Abstract
IHC4 and PAM50 assays have been shown to provide additional prognostic information for patients with early breast cancer. We evaluated whether incorporating TP53 mutation analysis can further enhance their prognostic accuracy. We examined TP53 mutation and the IHC4 score in tumors of 605 patients diagnosed with stage I–III breast cancer at National Taiwan University Hospital (the NTUH cohort). We obtained information regarding TP53 mutation and PAM50 subtypes in 699 tumors from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort. We found that TP53 mutation was significantly associated with high-risk IHC4 group and with luminal B, HER2-enriched, and basal-like subtypes. Despite the strong associations, TP53 mutation independently predicted shorter relapse-free survival (hazard ratio [HR] = 1.63, P = 0.007) in the NTUH cohort and shorter breast cancer-specific survival (HR = 2.35, P = <0.001) in the METABRIC cohort. TP53 mutational analysis added significant prognostic information in addition to the IHC4 score (∆ LR-χ2 = 8.61, P = 0.002) in the NTUH cohort and the PAM50 subtypes (∆ LR-χ2 = 18.9, P = <0.001) in the METABRIC cohort. We conclude that incorporating TP53 mutation analysis can enhance the prognostic accuracy of the IHC4 and PAM50 assays.
Breast cancer is one of the leading malignancies in women, with an increasing incidence over the past 2 decades. With the advent of screening, a large number of patients are being diagnosed at an early stage and have a favorable prognosis. However, these patients have a certain recurrence rate, depending on the clinicopathological features. Adjuvant chemotherapy can reduce the recurrence risk, but it has moderate adverse effects. The established clinicopathological features are insufficient to guide patients for adjuvant chemotherapy, and therefore substantial over- or under-treatment can occur.
Several multi-gene tests, such as MammaPrint1,2, Oncotype DX3,4, and PAM505,6, have been shown to provide additional prognostic information over classical clinicopathological factors. To improve the applicability, Cuzick et al. constructed the IHC4 score by combining 4 widely examined immunohistochemical markers, namely, estrogen receptor (ER), progesterone receptor (PgR), human epidermal growth factor receptor 2 (HER2; including fluorescent in situ hybridization in the IHC 2+ group), and Ki-677. Previous studies have shown that the IHC4 score provides similar prognostic information compared with the Oncotype DX recurrence score (RS)7 and PAM50 risk of recurrence score (ROR)8, and one study suggested that IHC4 has the potential to be the most cost-effective prognosis tool9.
Spearman correlations of risk groups defined by the RS, ROR, or IHC4 score are only modest to moderate (RS and IHC4 scores: r = 0.72; ROR and IHC4 scores: r = 0.48; RS and ROS scores: r = 0.39)7,8. Although a considerable difference exists among these 3 assays, combinations of 2 assays out of the 3 assays did not significantly improve the prognostic value7,8. A similar prognostic power, modest-to-moderate concordance, and the absence of a significant additive effect of these assays suggest the need for incorporating other factors to improve the prognostic accuracy.
The PIK3CA and TP53 somatic mutations are the 2 most frequently mutated genes in breast cancer, and their frequencies are much higher than other somatic mutations10,11. In contrast to the conflicting findings about prognostic value of PIK3CA mutation12,13,14,15, TP53 mutation has been consistently shown to predict poor outcomes in 2 meta-analyses (hazard ratio [HR] = 2.0 and 2.27)16,17. Mutant P53 in breast cancer may act at various cancer stages, such as early tumorigenesis, tumor growth, and metastasis. TP53 mutation in breast cancer is associated with high-grade tumor behavior, and is molecularly distinct from wild type tumor18,19. MammaPrint, Oncotype DX, PAM50, and IHC4 mainly focus on measuring tumor proliferation on the basis of protein or mRNA expressions. The broad-spectrum effects of TP53 mutation may be more likely to show additive effect on these assays. We determined whether incorporating TP53 mutation can enhance the prognostic accuracy of the IHC4 and PAM50 assays.
Methods
Patients and sample collection
We evaluated the prognostic effect of TP53 mutation in one retrospective cohort by using IHC4 scores and in one public data set by using PAM50 scores. The retrospective cohort included 659 patients with stage I–III breast cancer diagnosed at National Taiwan University Hospital (NTUH) from January 1997 to December 2005. Among them, 605 patients with adequate tumor DNAs for the TP53 mutation analysis were enrolled in this study. This study was approved by the Ethics Committee of NTUH (201112010RIC). The informed consent was obtained from all subjects, and the methods used in this study were carried out in accordance with the approved guidelines. In this cohort, the clinicopathological data were extracted from medical charts. The relapse-free survival (RFS) data used in this study were current as of December 31, 2011. The RFS was defined as the duration from diagnosis to the confirmation of disease recurrence, including local, regional, and distant recurrences.
The Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) dataset comprised 1992 patients with breast cancer from the United Kingdom and Canada, and data regarding clinicopathological features and PAM50 classification were publicly available for all patients20. From this dataset, we analyzed 699 patients with stage I–III breast cancer and publicly available TP53 mutation status (METABRIC cohort).
Classification of patients into risk groups according to the IHC4 scores in the NTUH cohort
In the NTUH cohort, tumors were stained for ER, PgR, and HER2 by using IHC as previously described21. The ER and PgR statuses were determined using the Ventana Benchmark system (Ventana Medical Systems Inc., Tucson, AZ, USA) and prediluted antibodies (anti-ER clone 6F11 and anti-PgR clone 16). ER and PgR were scored as percentage of tumor cells positively staining nuclei, and tumors with ≥10% positively stained cells were considered positive. The HER2 status was determined according to the American Society of Clinical Oncology/College of American Pathologists updated guideline22. Briefly, scores of 0 and 1+ by IHC were considered negative, and 3 + was considered positive. Cases with a score of 2+ were tested for gene amplification by dual probe fluorescence in situ hybridization. HER2/CEP17 ratio ≥2.0 and/ or an average HER2 copy number ≥6.0 signals/cell were considered positive. The primary antibody for staining Ki67 was anti-Ki67 (1:200 dilution, clone MIB-1, DakoCytomation, Denmark)23,24, and tumors with ≥13.25% positively stained nuclei were considered as highly expressed25.
According to the study by Cuzick et al.7, the IHC4 score of each tumor was computed as IHC4 = 94.7 × (− 0.100 ∙ ER10 − 0.079 ∙ PgR10 + 0.586 ∙ HER2 + 0.240 ln [1 + 10 ∙ Ki67]). To avoid the bias caused by the differences in methodology and the antibodies between the present study and the study by Cuzick et al.7, we categorized our study participants into low, intermediate, and high risk groups according to the IHC4 scores of <25th, 25th–75th, and >75th percentiles, respectively.
TP53 mutational analysis in the NTUH cohort
In the NTUH cohort, TP53 exons 4–9 were sequenced for each tumor, as previously described26. The hematoxylin and eosin stained slides of the tumors were examined, and the tumor areas were marked for macrodissection to enrich tumor DNAs. The genomic DNA of the macrodissected tumor specimens was isolated using the QIAamp DNA Mini Kit (Qiagen Inc., Valencia, CA, USA) and amplified using PCR. Forward and reverse sequencing of the amplified DNA was performed for the TP53 exons 4–9 in an autosequencer (Applied Biosystems, USA) using sequencing or corresponding PCR primers.
Clinicopathological data, PAM50 classification, and TP53 mutational status in the METABRIC cohort
We extracted the data on demographics, survival, PAM50 classification, and the TP53 mutation status from the METABRIC cohort. These data are listed in the supplementary Table 2 in the study by Curtis et al.20. Among the 1992 tumors, 820 tumors had TP53 mutation. Because the METABRIC cohort did not have information on cancer relapse, we used breast cancer-specific survival (BCSS) as the endpoint. After excluding 9 patients with stage IV breast cancer, 17 patients without survival information, and 95 patients who died of conditions other than breast cancer, 699 patients were finally included in the analysis. In the METABRIC cohort, TP53 mutations in all of the 11 exons were sequenced in forward and reverse directions as previously described27 (with the exception that exon 7 was sequenced in only one direction). According to the study by Parker et al., the risk of relapse (ROR) score of each tumor was computed as per the following formula: 0.05 ∙ basal + 0.12 ∙ HER2 enriched − 0.34 ∙ luminal A + 0.23 ∙ luminal B 5.
Statistical analysis
The distributional properties of categorical variables were presented as the frequency and percentage. The differences in the distributions of categorical variables between the TP53 wild and mutant tumors of patients with breast cancer were examined using the chi-square test. The survival outcomes were estimated using the Kaplan–Meier method. In the univariate analysis, the effects of each potential predictive factor for the RFS outcome in the NTUH cohort and BCSS in the METABRIC cohort were examined using the log-rank test. Next, multivariate analysis was conducted by fitting Cox proportional hazards models to estimate the adjusted effects of predictors on the RFS and BCSS outcomes. Specifically, the stepwise variable selection procedure (with iterations between the forward and backward steps) was applied to obtain the most appropriate candidate for the final Cox proportional hazards model. To ensure quality, basic model-fitting techniques for (1) variable selection, (2) goodness-of-fit (GOF) assessment, and (3) regression diagnostics and remedies were performed in our regression analyses. To assess the prognostic effect of adding TP53 mutation as a variable, we used changes in the likelihood ratio (LR) values (∆ LR-χ2) to quantitatively measure the relative score with TP53 mutation information compared with that without TP53 mutation information in the Cox model. A P value ≤ .05 was used to indicate statistical significance, and all tests were 2-tailed. Complete details of the statistical analysis are provided in the supplementary materials.
Results
Clinical and pathological characteristics of patients
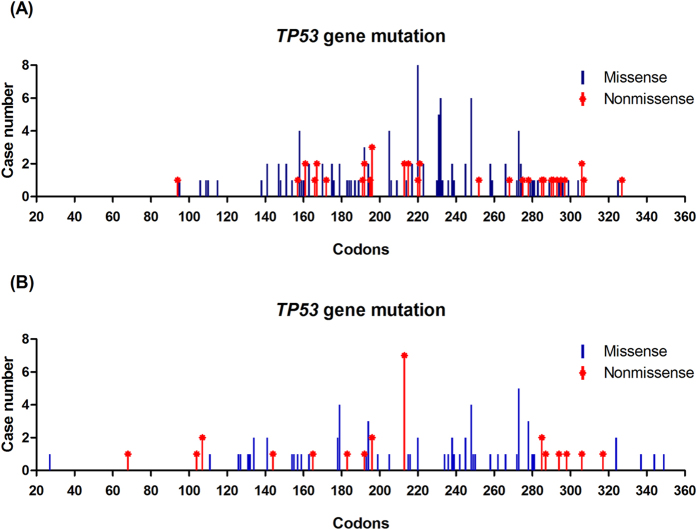
The mutation distribution in the TP53 coding regions in both cohorts are presented in Fig. 1. In the NTUH cohort, 153 mutations including 116 missense and 37 nonmisssense mutations were identified in 132 tumors. Some recurrently mutated codons, such as codons 220 (5.3%), 232 (3.9%), 248 (3.9%), and 231 (3.3%) were observed. In the METABRIC cohort, 90 mutations including 64 missense and 26 nonmisssense mutations, were identified in 90 tumors. Some recurrently mutated codons, such as codons 213 (7.6%), 273 (5.4%), 248 (4.3%), and 179 (4.3%) were observed. The clinical and pathological data of patients based on the TP53 mutation status in the NUTH and the METABRIC cohorts are listed in Table 1. TP53 mutation was associated with a high histological grade, ER negativity, and HER2 overexpression in both cohorts. In addition, TP53 mutation was associated with PR negativity and a high Ki67 expression in the NUTH cohort and a high frequency of chemotherapy use in the METABRIC cohort. TP53 mutation was not significantly associated with age, tumor size, or the axillary lymph node status of patients.
Figure 1.
Distribution of missense and nonmissense mutations along the coding region of TP53 in the NTUH cohort (A) and METABRIC (B) cohort.
Table 1. Correlations of TP53 mutation status with clinicopathological characteristics (A) and IHC4 risk group and PAM50 subtypes (B).
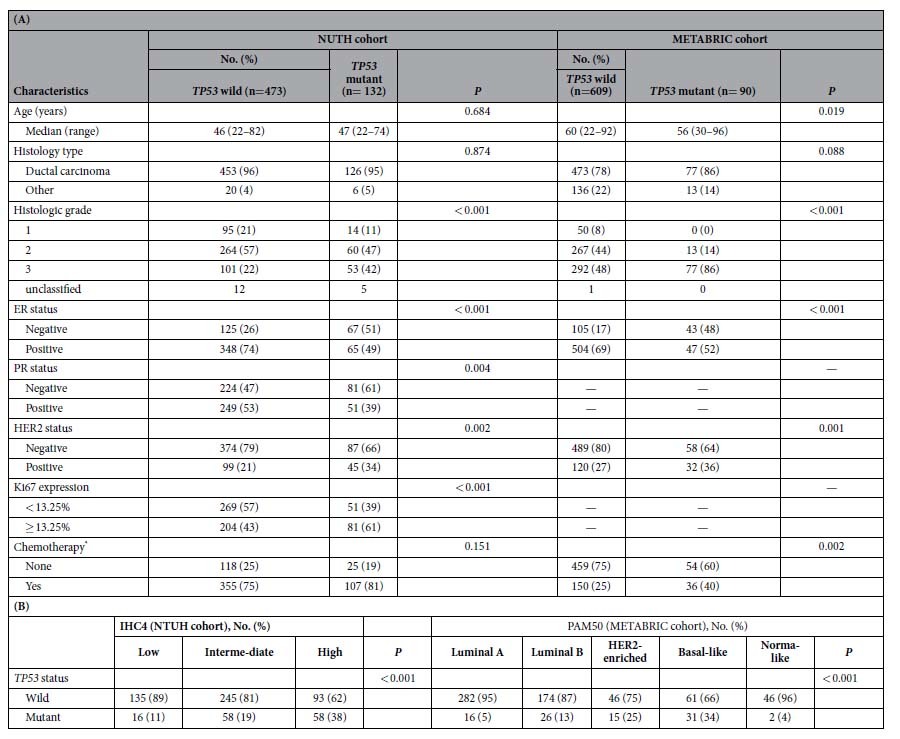
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2. *Neoadjuvant and/or adjuvant therapy.
In the NTUH cohort, low, intermediate, and high risk groups were defined according to IHC4 scores of <−16.0, −16.0 to 119.7, and >119.7, respectively (Fig. S1). TP53 mutation was significantly associated with higher risk groups on the basis of the IHC4 score (high vs. intermediate vs. low risk, 38% vs. 19% vs. 11%, P < 0.001). In the METABRIC cohort, the TP53 mutation was significantly more common in HER2-enriched (25%) and basal-like (34%) subtypes than in luminal A (5%) and luminal B (13%) subtypes. Compared with luminal A subtype, luminal B subtype had significantly higher TP53 mutation frequency. TP53 mutation in the normal breast subtype was low (4%), but this mutation was observed in a relatively limited number of cases (Table IB). Furthermore, TP53 mutation was significantly associated with a high ROR score (mean rank, TP53 mutant vs. wild, 383.2 vs. 317.1, P = 0.001, according to a Mann–Whitney U test).
Univariate survival analyses of prognostic factors
In the NTUH cohort, the median follow-up duration was 77.4 months (95% confidence interval [CI], 75.1–79.7 mo), and breast cancer relapse was observed in 144 patients. In the METABRIC cohort, the median follow-up duration was 120.5 months (95% CI, 111.9–129.1 mo) and 196 patients died of breast cancer. In the univariate analysis, the conventional clinicopathological factors including higher histological grade, higher T stage, higher N stage, and ER negativity were significantly associated with poor outcomes in the NTUH and the METABRIC cohorts. HER2 overexpression was marginally significantly and significantly associated with poor outcomes in the NTUH and the METABRIC cohorts, respectively. PR negativity and high Ki67 staining, which were unavailable in the METABRIC cohort, were significantly associated with shorter RFS in the NTUH cohort (Table S1).
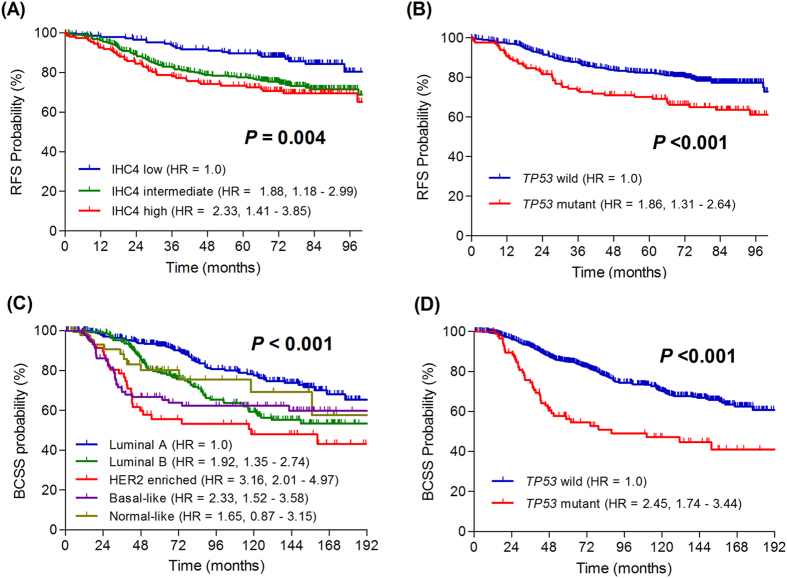
In the NTUH cohort, IHC4 intermediate (HR = 1.88, 95% CI = 1.18–2.99) and high risk (HR = 2.33, 95% CI = 1.41–3.85) groups were significantly associated with shorter RFS compared with the low risk group. In the METABRIC cohort, luminal B (HR = 1.92, 95% CI = 1.35–2.74), HER2-enriched (HR = 3.16, 95% CI = 2.01–4.97), and basal-like (HR = 2.33, 95% CI = 1.52–3.58) subtypes were associated with shorter BCSS compared with the luminal A subtype. The survival of normal breast subtype was not significantly different from that of the luminal A subtype. TP53 mutation was significantly associated with shorter RFS (HR = 1.86, 95% CI = 1.31–2.64) in the NTUH cohort and with shorter BCSS (HR = 2.45, 95% CI = 1.74–3.44) in the METABRIC cohort (Fig. 2 and Table S1).
Figure 2.
Kaplan-Meier plots of relapse-free survival by IHC4 risk classification (A) and TP53 mutational status (B) in NTUH cohort, and breast cancer-specific survival by PAM50 classification (C) and TP53 mutational status (D) in METABRIC cohort. (unadjusted analysis).
Multivariate survival analyses of prognostic factors
In the NTUH cohort, we conducted the multivariate analyses separately by including each IHC4 marker or IHC4 risk group as a variable. Higher T and N stages were associated with shorter RFS in both analyses. When the IHC4 risk groups were used as a variable, both of the IHC4 high risk group (HR = 1.90, 95% CI = 1.32–2.73) and TP53 mutation (HR = 1.63, 95% CI = 1.14–2.32) independently predicted shorter RFS (Table 2A). When each IHC4 marker was used as a variable and its cutoff was determined using a stepwise variable selection procedure, PR staining <10% (HR = 1.58, 95% CI = 1.11–2.24), Ki67 staining ≥10% (HR = 1.91, 95% CI = 1.33–2.76), and TP53 mutation (HR = 1.53, 95% CI = 1.07–2.19) independently predicted shorter RFS (Table S2A).
Table 2. Multivariate Cox’s proportional hazards models for relapse-free survival in NTUH cohort (A) and breast cancer-specific mortality in METABRIC cohort (B) and comparison of added prognostic information by TP53 in the two cohorts (C).
| (A) Relapse-free survival | |||
|---|---|---|---|
| Characteristic | HR | 95% CI | P |
| T stage | |||
| T3 v T1 / T2 | 2.65 | 1.78–3.94 | <0.001 |
| N stage | |||
| N0 v N1 / N2 | 0.47 | 0.32–0.70 | <0.001 |
| N3 v N1 / N2 | 1.64 | 1.05–2.57 | 0.030 |
| IHC4 score | |||
| High/intermediate v low | 1.90 | 1.32–2.73 | <0.001 |
| TP53 status | |||
| Mutant v wild | 1.63 | 1.14–2.32 | 0.007 |
| (B) Breast cancer-specific mortality | |||
| Age | |||
| <45/ >65 v 45–65 years | 1.71 | 1.27–2.29 | <0.001 |
| T stage (ordinal) | |||
| Increased one unit (1 v 2 v 3) | 1.45 | 1.11–1.91 | 0.007 |
| N stage (ordinal) | |||
| Increased one unit (0 v 1 v 2. v 3) | 1.58 | 1.33–1.87 | <0.001 |
| HER2 overexpression | |||
| Yes v no | 1.58 | 1.12–2.23 | 0.010 |
| PAM50 | |||
| Luminal B v luminal A | 1.38 | 0.95–1.99 | 0.090 |
| HER2 enriched v luminal A | 1.73 | 1.04–2.87 | 0.036 |
| Basal-like v luminal A | 1.74 | 1.12–2.70 | 0.013 |
| Normal-like v luminal A | 1.90 | 0.99–3.65 | 0.054 |
| TP53 status | |||
| Mutant v wild | 2.35 | 1.64–3.36 | <0.001 |
| (C) | |||
| Cohort | ∆ LR-χ2 | P | |
| NTUH | |||
| T stage + N stage + IHC4 + TP53 v T stage + N stage + IHC4 (df=1) | 1668.3 − 1659.7 = 8.6 | 0.002 | |
| METABRIC | |||
| age + T stage + N stage + HER2 + PAM50 + TP53 v age + T stage + N stage + HER2 + PAM50 (df=1) | 2261.3 − 2242.4 = 18.9 | < 0.001 | |
In the METABRIC cohort, we conducted the multivariate analyses separately by including each PAM50 subtype or a continuous variable of ROR score as a variable. In both analyses, age <45 or >65 years, higher T stage, higher N stage, and HER2 overexpression were associated with shorter BCSS. When PAM50 subtypes were used as a variable, HER2-enriched (HR = 1.73, 95% CI = 1.04–2.87) and basal-like (HR = 1.74, 95% CI = 1.12–2.70) subtypes were significantly associated with shorter BCSS, and luminal B and normal breast subtypes were marginally associated with shorter BCSS compared with the luminal A subtype. TP53 mutation independently predicted shorter BCSS (HR = 2.35, 95% CI = 1.64–3.36) (Table 2B). When ROR score (the continuous variable) was used as a variable, the multivariate analysis showed that an increased ROR score was significantly associated with shorter BCSS (HR = 2.03, 95% CI = 1.09–3.77). TP53 mutation remained an independent predictive factor of shorter BCSS (HR = 2.46, 95% CI = 1.72–3.51) (Table S2B).
TP53 mutational analysis added significant prognostic information in addition to the IHC4 score (∆LR-χ2 = 8.61, P = 0.002) in the NTUH cohort, and PAM50 subtypes (∆ LR-χ2 = 18.9, P ≤ 0.001) in the METABRIC cohort (Table 2C). In view of the importance of lymph node status, we conducted subgroup analyses to assess whether the predicting effects of TP53 mutation were different in lymph node negative and positive subgroups by adding two-way interactions of N stage (N0 vs. N1/ N2/ N3) and TP53 mutation (mutant vs. wild) to the variable lists of the stepwise variable selection procedures in regression analyses in the two cohorts respectively. The interactive effects were not statistically significant in both Cox’s proportional hazards models reported in Tables 2A and 2B. It indicates that the predictive value of TP53 mutation was not affected by lymph node status. To demonstrate the potential clinical utility, we drew covariate-adjusted survival curves that could facilitate fine tuning according to the association between the TP53 mutational status and the IHC4 risk groups (Fig. 3A) or the PAM50 subtypes (Fig. 3B).
Figure 3.
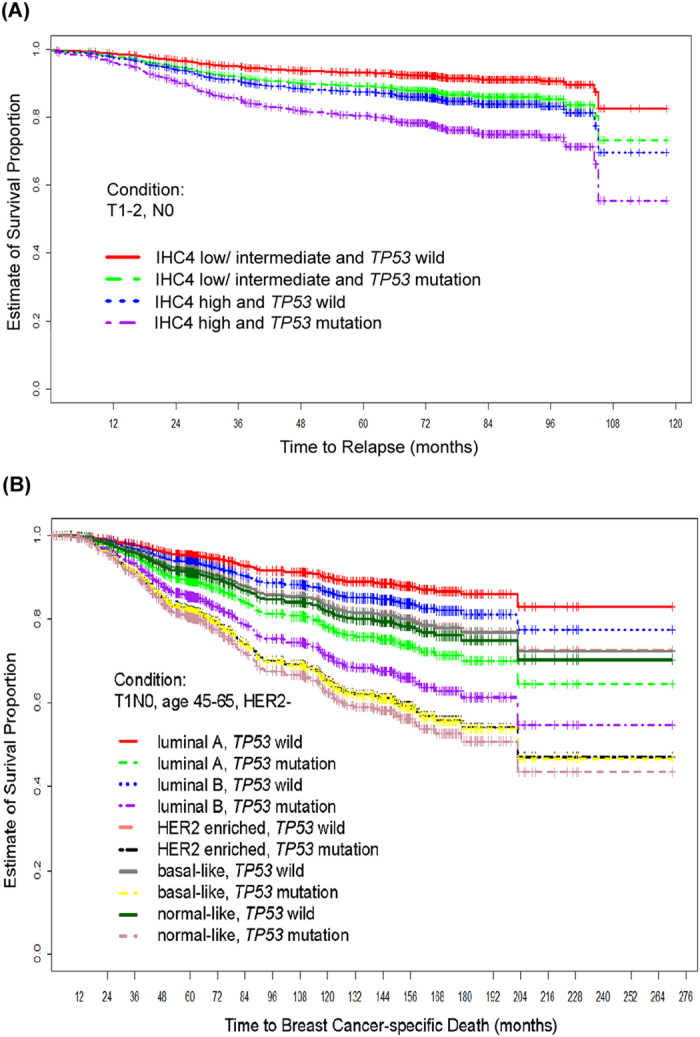
Covariate-adjusted survival curves for time to relapse in the NTUH (A) and METABRIC (B) cohorts.
Discussion
This study shows that TP53 mutation is an independent prognostic factor beyond the IHC4 and PAM50 assays. Incorporating TP53 mutation into these 2 assays can significantly enhance their prognostic accuracy.
In the present study, TP53 mutation was significantly associated with IHC4 risk groups and PAM50 subtypes, but the Spearman correlations between TP53 mutation and IHC4 or PAM50 (TP53 mutation and IHC4: r = 0.24; TP53 mutation and PAM50: r = 0.21) were lower than those between IHC4 and RS, ROR, and IHC4 in previous reports (RS and IHC4: r = 0.72; ROR and IHC4: r = 0.48; RS and ROS: r = 0.39)7,8. The relatively low Spearman correlations of TP53 mutation with IHC4 and PAM50 and the independent prognostic value of TP53 mutation strongly suggest that TP53 mutation has a biology distinct from that of ER, PR, HER2, and markers of cell proliferation or PAM50 subtypes. Several previous studies have shown that in addition to the increase in cancer cell proliferation, mutant P53 can cause other effects, such as genomic instability, inflammation, angiogenesis, epithelial-to-mesenchymal transition, and stemness18,28,29,30,31,32,33.
Although the IHC4 score is widely applicable and its prognostic accuracy is similar to that of RS7 and ROR8 scores, the major drawback of using the IHC4 score is the lack of standardization of the Ki67 assay and its interpretation. To evaluate Ki67 expression, we used the well-known MIB-1 antibody34. A prior study showed a strong correlation between MIB-1 and SP6 antibodies, and no adjustment was required35. However, Cuzick et al. used the SP6 antibody to generate an IHC4 score in an exploratory cohort (ATAC trial) and the MIB-1 antibody in a validation cohort (Nottingham data) and reported that the adjustment should be made using a factor of approximately 2.5 when the MIB-1 antibody is used7. Another limitation of the Ki67 assay is the lack of consistency across laboratories; for example, an international Ki67 reproducibility study reported high intralaboratory reproducibility but only moderate interlaboratory reproducibility36. To reduce these biases, we conducted multivariate analyses separately by including the IHC4 risk group or each IHC4 marker as a variable. Both of the multivariate analyses consistently showed that TP53 mutation was an independent prognostic factor (HR = 1.63 when the IHC4 risk group was considered a variable and HR = 1.53 when each IHC4 marker was considered a variable) (Table 2A and Table S2A).
Among the 5 PAM50 subtypes, the normal-like subtype represented the gene signature close to true “normals”, resulting from reduction mammoplasty or grossly uninvolved tissue5. Because the normal-like subtype was considered a poor quality-control measure, it was excluded from the ROR score. Conservatively, we conducted the multivariate analyses separately by including each subtype or the ROR score as a variable in the METABRIC cohort. Both of the multivariate analyses consistently showed that TP53 mutation was an independent prognostic factor (HR = 2.35 when each subtype was considered a variable and HR = 2.46 when the ROR score was considered a continuous variable, Table 2B and Table S2B). Recently, Silwal-Pandit et al. obtained data on 1460 tumors from a METABRIC cohort and sequenced all of the TP53 exons. Their findings were consistent with those of our study, in which TP53 mutation caused significantly inferior BCSS (HR = 2.03). Conversely, the PAM50 subtype was not included as a variable in the multivariate analysis in their study37.
Different types of TP53 mutations have been reported as having different functional effects and prognostic values38,39. However, reports of these associations are inconsistent17,37,40,41,42,43. In our study, missense and nonmissense mutations were significantly associated with RFS (missense mutation vs. wild; HR = 1.80; nonmissense mutation vs. wild, HR = 2.27) in the NTUH cohort (Fig. S2A) and with BCSS (missense mutation vs. wild, HR = 2.42; nonmissense mutation vs. wild, HR = 2.51) in the METABRIC cohort (Fig. S2B). We examined the prognostic value of recurrently mutated codons 220, 232, 248, and 231 in the NTUH cohort and 213, 273, 248, and 231 in the METABRIC cohort. None of these codons reached the statistical significance to predict patient outcomes when compared with the mutations of a pool of other codons (data not shown). Because the case number was limited by each codon, we could not exclude the possibility of different prognostic effects exerted by specific mutated codons.
In conclusion, the novel finding of the present study is that TP53 mutation has additional predictive value that isn’t captured in proliferation-oriented IHC4 and PAM50 platforms. To confirm the findings of this study is crucial for supporting the routine clinical application of TP53 mutation as outcome predictor for patients with early breast cancer. Furthermore, the findings warrant conducting studies to develop new platforms by incorporating TP53 mutation to IHC4 and PAM50 respectively, and to test the prognostic value of TP53 mutation when other assays, such as Oncotype DX and MammaPrint, are used.
Additional Information
How to cite this article: Lin, C.-H. et al. TP53 Mutational Analysis Enhances the Prognostic Accuracy of IHC4 and PAM50 Assays. Sci. Rep. 5, 17879; doi: 10.1038/srep17879 (2015).
Supplementary Material
Acknowledgments
We thank the members of the Office of Medical Records at the NTUH for their assistance in assessing the clinical data. We acknowledge the statistical assistance provided by the Taiwan Clinical Trial Bioinformatics and Statistical Center, founded by National Research Program for Biopharmaceuticals of the Ministry of Science and Technology, Taiwan (program number: 103-2325-B-002-033). This study used the data generated by the Molecular Taxonomy of Breast Cancer International Consortium, which was supported by Cancer Research UK and the British Columbia Cancer Agency Branch. We thank the members of the consortium for clarifying the definition of breast cancer specific survival. This work was supported by grants from National Taiwan University, Taiwan (grant number NTU-ICRP-103R7557) and National Center of Excellence for Clinical Trial and Research, Taiwan (grant number DOH103-TD-B-001).
Footnotes
Author Contributions C.H.L, I.C.C., Y.S.L. and A.L.C. conceived and designed the experiments. C.H.L., F.C.H., C.H.C. and Y.S.L. analyzed the data. C.H.L., C.S.H., W.H.K., K.T.K., C.C.W., P.F.W., D.Y.C., M.Y.W., W.W.C. and A.L.C. contributed reagents/ materials/ analysis tools. C.H.L., I.C.C., Y.S.L. and A.L.C. wrote the paper.
References
- van de Vijver M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347, 1999–2009 (2002). [DOI] [PubMed] [Google Scholar]
- Glas A. M. et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics 7, 278 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351, 2817–2826 (2004). [DOI] [PubMed] [Google Scholar]
- Dowsett M. et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: a TransATAC study. J Clin Oncol 28, 1829–1834 (2010). [DOI] [PubMed] [Google Scholar]
- Parker J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27, 1160–1167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen T. O. et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res 16, 5222–5232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J. et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol 29, 4273–4278 (2011). [DOI] [PubMed] [Google Scholar]
- Dowsett M. et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol 31, 2783–2790 (2013). [DOI] [PubMed] [Google Scholar]
- Ward S. et al. Gene expression profiling and expanded immunohistochemistry tests to guide the use of adjuvant chemotherapy in breast cancer management: a systematic review and cost-effectiveness analysis. Health technology assessment 17, 1–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes S. A. et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39, D945–950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. Y., Rong M., Grieu F. & Iacopetta B. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat 96, 91–95 (2006). [DOI] [PubMed] [Google Scholar]
- Perez-Tenorio G. et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res 13, 3577–3584 (2007). [DOI] [PubMed] [Google Scholar]
- Maruyama N. et al. Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res 13, 408–414 (2007). [DOI] [PubMed] [Google Scholar]
- Kalinsky K. et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res 15, 5049–5059 (2009). [DOI] [PubMed] [Google Scholar]
- Pharoah P. D., Day N. E. & Caldas C. Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer 80, 1968–1973 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M. et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12, 1157–1167 (2006). [DOI] [PubMed] [Google Scholar]
- Walerych D., Napoli M., Collavin L. & Del Sal G. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 33, 2007–2017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. D. et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA 102, 13550–13555 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H. et al. Molecular subtypes of breast cancer emerging in young women in Taiwan: evidence for more than just westernization as a reason for the disease in Asia. Cancer Epidemiol Biomarkers Prev 18, 1807–1814 (2009). [DOI] [PubMed] [Google Scholar]
- Wolff A. C. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31, 3997–4013 (2013). [DOI] [PubMed] [Google Scholar]
- de Azambuja E. et al. Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br J Cancer 96, 1504–1513 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi R., Woods R., Ravdin P. M., Hayes M. M. & Gelmon K. A. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol 11, 174–183 (2010). [DOI] [PubMed] [Google Scholar]
- Cheang M. C. et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101, 736–750 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G. et al. Loss of heterozygosity and p53 gene mutations in breast cancer. Cancer Res 54, 499–505 (1994). [PubMed] [Google Scholar]
- Zhou W. et al. Full sequencing of TP53 identifies identical mutations within in situ and invasive components in breast cancer suggesting clonal evolution. Molecular oncology 3, 214–219 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Hollstein M. & Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nature cell biology 9, 573–580 (2007). [DOI] [PubMed] [Google Scholar]
- Jong Y. J. et al. Chromosomal comparative genomic hybridization abnormalities in early- and late-onset human breast cancers: correlation with disease progression and TP53 mutations. Cancer genetics and cytogenetics 148, 55–65 (2004). [DOI] [PubMed] [Google Scholar]
- Yeudall W. A. et al. Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis 33, 442–451 (2012). [DOI] [PubMed] [Google Scholar]
- Fontemaggi G. et al. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nature structural & molecular biology 16, 1086–1093 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yan W. & Chen X. Mutant p53 disrupts MCF-10A cell polarity in three-dimensional culture via epithelial-to-mesenchymal transitions. J Biol Chem 286, 16218–16228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicalese A. et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138, 1083–1095 (2009). [DOI] [PubMed] [Google Scholar]
- Stuart-Harris R., Caldas C., Pinder S. E. & Pharoah P. Proliferation markers and survival in early breast cancer: a systematic review and meta-analysis of 85 studies in 32,825 patients. Breast 17, 323–334 (2008). [DOI] [PubMed] [Google Scholar]
- Zabaglo L. et al. Comparative validation of the SP6 antibody to Ki67 in breast cancer. J Clin Pathol 63, 800–804 (2010). [DOI] [PubMed] [Google Scholar]
- Polley M. Y. et al. An international Ki67 reproducibility study. J Natl Cancer Inst 105, 1897–1906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silwal-Pandit L. et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res 20, 3569–3580 (2014). [DOI] [PubMed] [Google Scholar]
- Petitjean A. et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 28, 622–629 (2007). [DOI] [PubMed] [Google Scholar]
- Brosh R. & Rotter, V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 9, 701–713 (2009). [DOI] [PubMed] [Google Scholar]
- Vegran F. et al. Only missense mutations affecting the DNA binding domain of p53 influence outcomes in patients with breast carcinoma. PloS one 8, e55103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong R. et al. Prognostic significance of TP53 gene mutation in 995 cases of colorectal carcinoma. Influence of tumour site, stage, adjuvant chemotherapy and type of mutation. Eur J Cancer 36, 2053–2060 (2000). [DOI] [PubMed] [Google Scholar]
- Fernandez-Cuesta L. et al. Prognostic and predictive value of TP53 mutations in node-positive breast cancer patients treated with anthracycline- or anthracycline/taxane-based adjuvant therapy: results from the BIG 02-98 phase III trial. Breast Cancer Res 14, R70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsner J., Yilmaz M., Guldberg P., Hansen L. L. & Overgaard J. Heterogeneity in the clinical phenotype of TP53 mutations in breast cancer patients. Clin Cancer Res 6, 3923–3931 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.