Abstract
1,2,4-Butanetriol (BT) is a valuable chemical with extensive applications in many different fields. The traditional chemical routes to synthesize BT suffer from many drawbacks, e.g., harsh reaction conditions, multiple steps and poor selectivity, limiting its industrial production. In this study, an engineered Escherichia coli strain was constructed to produce BT from xylose, which is a major component of the lignocellulosic biomass. Through the coexpression of a xylose dehydrogenase (CCxylB) and a xylonolactonase (xylC) from Caulobacter crescentus, native E. coli xylonate dehydratase (yjhG), a 2-keto acid decarboxylase from Pseudomonas putida (mdlC) and native E. coli aldehyde reductase (adhP) in E. coli BL21 star(DE3), the recombinant strain could efficiently convert xylose to BT. Furthermore, the competitive pathway responsible for xylose metabolism in E. coli was blocked by disrupting two genes (xylA and EcxylB) encoding xylose isomerase and xyloluse kinase. Under fed-batch conditions, the engineered strain BL21ΔxylAB/pE-mdlCxylBC&pA-adhPyjhG produced up to 3.92 g/L of BT from 20 g/L of xylose, corresponding to a molar yield of 27.7%. These results suggest that the engineered E. coli has a promising prospect for the large-scale production of BT.
1,2,4-Butanetriol (BT) is a four-carbon polyol with three hydrophilic hydroxyl groups. As an important fine chemical, BT has versatile applications in many different fields and attracted considerable interest in the past few years. For instance, BT can be used for making polyurethane foams. These foams have the same compression-bending characteristics as natural rubber1. BT severs as the recording agent of high-quality inks and renders the inks having well balanced anti-feathering and penetrability effects2. Optically active BT is also a potential building block for the synthesis of various pharmaceuticals, such as Crestor and Zetia3. Finally, BT is the direct precursor for the manufacture of butanetriol trinitrate, an excellent energetic plasticizer to replace nitroglycerin, which is less volatile, thermally more stable and offers better low-temperature properties4,5.
BT is traditionally manufactured through chemical routes using glycidol6, 2-butene-1,4-diol7, 3,4-dihydroxybutanoate8 or malate9,10 as the starting materials. Among them, the high pressure catalytic hydrogenation of esterified malate is currently commercially available. However, this process requires NaBH4 as the reducing agent. For each ton of BT to be synthesized, multiple tons of borate salts are generated as the by-products, thus resulting in high production costs and severe environmental pollution11. In addition, all of the chemical synthetic routes suffer from similar drawbacks, e.g., harsh reaction conditions, multiple steps and poor selectivity. These difficulties create hurdles that hamper further applications of the petrochemical-based BT.
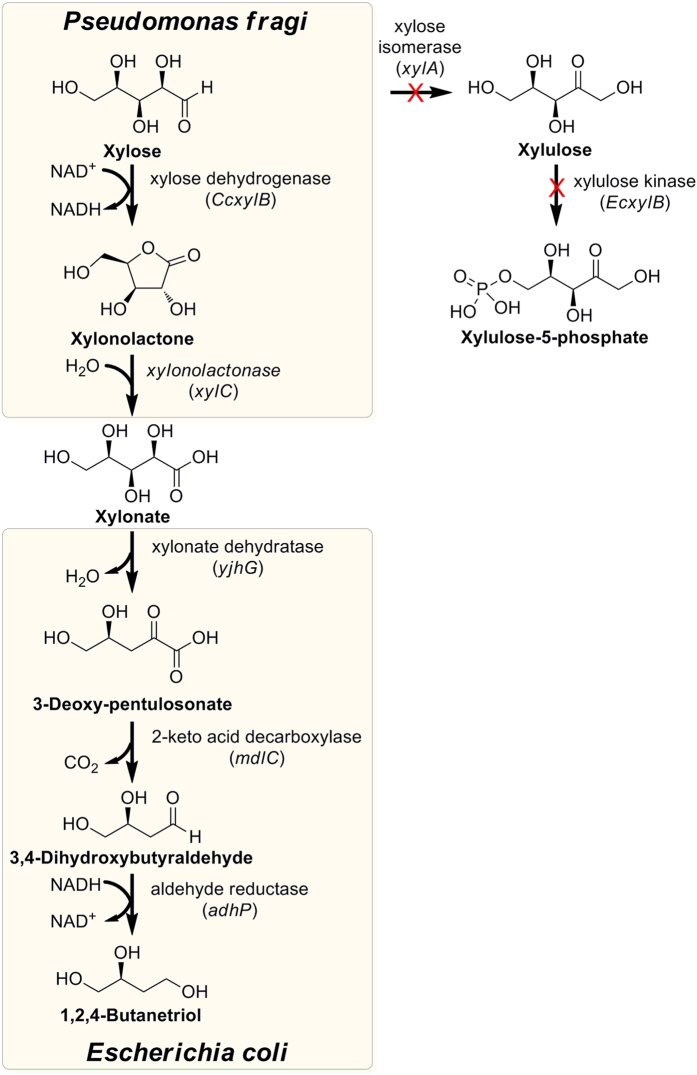
In recent years, biomass utilization for the production of value-added chemicals is attracting considerable interest12,13. Xylose is the second most abundant sugar in nature and a major constituent of hemicellulose in lignocellulosic biomass14. A number of wild-type microbial strains or metabolically engineered strains have been isolated or constructed to ferment xylose for the production of ethanol15, xylitol16, succinate17, lactate18 and other important products. The utilization of biomass-derived xylose to produce bio-based chemicals offers many advantages over the traditional petrochemical routes in that it is made from renewable resources, possesses mild transformation conditions and reduces the environmental pollution. Researchers also explored new routes for BT production and finally they established a new process to produce BT from xylose by microbial conversion19. Through the introduction of a 2-keto acid decarboxylase (mdlC) from Pseudomonas putita20 to Escherichia coli, the recombinant strain could efficiently covert xylonate to BT. Meanwhile, the feedstock xylonate could be obtained by microbial oxidation of xylose by Pseudomonas fragi21. The entire metabolic pathway from xylose to BT is proposed in Fig. 1. Although many wild-type bacterial species can produce xylonate in high yields21,22,23, these natural producers always require expensive nutrient media and take several days to reach the maximum titer. Meanwhile, separation and purification of xylonate from the fermentation broth is also a difficult task. As is known to all, the downstream processing of organic acids is estimated to make up the major cost in its microbial production24. Therefore, the two-step fermentation process would be economically unfeasible for large-scale production. A one-pot conversion from xylose to BT would be more reliable for industrial applications.
Figure 1. The metabolic pathway from xylose to BT.
Xylose is oxidized to xylonolactone by the xylose dehydrogenase (encoded by CcxylB). Xylonolactone is hydrolyzed to xylonate by the xylonolactonase (encoded by xylC). Xylonate dehydratase (encoded by yjhG) catalyzes the dehydration of xylonate to produce 3-deoxy-pentulosonate. Then 3-deoxy-pentulosonate is decarboxylated by 2-keto acid decarboxylase (encoded by mdlC) to form 3,4-dihydroxybutyraldehyde. At last, 3,4-dihydroxybutyraldehyde is reduced by the aldehyde reductase (encoded by adhP) to generate BT. Xylose isomerase (encoded by xylA) and xylulose kinase (encoded by EcxylB) responsible for xylose metabolism are knocked out to block the branched pathways.
The construction of a direct BT-producing E. coli strain from xylose was presented in this study. The Caulobacter crescentus xylose dehydrogenase (CCxylB), xylonolactonase (xylC) and P. putita mdlC were coexpressed with native yjhG (encoding xylonate dehydratase) and adhP (encoding aldehyde reductase/alcohol dehydrogenase) in E. coli BL21 star(DE3). To avoid the consumption of xylose for cell growth, the endogenous xylose catabolic pathway was blocked by disrupting two genes, xylA (encoding xylose isomerase) and EcxylB (encoding xylulose kinase) responsible for xylose utilization. The engineered strain was cultured under fed-batch conditions to evaluate the potential for large-scale BT production.
Results
Identification of recombinant enzymes expressed in E. coli
E. coli BL21 star(DE3) was chosen as the host for recombinant proteins expression and BT production in this work. This strain has a genotype that promotes mRNA stability and protein yield and thus is ideal for use with low copy number, T7 promoter-based plasmids. In order to express the CCXylB, XylC, YjhG, MdlC and AdhP enzymes which constitute the entire metabolic pathway from xylose to BT, we cloned the coding regions of these genes into the expression vectors pET28a, pTrcHis2B, pETDuet-1 and pACYCDuet-1, respectively. The expression constructs were checked by restriction enzyme digestion and DNA sequencing. To verify the expression levels of these recombinant proteins, E. coli BL21 star(DE3) was transformed with the expression vectors and grown in liquid LB medium followed by induction with 0.5 mM IPTG. Figure 2 showed the gel electrophoresis patterns of protein samples visualized by Coomassie Brilliant Blue staining. We noted distinct bands of the expected sizes from crude extracts of the recombinant strains when compared with the control strain BL21/pET28a. SDS-PAGE analysis of the recombinant strain carrying pET-mdlC showed a band corresponding to the molecular weight of 56.4 kDa while strain BL21/pA-yjhGadhP revealed two protein bands (35.4 kDa and 70.0 kDa). The recombinant strain harboring both of the two plasmids gave all three bands. Strain BL21/pA-xylBC revealed the recombinant proteins corresponding to the molecular weights of 26.6 kDa and 31.6 kDa. And the final strain harboring both pE-mdlCxylBC and pA-yjhGadhP overexpressed all of the five enzymes.
Figure 2. Validation of the expression of different recombinant enzymes in E. coli through SDS-PAGE analysis.

Recombinant protein expression was induced using 0.5 mM IPTG for a cultivation time of 4 h at 37 °C. Lane M, prestained protein molecular weight marker; lane 1, crude cells extracts from E. coli BL21 star(DE3) harboring pET28a; lane 2, crude cells extracts from strain BL21/pET-mdlC; lane 3, crude cells extracts from strain BL21/pA-adhPyjhG; lane 4, crude cells extracts from strain BL21/pET-mdlC&pA-adhPyjhG; lane 5, crude cells extracts from strain BL21/pA-xylBC; lane 6, crude cells extracts from strain BL21/pE-mdlCxylBC&pA-adhPyjhG. The bands corresponding to the individual proteins were indicated by an arrow.
Enhancement of BT production from xylonate by coexpression of the entire pathway
It has been demonstrated that heterologous expression of P. putida mdlC gene in E. coli DH5α led to the synthesis of BT from xylonate19. However, this biotransformation process could not be achieved using E. coli BL21(DE3) as the host strain. Frost and co-workers have postulated a potential metabolic pathway from xylonate to BT (Fig. 1). Two enzymes, xylonate dehydratase (encoded by yjhG) and aldehyde reductase (encoded by adhP) are required for the necessary steps of the bioconversion. E. coli BL21(DE3) genome does not carry any xylonate dehydratase25 and thus cannot metabolize xylonate. To render strain BL21 the ability of producing BT from xylonate, we cloned the yjhG and adhP genes from strain DH5α into the expression vector pACYCDuet-1 sequentially, resulting pA-yjhG and pA-adhPyjhG. To evaluate the effects of coexpression of the entire pathway on BT production, E. coli DH5α, W3110, JM109(DE3) and BL21 star(DE3) was transformed with pTrc-mdlC, pET-mdlC, pA-yjhG, pA-adhPyjhG or the combination of these plasmids. The recombinant strains were cultured using liquid LB medium supplemented with 5 g/L of potassium xylonate under shake-flask conditions. After being induced for 48 h, the bacterial cells were separated by centrifugation and BT produced in the supernatant was derivatized and analyzed by gas chromatography—mass spectrometry (GC-MS). Figure 3 showed the total ion current (TIC) chromatogram and mass spectrum of the silylation products of the fermentation supernatant from strain BL21/pET-mdlC&pA-adhPyjhG. 1,2,4-Tri(trimethylsilyl)butanetriol (corresponding to the retention time of 4.53 min) was identified by matching a standard NIST library. BT production of different engineered strains in the culture supernatant was then directly determined by high-performance liquid chromatography (HPLC). As shown in Fig. 4, cell growth was comparable between these recombinant strains. The titer of BT for strain DH5α/pTrc-mdlC reached 0.11 g/L after 48 h of induction. Similar results were obtained by strain W3110/pTrc-mdlC and JM109/pTrc-mdlC. When the yjhG gene was coexpressed with mdlC, BT production was enhanced to 0.17 g/L. In the strain JM109/pET-mdlC&pA-adhPyjhG and BL21/pET-mdlC&pA-adhPyjhG coexpressing the entire BT biosynthesis pathway, the final titer of BT was further elevated to 0.31 g/L, which is 1.8-fold higher than the original strain. It has been demonstrated that the aldehyde reductase AdhP could convert aldehydes to alcohols26 and reduce the toxicity of the intermediate 3,4-dihydroxybutanal, thus leading to an enhanced production of BT.
Figure 3. Identification of BT accumulated in the culture broth by GC-MS.
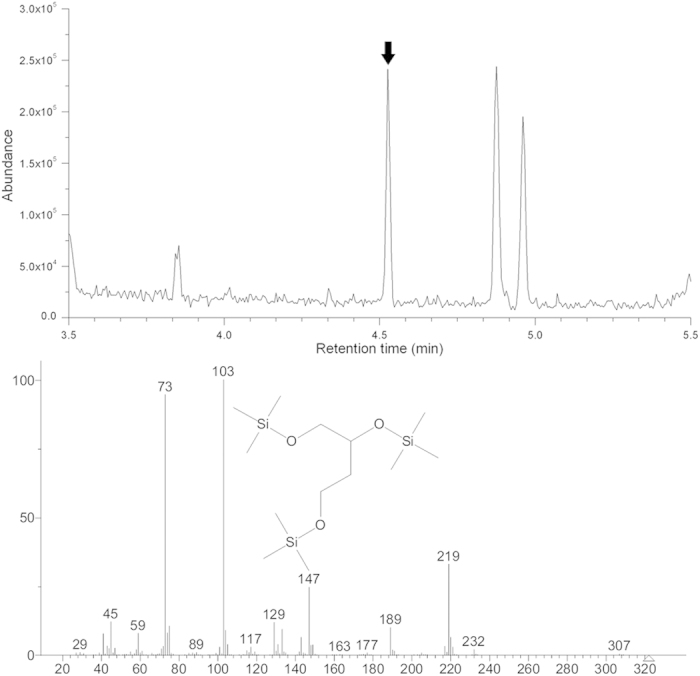
(A) total ion current (TIC) chromatogram of the silylation products of the fermentation supernatant from E. coli BL21 star(DE3) harboring pET-mdlC and pA-adhPyjhG after being induced for 48 h. (B) mass spectrum of silylation product of BT (corresponding to the retention time of 4.53 min).
Figure 4. Comparison of BT productions of several different strains.
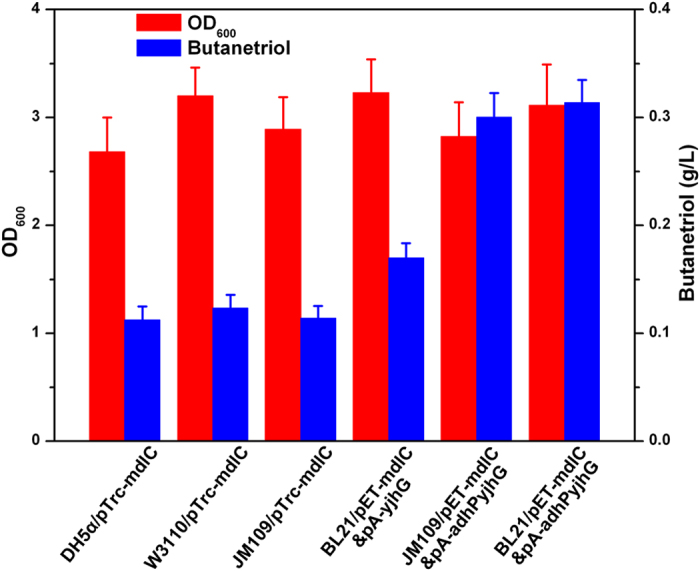
DH5α/pTrc-mdlC, strain DH5α P. putida 2-keto acid decarboxylase; BL21/pET-mdlC&pA-yjhG, strain BL21 star(DE3) expressing P. putida 2-keto acid decarboxylase and native E. coli dehydratase; BL21/pET-mdlC&pA-adhPyjhG, strain BL21 star(DE3) expressing P. putida 2-keto acid decarboxylase and native E. coli dehydratase and aldehyde reductase. Data were obtained after each strain was induced for 48 h in liquid LB medium supplemented with 5 g/L of potassium xylonate as the substrates for BT production.
Direct bioconversion of xylose to BT
Previous studies employed a two-step process to synthesize BT from xylose because E. coli was incapable of producing xylonate from xylose. The conversion of xylose to xylonate requires two enzymes, xylose dehydrogenase and xylonolactonase. Recently, the xylose dehydrogenase (CCxylB) and xylonolactonase (xylC) were identified from the freshwater bacterium C. crescentus27. Here, the two genes were coexpressed in E. coli BL21 star(DE3) using the recombinant vector pA-xylBC. Accumulations of xylonolactone and xylonate were detected in the cultures of the recombinant strain. As shown in Fig. 5, 0.38 g/L of xylonolactone and 3.19 g/L of xylonate were produced from 5 g/L of xylose after being induced for 24 h. E. coli could utilize xylose via the pentose phosphate pathway for cell growth. The first two genes responsible for xylose catabolism have been recognized as xylose isomerase (encoded by xylA) and xylulose kinase (encoded by EcxylB)28. In order to block the native xylose catabolic pathway, we disrupted the xylA and EcxylB genes in strain BL21 star(DE3) to create BL21ΔxylAB. The final titers of xylonolactone and xylonate in strain BL21ΔxylAB/pA-xylBC were enhanced to 0.49 g/L and 4.07 g/L, respectively. The molar yield of xylonolactone and xylonate on xylose also reached 83.5%, which is 18.1% higher than strain BL21/pA-xylBC.
Figure 5. Production of xylonate and xylonolactone through the upstream pathway.
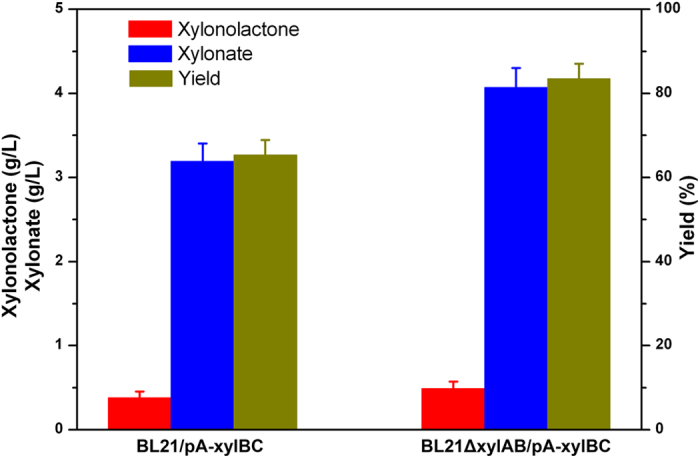
BL21/pA-xylBC, strain BL21 star(DE3) expressing C. crescentus xylose dehydrogenase and xylonolactonase; BL21ΔxylAB/pA-xylBC, knockout of native xylA and EcxylB while coexpressing C. crescentus xylose dehydrogenase and xylonolactonase. Data were obtained after each strain was induced for 24 h in liquid LB medium supplemented with 5 g/L of xylose.
In order to integrate the xylonate and BT producing pathway, the CCxylB and xylC genes were coexpressed with mdlC using the recombinant vector pE-mdlCxylBC. Both pE-mdlCxylBC and pA-adhPyjhG were co-transformed to BL21ΔxylAB competent cells. The resulting strain BL21ΔxylAB/pE-mdlCxylBC&pA-adhPyjhG was grown in LB medium containing 5 g/L of xylose. As expected, accumulation of BT was found in the culture of this engineered strain. BT production after being induced for 48 h reached 0.30 g/L, which is comparable with the strain converting xylonate to BT. The molar yield of BT to xylose reached 8.5%. As xylose, xylonate and many other organic acids shared a similar retention time in the HPLC chromatograph, we use the ion chromatography (IC) to separate these metabolites. IC analysis showed that the initial xylose was completely exhausted at 48 h post-induction, but intermediate metabolites of the BT biosynthesis pathway were detected in the culture broth. This fact indicated that the downstream pathway (xylonate to BT) instead of the upstream pathway (xylose to xylonate) was the rate-limiting step for BT biosynthesis.
BT production in fed-batch cultivation
In order to test the suitability of the recombinant E. coli strains for larger-scale production of BT, we established fed-batch fermentation based on the results obtained with shake-flask cultures. Strain BL21ΔxylAB/pE-mdlCxylBC&pA-adhPyjhG was cultured in a 5 L-scale laboratory fermenter. Glycerol was selected as the carbon source instead of glucose to avoid catabolite repression of xylose uptake. Cell density, residual xylose and BT accumulation were monitored over the course of the experiment. The residual glycerol concentration was controlled at a low level to avoid acetate formation, which might hamper cell growth and BT detection. Figure 6 showed the time profiles of cell density and concentrations of different metabolites during the whole fermentation period. For approximately 32 h post-induction, the bacteria grew rapidly to an OD600 of 75 or so. Most of the xylose was utilized in the first 18 h of fermentation. The intermediate metabolite xylonate reached a maximum titer of 10.2 g/L at 10 h post-induction and was then gradually consumed by the downstream pathway. BT also accumulated rapidly in the culture broth during the first 40 h post-induction and then gradually to a stable value. The highest BT production was obtained after being induced for 56 h, that is, 3.92 g/L, corresponding to a volumetric productivity of 0.89 mg/(L·h·OD600). The final molar yield of BT on xylose reached 27.7%. The above results achieved at the fermenter level greatly enhanced the titer of BT in the fermentation broth and demonstrated that this engineered E. coli strain had the potential to produce BT in a large scale.
Figure 6. Time profiles for cell density (OD600), residual xylose and BT concentrations in the culture broth during fed-batch culture of the engineered strain BL21ΔxylAB/pE-mdlCxylBC&pA-adhPyjhG.
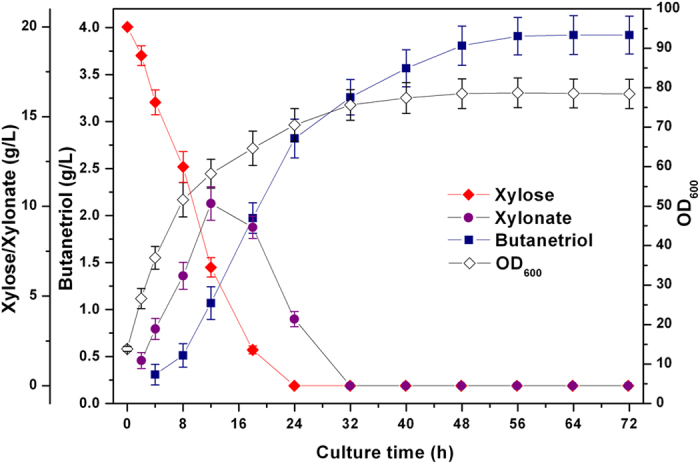
Cultures were performed in a 5L laboratory fermentor. 20 g/L of xylose was used as the substrates for BT production. Error bars represent the range of three independent fermentations.
Discussion
In this work, we integrated the two steps for biotechnological BT production in a single E. coli host and the metabolic pathway was further engineered to enhance the carbon flux to BT. Both the titer and yield of BT were enhanced to some extent compared with the original study19. These results demonstrated the power of rational design to improve the ability of microorganisms to produce bio-based chemicals. The one-step conversion process also makes the cells maintain redox balance. Reducing equivalent supply is crucial for the biosynthesis of value-added chemicals29. The metabolic pathway from xylonate to BT consists of three steps: dehydration, decarboxylation and reduction. One mole of NADH is required for each mole of BT formed. Therefore, the cells should provide additional NADH to facilitate the reduction of 3,4-dihydroxybutanal. The NADH balance forces more carbon flux to be channeled into the TCA cycle, leading to excessive consumption of the carbon sources. The upstream metabolic pathway from xylose to xylonate could produce one mole of NADH for each mole of xylose to be oxidized30. NADH consumed by the production of BT could be regenerated through the oxidation of xylose. Therefore, the integration of the upstream pathway and the downstream pathway in a single host could provide appropriate reducing power, which might contribute to BT biosynthesis.
The engineered strain in current study has greatly improved BT production. However, the yield of BT on xylose is still far from the theoretical limits. This result might be due to the following issues. First of all, the rate-determining step for BT biosynthesis is the decarboxylation of 3-deoxy-pentulosonate catalyzed by MdlC. The original activity of this enzyme is a benzoylformate decarboxylase participating in mandelate degradation20. 3-Deoxy-pentulosonate is not the natural substrate and thus it shows a much lower substrate specificity and catalytic activity. Therefore, protein engineering of the MdlC decarboxylase to improve its catalytic activity towards 3-deoxy-pentulosonate would be helpful to BT production31. On the other hand, there are many competing pathways associated with the BT biosynthesis pathway. For instance, 3-deoxy-pentulosonate can be split to pyruvate and glycolaldehyde32 as well as forming 2-amino-4,5-dihydroxypentanoate catalyzed by aminotransferase33. The oxidation of 3,4-dihydroxybutanal to 3,4-dihydroxybutyrate might also compete with its reduction to BT. Inactivation of these branched metabolic pathways would be expected to further improve BT production. However, these reactions are always catalyzed by multiple enzymes or key enzymes essential for the cell’s normal metabolism. It is difficult to completely block these branched pathways without affecting cell viability. Therefore, regulating the metabolic flux to BT biosynthesis would be critical to enhance its production.
Materials and Methods
Bacterial strains and plasmids construction
A list of bacterial strains and recombinant plasmids was presented in Table 1. Primers used for plasmids construction was provided in Table 2. E. coli DH5α was used for gene cloning and E. coli BL21 star(DE3) was used as the host for the expression of the recombinant proteins. The chromosomal xylA and EcxylB genes of strain BL21 star(DE3) responsible for xylose utilization were knocked out using the λ-Red recombination strategy in a previous study, resulting strain BL21ΔxylAB34.
Table 1. Strains and plasmids used in this study.
| Strains or plasmids | Genotype/Description | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− recA endA1 Φ80dlacZΔM15 hsdR17(rK− mK+) λ− | Invitrogen |
| E. coli W3110 | F− λ− rph-1 INV(rrnD rrnE) | Coli Genetic Stock Center |
| E. coli JM109 (DE3) | recA endA1 lacZΔM15 (lac-proAB) hsdR17(rK− mK+) IDE3 | Promega |
| E. coli BL21 star (DE3) | F− ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | Invitrogen |
| E. coli BL21 star (DE3) ΔxylAB | Knockout of xylA and EcxylB encoding xylose isomerase and xylulose kinase | 34 |
| Plasmids | ||
| pET28a | Kanr oripBR322 lacIq T7p | Novagen |
| pTrcHis2B | Ampr oripBR322 lacIq Trcp | Invitrogen |
| pETDuet-1 | Ampr oripBR322 lacIq T7p | Novagen |
| pACYCDuet-1 | Cmr oriP15A lacIq T7p | Novagen |
| pET-mdlC | pET28a harboring P. putida mdlC | This study |
| pTrc-mdlC | pTrcHis2B harboring P. putida mdlC | This study |
| pA-xylBC | pACYCDuet-1 harboring C. crescentus CCxylB and xylC | 34 |
| pE-xylB | pETDuet-1 harboring C. crescentus CCxylB | This study |
| pE-mdlCxylB | pETDuet-1 harboring P. putida mdlC and C. crescentus CCxylB | This study |
| pE-mdlCxylBC | pETDuet-1 harboring P. putida mdlC and C. crescentus CCxylB and xylC | This study |
| pA-yjhG | pACYCDuet-1 harboring E. coli yjhG | This study |
| pA-adhPyjhG | pACYCDuet-1 harboring E. coli yjhG and adhP | This study |
Table 2. Primers used in this study for plasmids construction or allele verification.
| Oligonucleotide primers | Description |
|---|---|
| xylB_F_NdeI | GGGAATTCCATATGTCCTCAGCCATCTATCCC |
| xylB_R_KpnI | CGGGGTACCTCAACGCCAGCCGGCGTCGAT |
| mdlC_F_NcoI | CATGCCATGGCTTCTGTTCACGGTACCACC |
| mdlC_R_EcoRI | CCGGAATTCTTATTTAACCGGAGAAACGGTAG |
| T7xylC_F_EcoRI | CCGGAATTCTAATACGACTCACTATAGGGGAATTG |
| T7xylC_R_NotI | AAGGAAAAAAGCGGCCGCTTAAACCAGACGAACTTCGTGCTG |
| yjhG_F_NdeI | GGAATTCCATATGTCTGTTCGCAATATTTTTGC |
| yjhG_R_XhoI | CCGCTCGAGTCAGTTTTTATTCATAAAATCGCG |
| adhP_F_NcoI | CATGCCATGGGCATGAAGGCTGCAGTTGTTACG |
| adhP_R_EcoRI | CCGGAATTCTTAGTGACGGAAATCAATCACC |
The 2-keto acid decarboxylase from P. putita (mdlC, GenBank accession no.: AY143338) was codon optimized, chemically synthesized and cloned into pET28a or pTrcHis2B vector between NcoI and EcoRI sites to create pET-mdlC or pTrc-mdlC. The CCxylB (GenBank Accession No.: NACL94329) and xylC (GenBank Accession No.: NACL94328) genes from C. crescentus were also cloned into pACYCDuet-1 vector simultaneously to generate plasmid pA-xylBC in our previous study34. Then CCxylB was PCR amplified from vector pA-xylBC and cloned into pETDuet-1 vector between NdeI and KpnI sites to create pE-xylB. The mdlC gene was digested from pET-mdlC using NcoI and EcoRI, and then cloned into the same restrict sites of pE-xylB, resulting pE-mdlCxylB. The xylC gene was amplified from pA-xylBC along with T7 promoter and the PCR product T7xylC was then cloned into pE-mdlCxylB between EcoRI and NotI sites, to create pE-mdlCxylBC. The coding sequences of native E. coli yjhG (Gene ID: 6060334) and adhP (Gene ID: 6059208) were generated by PCR using the primers based on the selected sequences around the start or stop codons of the genes and with restriction enzyme sites at 5′ ends. The resulting PCR product of yjhG was cloned into pACYCDuet-1 between NdeI and XhoI sites leading to plasmid pA-yjhG. The adhP gene was then cloned into pA-yjhG between NcoI and EcoRI sites to create pA-adhPyjhG. Successful gene cloning was verified by colony PCR, restriction mapping and direct nucleotide sequencing.
Protein expression and gel electrophoresis analysis
For the expression of different recombinant proteins, single colonies of E. coli BL21 star(DE3) harboring different recombinant plasmids were used to inoculate Luria-Bertani (LB) medium containing appropriate antibiotics and grown at 37 °C overnight. The saturated culture was diluted 1:100 into fresh LB medium and incubated under the same conditions. When the optical density at 600 nm (OD600) of the culture reached about 0.6, recombinant protein expression was induced by 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and growth was continued for 4 h. Cells were collected from 5 ml of bacteria cultures by centrifugation and washed with sterile distilled water. The washed pellets were suspended in 500 μl Tris-HCl buffer (pH 8.0) and subject to ultrasonication. The cell lysates were centrifuged and the supernatant obtained was mixed with 2 × sodium dodecyl sulfate (SDS) sample buffer, heated at 100 °C for 10 min and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) according to standard protocols35.
Shake-flask cultivation
To evaluate the BT producing ability of different engineered strains, shake-flask experiments were carried out in triplicate series of 100 ml Erlenmeyer flasks containing 20 ml of liquid LB medium supplemented with appropriate antibiotics. E. coli strains were inoculated to the culture medium and incubated in a gyratory shaker incubator at 37 °C and 180 rpm. When the OD600 of the culture reached about 0.6, IPTG was added to a final concentration of 0.5 mM to induce the expression of different enzymes. 5 g/L of potassium xylonate or xylose was added to the culture at the same time as the substrates for BT production. Samples were taken at different intervals to determine cell density, residual xylose and BT accumulated in the culture broth during the whole fermentation courses.
Fed-batch fermentation
For large-scale production of BT, fed-batch cultures were performed in a Biostat B plus MO5L fermentor (Sartorius, Germany) containing 3 L of growth medium (20 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl and 5 g/L K2HPO4·3H2O) that was sterilized at 121 °C for 20 min. Glycerol (10 g/L), MgSO4 (0.12 g/L) and trace elements (1 ml per liter, 3.7 g/L (NH4)6Mo7O24·4H2O, 2.9 g/L ZnSO4·7H2O, 24.7 g/L H3BO3, 2.5 g/L CuSO4·5H2O, 15.8 g/L MnCl2·4H2O) were autoclaved or filter-sterilized separately and added prior to initiation of the fermentation. Fermentation was started by inoculating 100 ml of overnight seed culture prepared in LB medium. During the fermentation process, continuous sterile air was supplied at a flow rate of 3 L/min. The temperature was controlled at 37 °C and the pH was controlled at 7.0 via automatic addition of ammonia. Antifoam was added to prevent frothing if necessary. The agitation speed was set at 400 rpm and then associated with the dissolved oxygen (DO) to maintain the DO level above 20% air saturation. Fermentation was first operated in a batch mode until the initial glycerol was nearly exhausted. Then fed-batch mode was commenced by feeding 70% of glycerol at appropriate rates. When the cells were grown to an OD600 of about 15, 0.5 mM of IPTG, 20 g/L of xylose and 0.1 g/L of thiamine hydrochloride were added to the culture broth to induce recombinant protein expression and BT production. Samples of the fermentation broth were determined the same as shake-flask cultivation.
Analytic methods
BT produced in the culture broth was identified using gas chromatography-mass spectrometry (GC-MS). The culture supernatant was mixed with 9 volume of ethanol to deposit proteins and concentrated by reduced pressure distillation. The concentrated sample was then derivatized by bis(trimethylsilyl)trifluoroacetamide (BSTFA): appropriate amounts of the samples were added 1 ml of BSTFA and 0.2 ml of pyridine, and then reacted at 70 °C for 30 min36. The reaction mixture was diluted by hexane and then subjected to GC-MS. GC-MS analysis was performed with an Agilent 7890 GC system coupled to a quadrupole rods detector. The GC-MS conditions were as follows: a 30 m HP-5 ms column (internal diameter 0.25 mm, film thickness 0.25 μm); an oven temperature program composed of an initial hold at 100 °C for 2 min, ramping at 10 °C per min to 250 °C, and a final hold at 250 °C for 3 min; an ion source temperature of 220 °C and EI ionization at 70 eV.
BT concentrations in the culture supernatant was determined by an Agilent 1200 series HPLC system equipped with an Aminex HPX-87H (Bio-Rad, Hercules, CA) column (300 × 7.8 mm). All samples were filtered through 0.22 μm syringe filter. Ultrapure water with 5 mM H2SO4 was used as the eluent at a flow rate of 0.5 ml/min. The oven temperature was maintained at 55 °C. Peaks were detected by a refractive index detector (RID). Quantitation of BT was performed by using the external standard method.
The determination of xylose in the culture broth was performed on an ICS-3000 (Dionex, Sunnyvale, CA) ion chromatography (IC) system. The IC was equipped with an IonPacAS11 anion chromatography column (4.0 mm × 250 mm) and an AG-11 guard column (4.0 mm × 50 mm). Suppression was achieved with anion suppressor (ASRS 300 4 mm). Peaks were detected using electrochemical detector. A mixture of 250 mM NaOH (2%) and H2O (98%) was used for elution at a flow rate of 1 ml/min. For sample analysis, another elution step with 80% of 250 mM NaOH was employed to remove the residual components. Data collection and handling were carried out by Dionex Chromeleon software.
Additional Information
How to cite this article: Cao, Y. et al. Biotechnological production of 1,2,4-butanetriol: An efficient process to synthesize energetic material precursor from renewable biomass. Sci. Rep. 5, 18149; doi: 10.1038/srep18149 (2015).
Acknowledgments
This work was sponsored by National Natural Science Foundation of China (No. 21202179, 21376255 and 31200030), Key Program of the Chinese Academy of Sciences (No. KGZD-EW-606-1-3) and Taishan Scholars Climbing Program of Shandong (No. tspd20150210).
Footnotes
Author Contributions M.X. and H.L. conceived the project; Y.C., M.X. and H.L. designed the experiments; Y.C. performed the experiments; Y.C., W.N., J.G. and M.X. analyzed the primary data; Y.C. and M.X. drafted the manuscript; W.N., J.G. and H.L. revised the manuscript.
References
- Gmitter G. T. inventors; Gen Tire & Rubber Co., assignee. Cellular polyetherurethanes and methods of making same. United States patent US 3149083. 1964 Sep 15.
- Shirota K., Fukushima K. & Koike S. inventors; Canon KK (JP), assignee. Ink, ink-jet recording process, and instrument using the ink. Europe patent EP 0445746. 1991 Sep 11.
- Zingaro K. A. & Papoutsakis E. T. GroESL overexpression imparts Escherichia coli tolerance to i-, n-, and 2-butanol, 1,2,4-butanetriol and ethanol with complex and unpredictable patterns. Metab. Eng. 15, 196–205, doi: 10.1016/j.ymben.2012.07.009 (2013). [DOI] [PubMed] [Google Scholar]
- Gouranlou F. & Kohsary I. Synthesis and characterization of 1,2,4-butanetrioltrinitrate. Asian J. Chem. 22, 4221–4228 (2010). [Google Scholar]
- Li X., Cai Z., Li Y. & Zhang Y. Design and construction of a non-natural malate to 1,2,4-butanetriol pathway creates possibility to produce 1,2,4-butanetriol from glucose. Sci. Rep. 4, doi: 10.1038/srep05541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. J. & Hinshaw J. C. inventors; Thiokol Corporation, assignee. Making 1,2,4-butanetriol by hydroformylation of glycidol. United States patent US 4410744. 1983 Oct 18.
- Kwak B. S., Kim T. Y., Lee S. I. & Kim J. W. inventors; SK Corporation, assignee. Continuous production method of 1,2,4-butanetriol. International patent WO 2005/061424. 2005 Jul 7.
- Ikai K., Mikami M., Furukawa Y., Urano T. & Ohtaka S. inventors; Daiso Co., Ltd., assignee. Process for preparing 1,2,4-butanetriol. United States patent US 6949684. 2005 Sep 27.
- Ritter S. K. Biomass or bust. Chem. Eng. News 82, 31–33, doi: 10.1021/cen-v082n022.p031 (2004). [DOI] [Google Scholar]
- Adkins H. & Billica H. R. The hydrogenation of esters to alcohols at 25–150°. J. Am. Chem. Soc. 70, 3121–3125, doi: 10.1021/ja01189a085 (1948). [DOI] [Google Scholar]
- Frost J. W. & Niu W. inventors; Board of Trustees of Michigan State University, assignee. Microbial synthesis of D-1,2,4-butanetriol. United States patent US 20110076730. 2011 Mar 31.
- Nielsen J. et al. Engineering synergy in biotechnology. Nat. Chem. Biol. 10, 319–322, doi: 10.1038/nchembio.1519 (2014). [DOI] [PubMed] [Google Scholar]
- Ragauskas A. J. et al. The path forward for biofuels and biomaterials. Science 311, 484–489, doi: 10.1126/science.1114736 (2006). [DOI] [PubMed] [Google Scholar]
- Lachke A. Biofuel from D-xylose—The second most abundant sugar. Resonance 7, 50–58, doi: 10.1007/bf02836736 (2002). [DOI] [Google Scholar]
- Nielsen J., Larsson C., van Maris A. & Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Curr. Opin. Biotechnol. 24, 398–404, doi: 10.1016/j.copbio.2013.03.023 (2013). [DOI] [PubMed] [Google Scholar]
- Akinterinwa O. & Cirino P. C. Anaerobic obligatory xylitol production in Escherichia coli strains devoid of native fermentation pathways. Appl. Environ. Microbiol. 77, 706–709, doi: 10.1128/aem.01890-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. et al. Fermentation of xylose to succinate by enhancement of ATP supply in metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 94, 959–968, doi: 10.1007/s00253-012-3896-4 (2012). [DOI] [PubMed] [Google Scholar]
- Shinkawa S. et al. Improved homo L-lactic acid fermentation from xylose by abolishment of the phosphoketolase pathway and enhancement of the pentose phosphate pathway in genetically modified xylose-assimilating Lactococcus lactis. Appl. Microbiol. Biotechnol. 91, 1537–1544, doi: 10.1007/s00253-011-3342-z (2011). [DOI] [PubMed] [Google Scholar]
- Niu W., Molefe M. N. & Frost J. W. Microbial synthesis of the energetic material precursor 1,2,4-butanetriol. J. Am. Chem. Soc. 125, 12998–12999, doi: 10.1021/Ja036391 (2003). [DOI] [PubMed] [Google Scholar]
- Tsou A. Y. et al. Mandelate pathway of Pseudomonas putida: sequence relationships involving mandelate racemase, (S)-mandelate dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli. Biochemistry 29, 9856–9862, doi: 10.1021/bi00494a015 (1990). [DOI] [PubMed] [Google Scholar]
- Buchert J., Viikari L., Linko M. & Markkanen P. Production of xylonic acid by Pseudomonas fragi. Biotechnol. Lett. 8, 541–546, doi: 10.1007/bf01028079 (1986). [DOI] [Google Scholar]
- Ishizaki H., Ihara T., Yoshitak. J., Shimamur. M. & Imai T. D-Xylonic acid production by Enterobacter cloacae. J. Agric. Chem. Soc. Jpn. 47, 755–761 (1973). [Google Scholar]
- Buchert J. & Viikari L. Oxidative D-xylose metabolism of Gluconobacter oxydans. Appl. Microbiol. Biotechnol. 29, 375–379, doi: 10.1007/bf00265822 (1988). [DOI] [Google Scholar]
- Cao Y. et al. Fermentative succinate production: An emerging technology to replace the traditional petrochemical processes. Biomed Res. Int. 2013, 12, doi: 10.1155/2013/723412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H. et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 394, 644–652, doi: 10.1016/j.jmb.2009.09.052 (2009). [DOI] [PubMed] [Google Scholar]
- Rodriguez G. M. & Atsumi S. Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab. Eng. 25, 227–237, doi: 10.1016/j.ymben.2014.07.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C. et al. Genetic analysis of a novel pathway for D-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 189, 2181–2185, doi: 10.1128/jb.01438-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J. D. & Wiesmeye H. Control of xylose metabolism in Escherichia coli. Biochim. Biophys. Acta 201, 497–499, doi: 10.1016/0304-4165(70)90171-6 (1970). [DOI] [PubMed] [Google Scholar]
- Chin J. W., Khankal R., Monroe C. A., Maranas C. D. & Cirino P. C. Analysis of NADPH supply during xylitol production by engineered Escherichia coli. Biotechnol. Bioeng. 102, 209–220, doi: 10.1002/bit.22060 (2009). [DOI] [PubMed] [Google Scholar]
- Johnsen U. & Schönheit P. Novel xylose dehydrogenase in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 186, 6198–6207, doi: 10.1128/jb.186.18.6198-6207.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y., Rodriguez G. & Atsumi S. 2-Keto acids based biosynthesis pathways for renewable fuels and chemicals. J. Ind. Microbiol. Biotechnol. 42, 361–373, doi: 10.1007/s10295-014-1547-8 (2015). [DOI] [PubMed] [Google Scholar]
- Dahms S. A. 3-Deoxy-D-pentulosonic acid aldolase and its role in a new pathway of D-xylose degradation. Biochem. Biophys. Res. Commun. 60, 1433–1439, doi: 10.1016/0006-291X(74)90358-1 (1974). [DOI] [PubMed] [Google Scholar]
- Kuramitsu S., Okuno S., Ogawa T., Ogawa H. & Kagamiyama H. Aspartate aminotransferase of Escherichia coli: Nucleotide sequence of the aspC gene. J. Biochem. 97, 1259–1262 (1985). [DOI] [PubMed] [Google Scholar]
- Cao Y., Xian M., Zou H. & Zhang H. Metabolic engineering of Escherichia coli for the production of xylonate. PLoS ONE 8, e67305, doi: 10.1371/journal.pone.0067305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. et al. Production of free monounsaturated fatty acids by metabolically engineered Escherichia coli. Biotechnol. Biofuels 7, 59, doi: 10.1186/1754-6834-7-59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schummer C., Delhomme O., Appenzeller B. M. R., Wennig R. & Millet M. Comparison of MTBSTFA and BSTFA in derivatization reactions of polar compounds prior to GC/MS analysis. Talanta 77, 1473–1482, doi: 10.1016/j.talanta.2008.09.043 (2009). [DOI] [PubMed] [Google Scholar]