it. To distinguish between T7 RNAP disintegrating Mcm2-7 DHs or pushing Mcm2-7 DHs off the ends of the DNA, we monitored Mcm2-7 DH persistence on circular DNA. Mcm2-7 DHs were retained on relaxed circular pARS305 both in the absence and presence of transcribing T7 RNAP, indicating that Mcm2-7 DHs do not disintegrate upon collision with T7 RNAP. To exclude the possibility that positive supercoils between the Mcm2-7 DH and advancing T7 RNAP may prevent collision of T7 RNAP with the Mcm2-7 DH we also tested Mcm2-7 DH stability on negatively supercoiled plasmids in the presence of Vaccinia topoisomerase I, which relaxes both positive and negative supercoils. Again Mcm2-7 DHs were stably maintained irrespective of T7 RNAP transcription (Figure S2C). These experiments demonstrate that elongating T7 RNAP can push Mcm2-7 DHs along DNA and off free DNA ends.
Figure 2. T7 RNAP can push Mcm2-7 DHs off the ends of linear DNA.
(A) Reaction scheme. (B) Pre-RCs were tested for displacement by T7 RNAP on linear or circular relaxed forms of pARS305 (9.8 kb) indicated on the right. Protein-DNA complexes were analyzed by gel-filtration; relative Mcm2-7 content in each fraction was determined after SDS-PAGE and silver stain (Figure S2). PT7, T7 promoter.
Mcm2-7 DH activity after collision with T7 RNAP
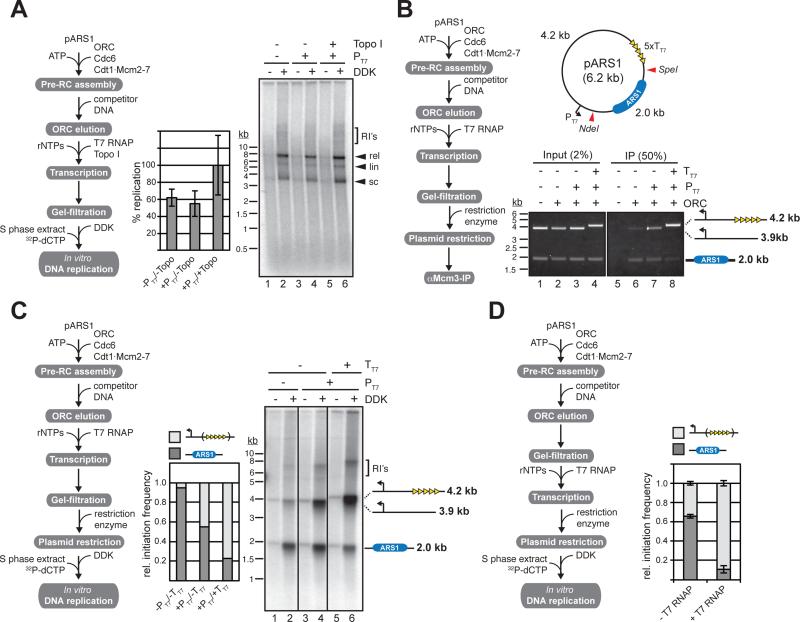
Next we asked if Mcm2-7 DHs continue to support DNA replication in vitro after collision with T7 RNAP. Transcription was initiated after pre-RC assembly and elimination of ORC; plasmid-bound Mcm2-7 DHs, purified by gel-filtration, were tested for their ability to support DNA replication as above. Intriguingly, T7 RNAP transcription did not interfere with the replication of pARS1, indicating that Mcm2-7 DHs remain functional after collision with T7 RNAP (Figure 3A).
Figure 3. Mcm2-7 DHs remain functional after displacement by T7 RNAP.
(A) Native agarose gel analysis of in vitro DNA replication products obtained with pre-RCs reconstituted on circular closed pARS1 (5.8 kb) treated with T7 RNAP in the absence or presence of a T7 promoter or Vaccinia Topo I as indicated. Averages and standard deviations of 32P-dCTP incorporation from three independent experiments are shown in the bar diagram. Reaction scheme on the left. (B) Mcm2-7 DH location analysis by co-immunoprecipitation of Mcm2-7 complexes and DNA fragments generated from pre-RCs assembled on circular closed pARS1 according to reaction scheme on left. Restriction sites used are indicated by red triangles in the plasmid map. PT7, T7 promoter; TT7, T7 terminator. DNA was analyzed by ethidium-bromide stain. (C) In vitro replication analysis using Mcm2-7/DNA fragment complexes obtained according to reaction scheme on left. Replication products were analyzed by native agarose gel-electrophoresis and autoradiography (right panel) and 32P incorporation into respective fragments was quantitated by phosphorimaging (middle graph). (D) In vitro replication analysis of Mcm2-7/DNA complexes obtained according to reaction scheme on left. The pARS1 template included T7 terminator sequences (see map in b); restriction was carried out with SpeI / NdeI. Replication products were analyzed by native agarose gel-electrophoresis and autoradiography and 32P incorporation into respective fragments was quantitated by phosphorimaging.
To determine Mcm2-7 DH position after collision with T7 RNAP we used a Mcm2-7 / DNA co-immunoprecipitation approach. For this we fragmented the template DNA by restriction endonuclease digestion after re-isolation from the pre-RC assembly / transcription reaction, and determined which fragments would co-immunoprecipitate with Mcm2-7 complexes under respective conditions (Figure 3B). Restriction digestion of pARS1 yielded a 2 kb ARS1-containing fragment, and a 3.9 kb vector backbone fragment, whose size was increased to 4.2 kb if T7 terminator sequences were included. In the absence of T7 transcription Mcm2-7 DHs associated primarily with the 2.0 kb ARS1-containing DNA fragment (Figure 3B, lane 6), consistent with ARS1 being the preferred site for pre-RC assembly on the plasmid. If T7 transcription was initiated upstream of ARS1 Mcm2-7 DHs were associated to roughly equal parts with both the ARS1-containing fragment and the vector backbone fragment (Figure 3B, lane 7). Termination of transcription downstream of ARS1 resulted in the majority of Mcm2-7 DHs being bound to the vector backbone fragment (Figure 3B, lane 8). These data demonstrate that Mcm2-7 DHs are initially loaded at ARS1, but are subsequently redistributed around the plasmid by T7 RNAP.
To determine if Mcm2-7 DHs dispersed by T7 RNAP can initiate DNA replication from sites distal to the loading site at the ARS we tested their ability to support replication of the bound restriction fragments (Figure 3C). Indeed restriction fragments were replicated in a manner that correlated with the Mcm2-7 DH distribution observed in figure 3B. On the promoterless template the majority of replication initiation events occurred on the ARS1-containing 2.0 kb fragment (Figure 3C, lanes 1-2). Transcription initiated upstream of ARS1 after Mcm2-7 loading increased the number of initiation events on the 3.9 kb vector backbone-containing fragment, resulting in an almost even distribution of initiation events between the 2.0 kb and the 3.9 kb fragments (Figure 3C, lanes 3-4). Termination of T7 transcription downstream of ARS1 essentially reversed the distribution of initiation events observed in the absence of transcription, with the majority of initiation events now occurring on the 4.2 kb vector backbone fragment (Figure 3C, lanes 5-6). Analogous results were also obtained with pARS305 (Figure S3A). To exclude the possibility that the redistribution of initiation events after T7 transcription resulted from redistributed Mcm2-7 loading by ORC that was not efficiently competed off the template DNA prior to transcription initiation, we isolated Mcm2-7 DHs from a pre-RC assembly reaction by gel-filtration prior to the initiation of transcription. Again DNA replication initiated preferentially on the ARS1-containing 2.0 kb DNA fragment in the absence of transcription, but was shifted to the T7 terminator-containing vector backbone by transcribing T7 RNAP (Figure 3D). The functional relocation of Mcm2-7 DHs on linear templates (Figure S3B-D) that are devoid of topological constraints, and therefore do not allow any potential DNA supercoils to accumulate between the advancing T7 RNAP and Mcm2-7 DH, indicates that T7 RNAP relocates Mcm2-7 DHs along DNA via direct physical contact. We conclude that relocated Mcm2-7 DHs remain competent for replication initiation at sites distal to the initial loading site. This is consistent with our previous study (Gros et al., 2014) showing that DNA sequences at the ARS promote preferential loading of Mcm2-7 DHs by the pre-RC in vitro, but that specific origin sequences are not required for activation of the Mcm2-7 DH during initiation. To test if the ability to slide upon collision with other proteins is a general feature of Mcm2-7 DHs, we also examined collisions between FtsK, a hexameric double-stranded DNA translocase, and Mcm2-7 DHs in vitro (Figure S4). Like T7 RNAP, FtsK was able to functionally displace Mcm2-7 DHs on DNA in vitro, demonstrating that Mcm2-7 DHs can be functionally relocalized along the DNA template by distinct classes of DNA translocases.
Mcm2-7 redistribution by RNA polymerase in vivo
Next we aimed to characterize collisions between the transcription machinery and replication origins in vivo. To this end we exploited the fact that ~ 90 % of budding yeast replication origins are located downstream of protein coding genes, i.e. either between convergent genes, or genes that are organized in tandem (MacAlpine and Bell, 2005). We reasoned that collisions between RNAP and pre-RCs may be enhanced in mutants defective for transcription termination and, therefore, decided to examine the positions of both ORC and Mcm2-7, as well as the sites of DNA replication initiation, genome wide in yeast cells that harbor a temperature-sensitive mutation in Rat1. Rat1 is a 5’-3’ ribo-exonuclease that promotes the release of RNA polymerase from the DNA template after cleavage of the nascent RNA transcript, in a process termed “torpedo” mechanism (Tollervey, 2004). rat1-1 cells exhibit pronounced termination defects during mRNA transcription by RNA polymerase II (RNAP II) and rRNA transcription by RNA polymerase I (RNAP I) at non-permissive temperature (El Hage et al., 2008; Kawauchi et al., 2008; Kim et al., 2004). Rat1 is essential for cell viability, yet genetic rescue experiments suggest that the essential function of Rat1 is distinct from its role in transcription termination (Luo et al., 2006). In the following experiments, asynchronous cultures of rat1-1 cells were shifted for two hours to non-permissive temperature (37°C), a condition that does not elicit a checkpoint response as judged by Rad53 phospho-shift analysis (Figure S5A), or induce gross changes in cell cycle stage as determined by FACS analysis of nuclear DNA content (Figure S5B).
To examine the consequence of collisions of RNAP with pre-RCs we used ChIP-seq to determine the chromosomal positions of both ORC and Mcm2-7 across the genome of rat1-1 cells. We identified 245 peaks of ORC binding, of which 202 overlapped with sites identified in an earlier study that analyzed ORC distribution in cells arrested in G2/M by nocodazole treatment (Figure S5C) (Eaton et al., 2010). For Mcm2-7 we identified 375 peak binding sites, 310 of which overlapped with sites identified in α-factor-arrested cells in a prior study (Figure S5C) (Blitzblau et al., 2012). The extensive overlap between the ORC and Mcm2-7 binding sites observed here and in prior studies validates our approach of assessing pre-RC positions in cultures of asynchronous yeast cells. 210 of the 245 ORC sites identified here overlap with Mcm2-7 binding sites, and also largely localize to annotated ARS sites (Figure S5C + D), thus fulfilling the criterion for origin-associated pre-RCs (Wyrick et al., 2001).
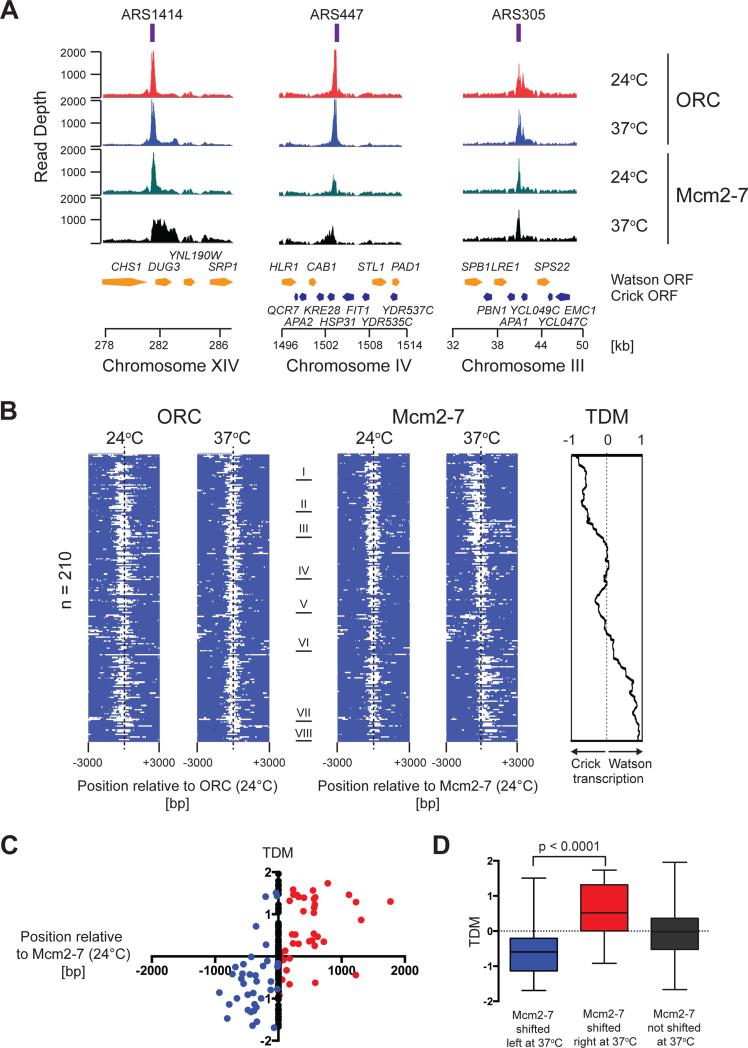
Inspection of the ORC binding peaks at individual origin sites indicates that ORC binding is not altered at elevated temperature in rat1-1 cells (Figure 4A and Figure S5D). On the contrary, at numerous origin sites the Mcm2-7 binding peaks are significantly broadened by several hundred base pairs and up to a few kilo base pairs at 37°C relative to those at 24°C (e.g. ARS1414 and ARS447, Figure 4A), while the Mcm2-7 binding pattern at some origins is unaffected by the temperature shift (e.g. ARS305, Figure 4A). Intriguingly, in those cases where the temperature shift induces a change in Mcm2-7 chromatin association, peak broadening occurs only in one direction, correlating with the orientation of the open reading frames (ORFs) around the origin (Figure 4A), which is consistent with RNAP collisions inducing the observed changes in Mcm2-7 binding around origins after defective transcription termination.
Figure 4. ChIP-seq analysis of ORC and Mcm2-7 in rat1-1 cells.
(A) ORC and Mcm2-7 enrichment around ARS1414, ARS447, and ARS305 at 24°C and 37°C is plotted over chromosomal position. Annotated ORFs on the Watson (yellow) and Crick (blue) strands are indicated. (B) Heat maps of ORC and Mcm2-7 enrichment at 210 origin sites where Mcm2-7 and ORC peaks coincide. Positive signal within 6 kb window is indicated in white. ORC maps are centered on ORC peak positions at 24°C; Mcm2-7 maps are centered on Mcm2-7 peak positions at 24°C. Mcm2-7 peaks obtained at 37°C were ordered into eight clusters (I-VIII). ORC (24°C and 37°C) and Mcm2-7 (24°C) maps were ordered according to Mcm2-7 (37°C). Panel on the right indicates average TDM at respective origin sites (see Figure S6B). (C) TDMs at the 210 ORC/Mcm2-7 peaks plotted as a function of Mcm2-7 peak position at 37°C vs. 24°C. red: Mcm2-7 shift to the right; blue: Mcm2-7 shift to the left; black: Mcm2-7 peak position unchanged. (D) Data from (C) summarized as box and whisker plots; significance of difference between origins at which Mcm2-7 at 37°C is displaced to the right and origins at which Mcm2-7 at 37°C is displaced to the left is indicated.
To examine the ORC and Mcm2-7 binding patterns at permissive and nonpermissive temperatures we aligned the 210 members of the ORC/Mcm2-7 set and plotted the ORC and Mcm2-7 ChIP-seq scores as heat maps centered on the peak positions of ORC and Mcm2-7 at 24°C, respectively. Mcm2-7 distributions at 37°C were ranked according to relative displacement from Mcm2-7 peak centers obtained at 24°C (clusters I – VIII), and the resulting origin order was subsequently imposed on the heat maps for ORC at 24°C and 37°C, and Mcm2-7 at 24°C (Figure 4B). While the ORC peaks at 37°C align with those obtained at 24°C, a large fraction of Mcm2-7 peak positions at 37°C deviates significantly from the Mcm2-7 peak centers obtained at 24°C, shifting either to the left or the right (Figure 4B and Figure S6A). To analyze if the direction of transcription around the respective origins correlates with the direction of the Mcm2-7 peak shift observed at 37°C, we determined the difference in transcript densities on the Watson and Crick strands (Xu et al., 2009) within a 4 kb window around each replication origin. The resulting transcript direction metric (TDM) serves as a measure for transcription direction, with TDM scores > 0 representing prevailing Watson strand (rightward) transcription, and TDM scores < 0 representing prevailing Crick strand (leftward) transcription (Figure S6B). Alignment of the average TDM scores with the ChIP-seq heat maps of the set of 210 ORC/Mcm2-7 peaks indicates that the shift in Mcm2-7 peak position at 37°C generally correlates with the direction of transcription around the origins (Figure 4B). This is confirmed by the strong enrichment of rightward Mcm2-7 peak shifts at origins with positive TDMs, and the converse enrichment of leftward shifted Mcm2-7 positions at origins with negative TDMs (Figure 4C + D). It is worth noting that this trend is maintained across the set of all 375 Mcm2-7 peaks identified here (Figure S6C + D). The observation that ORC positions do not change upon defective termination of RNAP II transcription implies that Mcm2-7 redistribution at origins under these conditions is not a consequence of alternative Mcm2-7 loading. Instead this data is consistent with Mcm2-7 complexes loaded at origins being pushed ahead of elongating RNAP along the chromatin lattice.
Redistribution of initiation sites by RNA polymerase in vivo
To examine origin activity genome wide we sequenced Okazaki fragments (McGuffee et al., 2013; Smith and Whitehouse, 2012) from yeast cells grown in conditions as above. Okazaki fragments were isolated from asynchronous cultures of rat1-1 or control cells after DNA ligase I depletion and 2 hours after growth at either permissive (24°C) or non-permissive (37°C) temperature. Remarkably, simple visual inspection reveals dramatic changes in the distribution of Okazaki fragments in rat1-1 cells at 37°C relative to RAT1 control cells, indicating major changes in both origin efficiency and origin distribution genome wide (Figure 5A and Figure S7A).
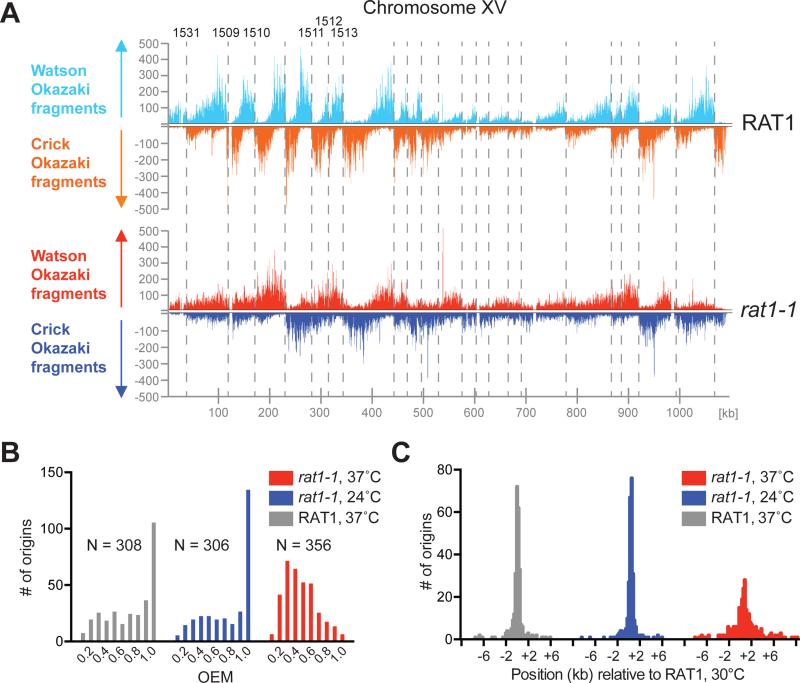
Figure 5. Okazaki fragment distribution in rat1-1 cells.
(A) Okazaki fragments mapping to Watson and Crick strands on S. cerevisiae chromosome XV in RAT1 or rat1-1 cells at 37°C. Select ARS IDs are indicated on top. (B) Distribution of origin efficiencies in rat1-1 and RAT1 cells. N = number of origins (OEM ≥ 0.1). (C) Distribution of origin positions in rat1-1 and RAT1 cells relative to those reported previously for RAT1 cells at 30°C (McGuffee et al., 2013).
Loss of origin efficiency is, for example, illustrated on the left arm of chromosome XV at ARS1531, ARS1509, ARS1510, and ARS1511 (Figure 5A), and more examples can be found across all chromosomes (e.g. Figure S7A). To quantitate these apparent changes we compared origin efficiencies in rat1-1 and control cells using the origin efficiency metric (OEM)(McGuffee et al., 2013). At 24°C origin efficiencies were very similar in rat1-1 and control cells (Figure 5B). In contrast, at 37°C the number of highly efficient (OEM = 1) origins was greatly reduced and the distribution of origin efficiencies was shifted towards lower OEM values in rat1-1 cells, but not in control cells, indicating that loss of Rat1 reduces origin efficiency at many sites genome wide.
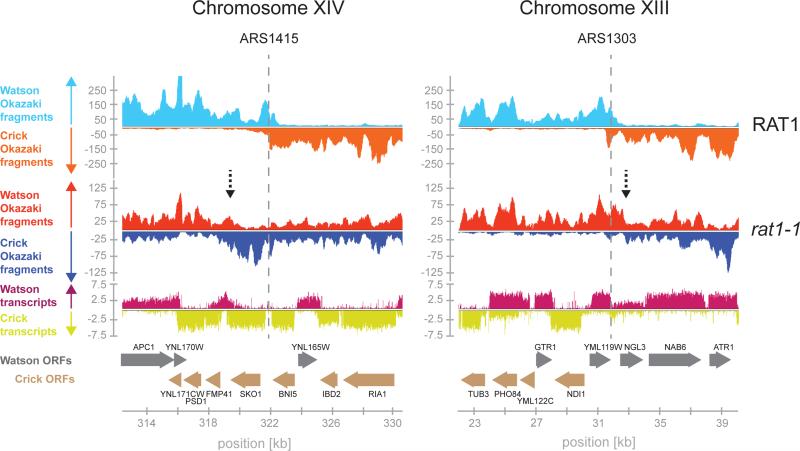
Origin position appears frequently shifted in rat1-1 cells. Genome wide we found that sites of initiation in rat1-1 cells at 24°C or control cells at 37°C were identical to those reported previously (McGuffee et al., 2013), whereas inactivation of Rat1 resulted in a significant positional shift at a large fraction of origin sites (Figure 5C). Initiation site shifts are illustrated at ARS1415 and ARS1303 (Figure 6). Intriguingly, the initiation site shift at these origin sites correlates with both the shift in Mcm2-7 distribution (Figure S7B) and with the prevailing direction of transcription around the respective origin (Figure 6). This led us to examine the correlation between the direction of transcription (TDM) and the direction of the shift in origin localization genome wide. We identified 58 origins associated with regions of prevailing Watson strand transcription (TDM +1 to +2), and 58 origins within regions of prevailing Crick strand transcription (TDM −1 to −2). Strikingly, within these origin clusters a significant fraction of the origins in rat1-1 cells at 37°C, but not in control cells or rat1-1 cells at 24°C, underwent a positional shift that correlates with the direction of transcription and with the degree of transcription bias (Figure S7C). In regions of prevailing Watson strand transcription, the average shift in origin position is ~ 2 kb to the right, whereas origins associated with predominant Crick transcription shift on average by ~0.5 kb to the left (Figure 7A), which is consistent with the extent of Mcm2-7 displacement observed above at 37°C. Moreover, we note that the magnitude of the origin shift is consistent with the extent of RNAP II elongation in rat1-1 cells (Kim et al., 2004). The reduced amplitude in density difference between Watson and Crick strand Okazaki fragments in rat1-1 cells indicates loss of origin efficiency, which may result from loss of Mcm2-7 DH function or dispersive origin redistribution. In addition, loss of origin activity after Mcm2-7 displacement would be expected based on our earlier observation that the initiation competence of Mcm2-7 DHs is limited by the DNA sequence or structure at the Mcm2-7 binding site (Gros et al., 2014). The non-random redistribution of initiation sites in regions of transcription bias, combined with the distinct co-transcriptional relocalization of Mcm2-7, but not ORC, in our ChIP-seq analysis, strongly argues against origin redistribution being a consequence of altered Mcm2-7 loading site selection. We conclude that Mcm2-7 DHs can be relocated by RNAP II in vivo and can remain competent for initiation at distal sites (Figure 7B).
Figure 6. Initiation site shift at ARS1415 and ARS1303 in rat1-1 cells at 37°C.
Okazaki fragments mapping around ARS1415 and ARS1303 in RAT1 (blue / orange) or rat1-1 (red / blue) cells at 37°C. Dashed vertical lines denote positions of ARS1415 and ARS1303, respectively; dashed vertical arrows denote main new initiation sites in rat1-1 cells. Transcript maps (Xu et al., 2009) and positions of annotated open reading frames (ORFs) are indicated below.
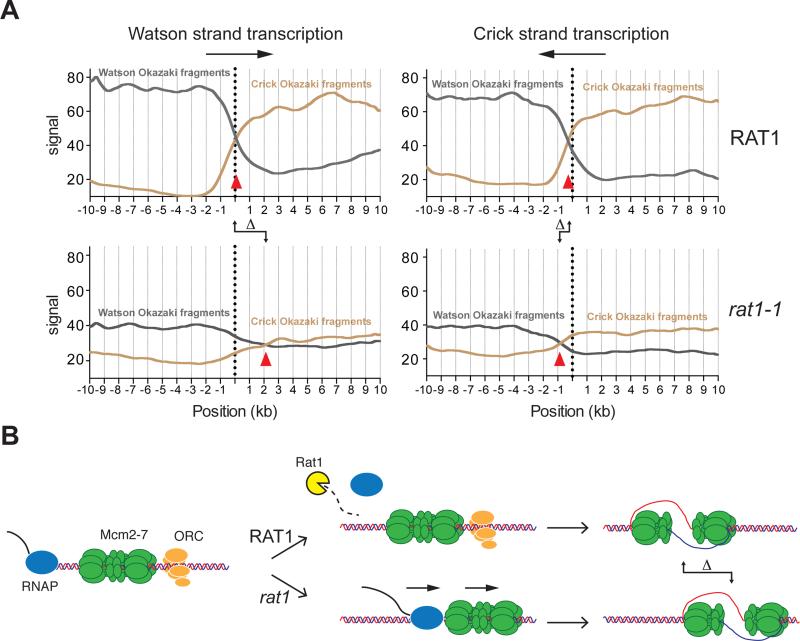
Figure 7. Initiation sites shift in the direction of transcription.
(A) Average Okazaki fragment densities at origins located in regions of prevailing Watson strand transcription (left panels; N = 58), or prevailing Crick strand transcription (right panels; N = 58) in Rat1 (top panels) or rat1-1 (bottom panels) cells at 37°C. Red triangles indicate origin position. Dotted vertical lines indicate previously reported positions for respective origins determined at 30°C (McGuffee et al., 2013). Δ indicates the shift in origin position. (B) Model for origin relocation during defective transcription termination in rat1-1 cells.
Discussion
Our observation that Mcm2-7 DHs support DNA replication initiation after displacement from the initial loading site suggests a general two-step mechanism for eukaryotic origin specification: Selection of the site of Mcm2-7 loading by the pre-RC in the first step, and refinement of the initiation site by the positioning of the Mcm2-7 DH in the second step. While the first step specifies origin sites globally and is mediated through the targeting of ORC to chromosomal sites, the second step affects origin position locally and helps sustain the replication competency of chromatin by promoting origin flexibility.
DNA and RNA polymerases share the same DNA template in vivo. Consequently, mechanisms that help avoid or resolve collisions between replication forks and transcription complexes are important for the maintenance of genome integrity (Aguilera and Garcia-Muse, 2013; Bermejo et al., 2012). However, the two-step mechanism of DNA replication initiation in eukaryotes suggests that transcription-replication conflicts can arise even before replication forks are established. Consistent with this notion, here we identify relocalization of Mcm2-7 DHs by RNA polymerase as a consequence of transcription-replication conflict.
It was previously shown that transcription through an origin can disrupt pre-RCs and inhibit origin function (Looke et al., 2010; Mori and Shirahige, 2007; Snyder et al., 1988; Tanaka et al., 1994). However, although Mcm2-7 complexes are lost from the original loading site after transcription through the origin, we find here that they can be functionally redistributed to downstream sites by elongating RNA polymerase. Plasmid loss over multiple generations in the aforementioned studies may thus in part be a consequence of interference of constitutive transcription across the replication origin with the establishment of pre-RCs at this site. Using a reconstituted in vitro DNA replication system we have found previously that the ability of Mcm2-7 DHs to be activated is partially limited by the DNA sequence or structure at the Mcm2-7 DH binding site (Gros et al., 2014). Similarly, here we observe at individual loci that Mcm2-7 redistribution in rat1-1 mutant cells is not always associated with a concomitant shift in the site of DNA replication initiation, but with a loss of replication initiation activity, thus contributing to the global loss in origin efficiency in these cells. Enhanced origin flexibility due to Mcm2-7 DH mobility may, therefore, be advantageous to promote efficient S phase progression under normal growth conditions when levels of transcriptional interference are low.
The incompatibility of transcription and replication origin function explains the intergenic localization of most budding yeast origins. Nonetheless, pervasive transcription (Jensen et al., 2013), heterogenous mRNA transcription (Pelechano et al., 2013), and imprecise termination of RNAP II elongation (Mischo and Proudfoot, 2013) may cause collisions between RNA polymerase and pre-RCs even in intergenic regions. The stochastic nature of these events may normally preclude the detection of their effect on origin activity by population-based origin mapping techniques.
Our data demonstrate that regulated transcription termination is essential to maintain the normal replication program in budding yeast. This highlights the importance of transcription termination not only to prevent interference with transcription from downstream promoters, but also to prevent interference with replication initiation from downstream replication origins. Active genes confine origin sites to non-transcribed intergenic regions, resulting in origins being frequently localized downstream of active genes also in higher eukaryotes (Hyrien, 2015). We, therefore, expect that ordered transcription termination is equally important to maintain origin activity in all eukaryotes. Moreover, Mcm2-7 DH mobility allows plasticity in initiation sites and may thus contribute to the establishment of the dispersive initiation zones found in higher eukaryotes (Mesner and Hamlin, 2005; Saha et al., 2004).
Mcm2-7 DHs are topologically bound around DNA by a specialized loading machinery. Mcm2-7, therefore, cannot spontaneously rebind DNA in the event of accidental displacement from the DNA template in the absence of the loading factors. While inactivation of ORC or Cdc6 induces rapid loss of Mcm2-7 complexes from chromatin during α-factor arrest in G1 phase and after cell cycle exit in G0 (Chen et al., 2007; Cocker et al., 1996; Diffley et al., 1994; Semple et al., 2006), cell cycle-dependent inhibition of pre-RC assembly in S phase does not induce loss of late and dormant origins, even after prolonged S phase arrest following checkpoint activation (Blow et al., 2011; Zegerman and Diffley, 2009). The mechanism of Mcm2-7 unloading in G1 and G0 phases is unknown, but we suggest that Mcm2-7 DH mobility contributes to the maintenance of origin function in S phase. It is noteworthy that cohesin complexes can be similarly pushed along the chromatin lattice by the transcription machinery (Lengronne et al., 2004), suggesting that the ability to slide is a general strategy to maintain topologically bound protein complexes around DNA when re-loading is not allowed. How nucleosomes may be bypassed or displaced during Mcm2-7 DH relocation through chromatin will be of interest for future studies.
Mechanisms that target ORC to specific sites on the chromosome have significantly diverged in evolution and are subject to modulation throughout development. While direct binding of ORC to the ARS consensus sequence (ACS) contributes to selection of the Mcm2-7 loading site in S. cerevisiae, ORC can be actively recruited to potential origin sites via alternative mechanisms in other organisms (Hyrien, 2015). The ability of Mcm2-7 DHs to support replication initiation from a variety of DNA sequence contexts may thus facilitate the evolution of mechanisms that modulate origin site selection during development.
The example of T7 RNAP demonstrates that specific protein-protein contacts are not required for functional displacement of Mcm2-7 DHs. This flexibility may promote the ability of Mcm2-7 DHs to maintain replication competence upon interference from diverse forms of genome traffic. In line with this notion we find that the double-stranded DNA translocase, FtsK, is similarly capable of relocating Mcm2-7 DHs along the DNA. It is worth noting that FtsK is capable of translocating along DNA at greater than 20 kb per second, demonstrating the remarkable robustness of the Mcm2-7 DH sliding mechanism (Lee et al., 2014).
Experimental procedures
DNA templates and proteins
DNAs and proteins are described in the supplementary methods.
Isolation of pre-RCs by gel-filtration
Pre-RC assembly was carried out in 30 μL reactions containing 45 mM Hepes-KOH pH 7.6 / 0.12 M KOAc / 0.02 % NP-40 / 5 mM Mg(OAc.)2 / 5 % glycerol / 5 mM ATP / 1 mM DTT / 0.5 pmol plasmid DNA / 2.5pmol ORC / 2.5 pmol Cdc6 / 3 pmol Cdt1•MCM2-7. Reactions were incubated for 30 minutes at 30°C. Subsequently, the reaction was fractionated by gravity flow on a ~2 mL Sephacryl S-1000 SF resin in buffer K (45 mM Hepes-KOH pH 7.6 / 0.1 M KOAc / 0.05 % NP-40 / 5 mM Mg(OAc.)2 / 10% glycerol) / 1 mM ATP / 2 mM β-mercaptoethanol. Fractions (~26 μL) were collected manually.
Isolation of Mcm2-7 DHs after transcription by T7 RNAP
Following pre-RC assembly, the reaction was supplemented with 5 pmol of a 296 bp ARS1-containing DNA fragment and incubated for a further 10 minutes at 30°C. Transcription by T7 RNAP was initiated by supplementing the reaction with 500 μM of each ATP / UTP / GTP / CTP, 10 mM DTT, 0.5 pmol of T7 RNA Polymerase (NEB) and incubation at 20°C for 30 minutes. The reaction was fractionated on a 2 mL S1000 column as above.
Co-immunoprecipitation of Mcm2-7 with DNA Fragments
70% of the Mcm2-7 DH / DNA peak fraction from an S1000 fractionation was digested with 5-10 U of restriction enzymes for 45 minutes at 30°C. Immunoprecipitation was carried out by incubation of 225 μg of Dynabeads Protein G (Life Technologies) coupled to M2 anti-FLAG antibody (Sigma) (0.5 μg antibody / 30 μg Dynabeads) for 1h at 30°C with agitation. Immunoprecipitates were washed with buffer K / 2 mM β-Mercaptoethanol and DNA eluted from the beads with 1.6U of Proteinase K (NEB) followed by phenol extraction.
In vitro replication using isolated pre-RCs
Approximately 15% of a pre-RC peak fraction was digested as described and simultaneously phosphorylated by addition of 0.75 pmol of purified DDK (Gros et al., 2014). 20μL of S phase extract prepared from YDR95 (Supplementary Table 1), supplemented with 5mM ATP / 0.1 mM each (dATP / dTTP/ dGTP) / 0.2 mM each (CTP / GTP/ UTP) / 5 μCi [α-32P]-dCTP (3000Ci / mmol) / 40 mM creatine phosphate / 0.01 mg / mL creatine kinase, was added to the reaction and incubation continued for 40 min at 30°C (40μL final reaction volume). The reaction was stopped by incubation with 15 mM EDTA / 0.2% SDS / 1.6U of Proteinase K at 37°C for 30 minutes. DNA was isolated by phenol extraction and ethanol precipitation, and analyzed by native agarose gel electrophoresis. Dried gels were analyzed by phosphor-imaging.
Chromatin immuno-precipitation and ChIP-seq data analysis
Cells (50 ml) were grown at 24°C to OD600 = 1, then continued to grow for two additional hours at either 24°C or 37°C. Cells were crosslinked with 1% formaldehyde, lysed, and sonicated to obtain a DNA shear size of 100-800 bp. Immunoprecipitation was carried out with polyclonal antibodies against ORC or Mcm2-7 (gift of Steve Bell, MIT). Fragments were sequenced on an Illumina platform and reads were mapped using Bowtie and Bedtools. MACS (Zhang et al., 2008) was used to identify ORC and MCM peak binding sites. No shifting model was built and a shift size twice the fragment length was utilized to call peaks. The midpoints of the Mcm2-7 peaks at 24°C, with a corresponding ORC peak (n=210), were used as reference to scan the Illumina reads within a 3 kb window upstream and downstream of every Mcm2-7 binding site. The resulting matrix was clustered using the Cluster 3.0 software and visualized using Java TreeView.
Purification of Okazaki Fragments
YJG15 (rat1-1) or YIW347 (wt) cells were grown at permissive temperature (24°C) to a density of 2.5 × 106 / ml and Cdc9 inactivation was induced by addition of doxycycline (75μg / mL final) and incubation at either permissive (24°C) or restrictive (37°C) temperature for two hours. Okazaki fragments were purified essentially as described (Smith and Whitehouse, 2012). Mapping data were confirmed with material isolated from two to three independent biological replicates.
Supplementary Material
Acknowledgements
We thank Ed Eng (NYSBC) for assistance with EM analysis, Scott Keeney (MSKCC) and Ken Marians (MSKCC) for critical reading of the manuscript, Steve Bell (MIT) for antibodies. This work was funded by National Institute of General Medical Sciences grants R01GM102253 (I.W.) and R01GM107239 (D.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.G. and G.L. performed and interpreted biochemical experiments; C.K. performed and analyzed ChIP-seq experiments. J.G., C.K., and T.Y., generated Okazaki fragment libraries. I.W. analyzed the Okazaki fragment sequencing data and edited the manuscript. D.R. conceived the project and wrote the paper.
Accession Numbers
Okazaki fragment and ChIP sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE69065.
References
- Aguilera A, Garcia-Muse T. Causes of genome instability. Annual review of genetics. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Molecular cell. 2012;45:710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Blitzblau HG, Chan CS, Hochwagen A, Bell SP. Separation of DNA replication from the assembly of break-competent meiotic chromosomes. PLoS genetics. 2012;8:e1002643. doi: 10.1371/journal.pgen.1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends in biochemical sciences. 2011;36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harbor symposia on quantitative biology. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Bogenschutz NL, Rodriguez J, Tsukiyama T. Initiation of DNA replication from non-canonical sites on an origin-depleted chromosome. PloS one. 2014;9:e114545. doi: 10.1371/journal.pone.0114545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes & development. 2007;21:2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Costa A, Hood IV, Berger JM. Mechanisms for initiating cellular DNA replication. Annual review of biochemistry. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershowitz A, Snyder M, Sbia M, Skurnick JH, Ong LY, Newlon CS. Linear derivatives of Saccharomyces cerevisiae chromosome III can be maintained in the absence of autonomously replicating sequence elements. Molecular and cellular biology. 2007;27:4652–4663. doi: 10.1128/MCB.01246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Donato JJ, Chung SC, Tye BK. Genome-wide hierarchy of replication origin usage in Saccharomyces cerevisiae. PLoS genetics. 2006;2:e141. doi: 10.1371/journal.pgen.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes & development. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes & development. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. In search of the holy replicator. Nature reviews. Molecular cell biology. 2004;5:848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Devbhandari S, Remus D. Origin plasticity during budding yeast DNA replication in vitro. The EMBO journal. 2014;33:621–636. doi: 10.1002/embj.201387278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;21:761–771. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. The Journal of cell biology. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. Peaks cloaked in the mist: The landscape of mammalian replication origins. The Journal of cell biology. 2015;208:147–160. doi: 10.1083/jcb.201407004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- Jacob FB, S., Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harbor symposia on quantitative biology. 1964;28:329–348. [Google Scholar]
- Jensen TH, Jacquier A, Libri D. Dealing with pervasive transcription. Molecular cell. 2013;52:473–484. doi: 10.1016/j.molcel.2013.10.032. [DOI] [PubMed] [Google Scholar]
- Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes & development. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kipling D, Kearsey SE. Reversion of autonomously replicating sequence mutations in Saccharomyces cerevisiae: creation of a eucaryotic replication origin within procaryotic vector DNA. Molecular and cellular biology. 1990;10:265–272. doi: 10.1128/mcb.10.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Calos MP. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Molecular and cellular biology. 1991;11:1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Finkelstein IJ, Arciszewska LK, Sherratt DJ, Greene EC. Single-molecule imaging of FtsK translocation reveals mechanistic features of protein-protein collisions on DNA. Molecular cell. 2014;54:832–843. doi: 10.1016/j.molcel.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looke M, Reimand J, Sedman T, Sedman J, Jarvinen L, Varv S, Peil K, Kristjuhan K, Vilo J, Kristjuhan A. Relicensing of transcriptionally inactivated replication origins in budding yeast. The Journal of biological chemistry. 2010;285:40004–40011. doi: 10.1074/jbc.M110.148924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes & development. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Bell SP. A genomic view of eukaryotic DNA replication. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- McGuffee SR, Smith DJ, Whitehouse I. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Molecular cell. 2013;50:123–135. doi: 10.1016/j.molcel.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Hamlin JL. Specific signals at the 3′ end of the DHFR gene define one boundary of the downstream origin of replication. Genes & development. 2005;19:1053–1066. doi: 10.1101/gad.1307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Proudfoot NJ. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochimica et biophysica acta. 2013;1829:174–185. doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Shirahige K. Perturbation of the activity of replication origin by meiosis-specific transcription. The Journal of biological chemistry. 2007;282:4447–4452. doi: 10.1074/jbc.M609671200. [DOI] [PubMed] [Google Scholar]
- Nieduszynski CA, Blow JJ, Donaldson AD. The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription. Nucleic acids research. 2005;33:2410–2420. doi: 10.1093/nar/gki539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- On KF, Beuron F, Frith D, Snijders AP, Morris EP, Diffley JF. Prereplicative complexes assembled in vitro support origin-dependent and independent DNA replication. The EMBO journal. 2014;33:605–620. doi: 10.1002/embj.201387369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harbor perspectives in biology. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. Journal of cell science. 1999;112(Pt 12):2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Shan Y, Mesner LD, Hamlin JL. The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes & development. 2004;18:397–410. doi: 10.1101/gad.1171404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Sawado T, Yamaguchi M, Shinomiya T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Molecular and cellular biology. 1999;19:547–555. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple JW, Da-Silva LF, Jervis EJ, Ah-Kee J, Al-Attar H, Kummer L, Heikkila JJ, Pasero P, Duncker BP. An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes. The EMBO journal. 2006;25:5150–5158. doi: 10.1038/sj.emboj.7601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Sapolsky RJ, Davis RW. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Molecular and cellular biology. 1988;8:2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb DT, Struhl K, Davis RW. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Sun J, Fernandez-Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, Speck C, Li H. Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function. Genes & development. 2014;28:2291–2303. doi: 10.1101/gad.242313.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes & development. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Halter D, Livingstone-Zatchej M, Reszel B, Thoma F. Transcription through the yeast origin of replication ARS1 ends at the ABFI binding site and affects extrachromosomal maintenance of minichromosomes. Nucleic acids research. 1994;22:3904–3910. doi: 10.1093/nar/22.19.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. Molecular biology: termination by torpedo. Nature. 2004;432:456–457. doi: 10.1038/432456a. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Walter JC. Prereplication-complex formation: a molecular double take? Nature structural & molecular biology. 2014;21:20–25. doi: 10.1038/nsmb.2738. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. DNA replication as a target of the DNA damage checkpoint. DNA repair. 2009;8:1077–1088. doi: 10.1016/j.dnarep.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS). Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.