Abstract
African green monkeys (AGMs) represent the most widely distributed non-human primates species in Africa. SIVagm naturally infects four of the 6 AGMs species at high prevalence in a species-specific manner. To date, only limited information is available on molecular characteristics of SIVagm infecting Chlorocebus tantalus.
Here, we characterized the full-length genome of a virus infecting a naturally infected captive C. tantalus from Cameroon by amplifying and sequencing sub-genomic PCR fragments. The isolate (SIVagmTAN-CM545) is 9923 bp long and contained all canonical genes of a functional SIV. SIVagmTAN-CM545 showed a mosaic structure, with gag, pol, nef and accessory genes closely related to SIVagmSAB infecting Chlorocebus sabaeus monkeys from west Africa, and the env gene, closely related to SIVagmTAN infecting tantalus monkeys from Central Africa. Thus SIVagmTAN-CM545 is SIVagmSAB/SIVagmTAN recombinant.
These unexpected findings suggest that the evolution of SIVagm is more complex than previously thought and warrant further studies.
Keywords: SIV, African green monkeys, Phylogeny, Cameroon
1. Introduction
Simian Immunodeficiency Viruses (SIVs), the closest relatives of Human Immunodeficiency Viruses (HIV-1 and HIV-2), have been identified in at least 45 different African non-human primate (NHP) species (Locatelli and Peeters, 2012). The genetic diversity of NHP lentiviruses is complex and includes examples of co-evolution between the virus and the host, cross-species transmission, recombination between distant SIV lineages and certain species can even harbor different SIV lineages. However, despite this complex evolutionary history, each NHP species is generally infected with a species-specific SIV which form monophyletic lineages in phylogenetic trees.
An increasing number of SIV lineages have been described recently, but our knowledge on genetic diversity, prevalence and SIV evolution in their natural hosts still remains limited (Liegeois et al., 2014). Only few animals have been tested and as a consequence few full-length SIV genomes have been characterized for each species and for some species no or only partial SIV sequence information is available. For example, for African green monkeys (AGMs) of the Chlorocebus genus, the observations of co-evolution between virus and host are based on full-length SIVagm genome sequences derived from eight animals. Based on morphological data, six geographically separated Chlorocebus species are described today across Africa and in four of them a species-specific SIV lineage has been identified: SIVagmSAB in Chlorocebus sabaeus (green monkey) from Senegal to the Volta river; SIVagmTAN in Chlorocebus tantalus (tantalus) in north central Africa from Ghana to Sudan; SIVagmGRI in Chlorocebus aethiops (grivet) in east Africa and SIVagmVER in Chlorocebus pygerythrus (vervet) in east and southern Africa (Allan et al., 1991; Hirsch et al., 1986, 1993; Lowenstine et al., 1986; Ohta et al., 1988) The remaining two species, Chlorocebus cynosuros (malbrouck) from the Albertine Rift in the Democratic Republic of Congo to the Atlantic coast and Northern Namibia and Zambia and finally, and Chlorocebus djamdjamensis (Bale Mountains vervet), located in a restricted area of Ethiopia have not been tested yet for SIV infection (Fig. 1). It has to be noted that the AGM classification is still on debate, and recent genetic studies challenged the actual classification.
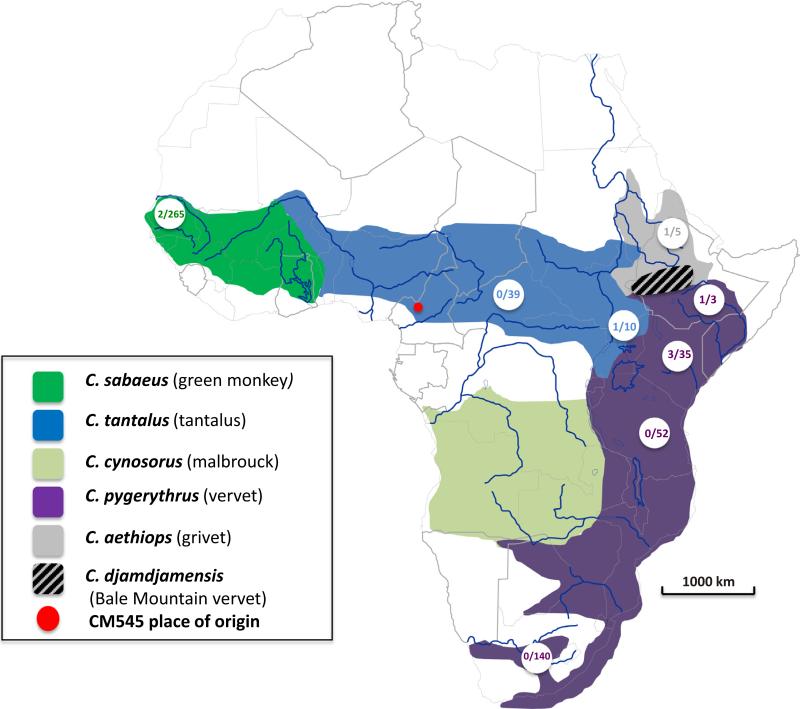
Fig. 1.
Distribution range of African green monkeys. The figure represents the distribution range of African green monkeys adapted from (Haus et al., 2013a). The range of C. sabaeus is colored in green, that of C. tantalus in blue, C. cynosuros in light-green, C. pygerythrus in magenta, C. aethiops in gray and C. djamdjamensis, dashed-gray. Circled numbers on the map indicate at the denominator the number of SIVagm from the given region/species found in the GenBank as of the 24th of July, 2014. The numerator gives the number of full-length genomes available at the same date. The red point on the map indicates the geographical origin of agm-CM545. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
African green monkeys represent the most widely geographically distributed NHP genus in Africa (VandeWoude and Apetrei, 2006) but the few SIV strains that have been characterized could represent a bias for the overall SIV genetic diversity across the wide geographic range of different Chlorocebus species. Moreover SIV seroprevalence are high in Chlorocebus species and can reach 50% in wild AGM populations (Hendry et al., 1986; Ma et al., 2013, 2014). Despite the fact that AGM represent potentially an important SIV reservoir, only around 400 wild animals have been tested across Africa and only 8 full-length SIVagm genome sequences are available.
In Cameroon, west Central Africa, all AGM classifications performed so far indicated that C. tantalus is the endemic species of the area, together with the neighboring countries of Nigeria in the west and Central African Republic in the South-East (Haus et al., 2013a). Despite a wealth of data on SIV prevalence and genetic diversity in NHP from Cameroon, there is no information on molecular characteristics of SIV infecting AGMs from this country and only one full-length SIVagmTAN sequence is available form a tantalus monkey from Uganda, almost at the extreme eastern part of the tantalus range. To fill this gap, we report here the characterization on an SIV infecting a C. tantalus from Cameroon.
2. Material and methods
2.1. Animal and serology
A female tantalus monkey (animal 1405, sample No. 545) was confiscated in April 2001 by authorities of the Cameroonian Ministry of Environment and Forestry in Bamenda (Fig. 1), in the North-West region of Cameroon, where the animal was kept as a pet. The animal was subsequently transferred to the Limbe Wildlife Center (LWC) (http://limbewildlifecentre.wildlifedirect.org/). During the quarantine period, venous blood was drawn for the routine veterinary checkup, which included HIV/SIV serology performed at the Centre Pasteur du Cameroun in Yaoundé as described earlier and the plasma of the animal showed cross-reactive antibodies with HIV antigens using commercial HIV screening assays (Ahuka-Mundeke et al., 2011). When the animal 1405 was transferred to the LWC, 5 other tantalus monkeys were already present and arrived at the zoo between January 1997 and August, 2001. They all tested SIV negative during the quarantine period and thence over. The SIV+ status was further confirmed with a previously described highly sensitive in-house Luminex Multiple Analyte Profiling (xMAP) assay using peptides representing the major HIV and SIV lineages, which has been shown to detect even genetically divergent SIVs (Ahuka-Mundeke et al., 2011). The animal 1405 had cross-reactive antibodies to SIVagm, SIVsmm, SIVrcm, SIVdeb, and SIVtal gp41 peptides from African green monkeys, sooty mangabey monkeys, red-capped mangabeys, De Brazza's monkeys and talapoins, respectively.
2.2. Species confirmation
We confirmed the monkey species by amplifying a 1200 bp fragment in mitochondrial DNA (mtDNA) coding for cytochromeb, as described previously (Haus et al., 2013a). Briefly, we used CytbAFnew (5′-CCACYGTTGTACTTCAACTAC-3′) and CytbBrev (5′-TAGTTTACAAGACTAGTGTATTAG-3′) to amplify the 1200 bp fragment from 5 μl of DNA. PCR reactions were performed with the Taq DNA polymerase Core kit (MP Biomedicals, Irvine, CA, USA), following manufacturers’ instructions. Thermocycling conditions using touch-down PCR strategy, were as follow: 10 cycles of denaturation at 92 °C for 20 s, annealing at 55 °C for 30 s (−0.5 °C/cycle) and elongation at 72 °C for 90 s; followed by 30 cycles of denaturation at 92 °C for 20 s, annealing at 55 °C for 30 s and elongation at 72 °C for 90 s, with a final elongation step at 72 °C for 5 min before a cool-down to 10 °C, in a thermal cycler. In addition, we included seven other monkeys from Cameroon, visually identified as AGMs, in the species’ molecular confirmation. These seven samples were collected from carcasses of animal sold as bushmeat in markets of Yaoundé (n = 5) or kept as pets (n = 2) in the framework of a study on SIV screening and characterization as reported previously (Aghokeng et al., 2010; Peeters et al., 2002). None of these seven samples was found SIV positive. The newly obtained AGM cytochrome b sequences from AGM from Cameroon were aligned with previously reported AGM sequences from the different Chlorocebus species from a wide geographic range published by Haus et al. (2013a) and Wertheim and Worobey (2007). Phylogenies were inferred by both Maximum likelihood (ML) and Bayesian methods using PhyML (Guindon and Gascuel, 2003) and MrBayes3.2 (Ronquist and Huelsenbeck, 2003) softwares, respectively.
2.3. Molecular characterization of the new SIVagm strain
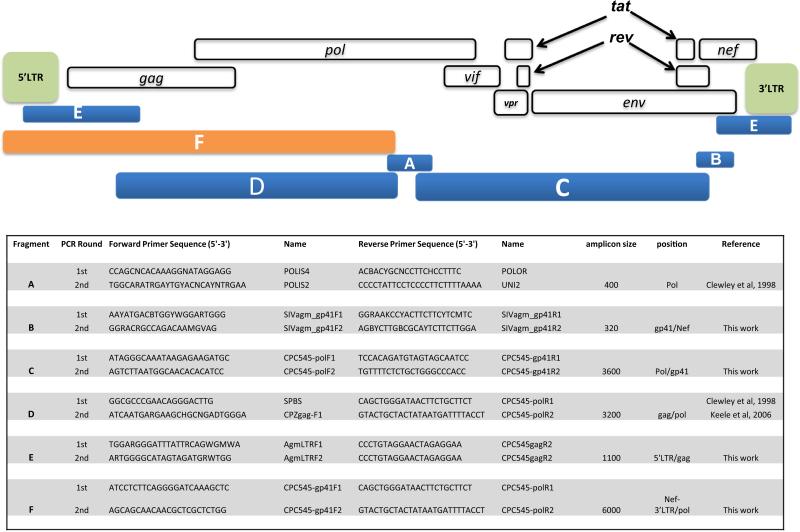
For molecular detection of SIV, proviral DNA was PCR-amplified from peripheral blood mononuclear cells using universal HIV/SIV primers (Clewley et al., 1998; Keele et al., 2006) targeting a 400 bp fragment in the integrase region of the pol gene. To study more in detail SIVagmTAN-CM545 viral isolate, we characterized the full genome of its proviral DNA by amplifying and sequencing overlapping PCR fragments. Primers used to obtain full-length SIVagm sequences are provided, together with the genomic architecture of a prototypic SIVagm in Fig. 2. Amplification and sequencing methods were done as previously described (Aghokeng et al., 2010; Liegeois et al., 2009; Ahuka-Mundeke et al., 2010). PCR amplifications were performed using the Taq DNA polymerase Core kit (MP Biomedicals, Irvine, CA, USA), following manufacturers’ instructions. Each amplification reaction included a manual hot-start at 94 °C for 3 min, followed by 35–45 cycles with a denaturation step at 92 °C for 20 s, an annealing temperature set according to the primers melting temperatures and a variable extension time depending on the size of the expected fragment (1 min/kb). Positive PCR products were purified on agarose gel (1%) and directly sequenced using cycle sequencing and dye terminator methodologies (ABI PRISM Big Dye Terminator Cycle sequencing Ready Reaction kit with amplitaq FS DNA polymerase) on an automated sequencer (ABI 3130XL, applied Biosystems, Courtaboeuf, France). The full-length genome sequence was reconstructed by assembling overlapping sequences into contiguous ones using SeqMan Pro, version 11.2.1 software (DNASTAR, Madison, WI).
Fig. 2.
Primers and strategy used to PCR-amplify sub-genomic fragments of SIVagmTAN-CM545. The figure shows, on the top, the genome architecture of a prototypic SIVagm with its different genes indicated. Beneath this scheme are represented the different sub-genomic fragments amplified and whose concatenation gave the full-length genome of SIVagmTAN-CM545. In the lower part, the primers used as well as the target region and the amplicon size are given.
2.4. Phylogenetic analysis of the new SIVagmTAN-CM545
Nucleotide and protein sequences were aligned using the online version of MAFFT (http://mafft.cbrc.jp/alignment/server/), with minor manual adjustments. Sites that could not be unambiguously aligned were excluded. The predicted protein sequences encoded by SIVagmTAN-CM545 were compared to other SIVagm strains and representatives of known HIV/SIV lineages. Evolutionary trees from Gag, Pol, and Env amino acid sequences were constructed to estimate the phylogeny of the new SIVagmTAN-CM545 strain in relation to other primate lentiviruses in these three main genome domains. Phylogenies were inferred by both Maximum likelihood (ML) and Bayesian methods using PhyML (Guindon and Gascuel, 2003) and MrBayes3.2 (Ronquist and Huelsenbeck, 2003) softwares, respectively. Detection of recombination events was performed with SimPlot version 3.5.1 (Lole et al., 1999) and RDP (Martin et al., 2010) 4.35 softwares.
3. Results
3.1. Confirmation of SIV infection and host species
To confirm the SIV positive status of agm545, we PCR-amplified and sequenced a 400-bp DNA fragment in the pol region. Maximum likelihood phylogenetic analysis using this 400-bp DNA fragment showed that the new SIVagmTAN-CM545 clustered with representatives of the SIVagmSAB lineage from C. sabaeus and not with previously documented SIVagmTAN strains from C. tantalus as would have been expected based on the generally assumed virus–host co-evolution of SIV in African green monkeys (not shown).
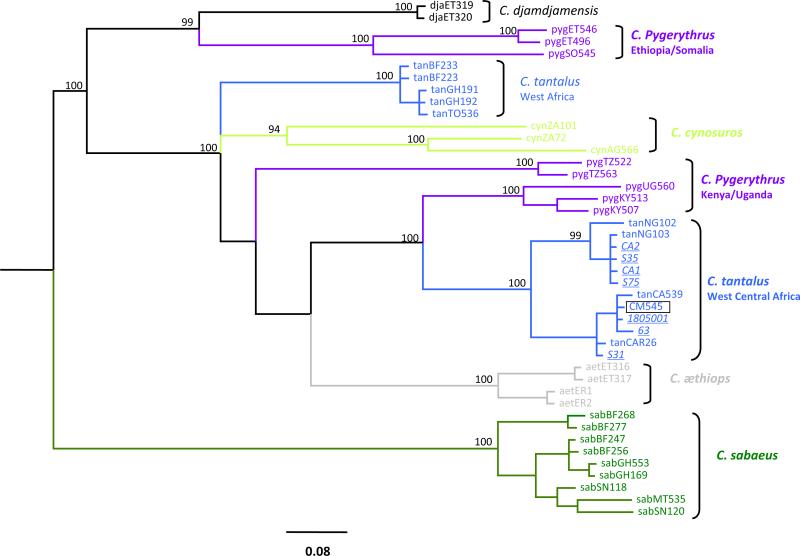
We then verified the host species through molecular phylogeny of mtDNA cytochrome-b fragment, together with the 7 other visually identified African green monkeys from Cameroon. Bayesian inference phylogeny derived from these mtDNA sequences showed that the 8 African green monkey samples from Cameroon clustered with other tantalus monkeys from Nigeria and the Central African Republic (Fig. 3).
Fig. 3.
Phylogenetic relationship of the 8 African green monkeys (AGM) from Cameroon to other AGM cytochrome b sequences using Bayesian inference. We used a 1200 bp mitochondrial cytochrome-b nucleotides alignment for Bayesian inference of phylogeny with MrBayes3.2 (Ronquist and Huelsenbeck, 2003) by sampling 10,000 trees with a Markov Chain Monte Carlo under GTR + G + I with 6 rates in 60,000,000 generations. Posterior probabilities ≥94% are shown. The scale bars indicate the number of nucleotide substitutions per site. Bayesian consensus trees were displayed with FigTree1.4 (http://tree.bio.ed.ac.uk/software/figtree/). The same color codes were used for AGM distribution range and on the phylogenetic tree. The 8 novel sequences are in italics and underlined. agmTAN-CM545 is boxed. All the 8 samples are Chlorocebus tantalus, based on this phylogeny. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Full-length sequence analysis of the new SIVagmTAN-CM545
We next sequenced and characterized the full-length genome of SIVagmTAN-CM545, which is 9923 bp long, and contained all the usual genes and organization of SIVagm genomes, with the absence of vpx and vpu genes. Similarly to all currently known primates lentiviruses, The Gag precursor possesses a p6 domain with conserved motifs important for viral budding and dissemination. The L-X-X-L-F motif near the C-terminus of p6gag, absolutely conserved, is present in the p6gag of SIVagmTAN-CM545 as L-R-S-L-F. The P-T/S-A-P motif, highly conserved in primates’ lentiviruses was also present in the p6gag of the novel SIVagm isolate as PSAP. Finally, the dileucine motif D-X-A-X-X-L-L found to govern viral particle association of vpx and vpr of SIVmac and SIVagm, respectively (Accola et al., 1999), was also present in the novel SIVagmTAN-CM545 as D-P-A-T-A-L-L. Finally, in Env, the 18 highly conserved cysteine residues are present in SIVagmTAN-CM545. The V3 loop is 34 amino acids long, with the crown consisting of M-A-G triplet, as for the other SIVagm known so far.
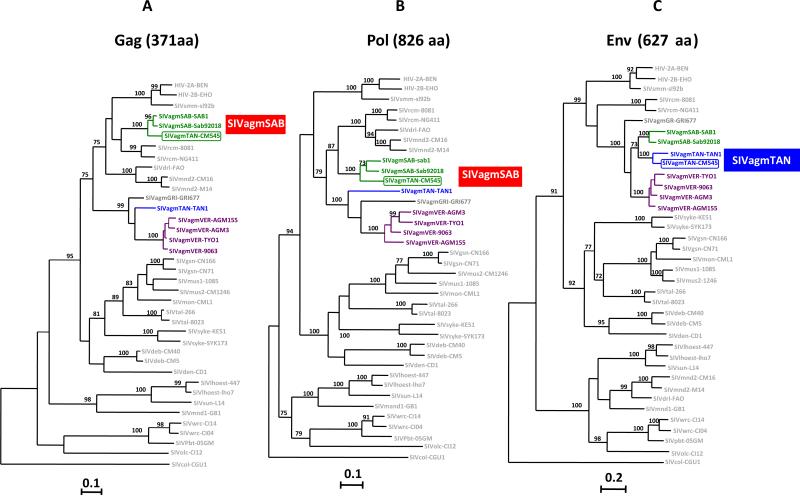
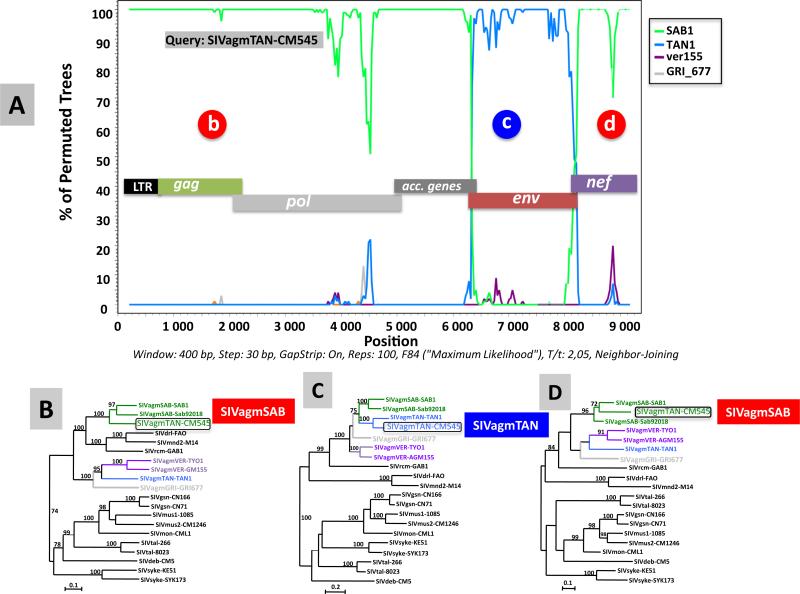
ML phylogenies of Gag, Pol and Env deduced amino-acids sequences (Fig. 4, panels A through C, respectively), revealed an usual structure of SIVagmTAN-CM545. Indeed, in Gag and Pol proteins, this novel isolate clustered with the prototypic SIVagmSAB strains, SAB1 and Sab92018 from 2 green monkeys (C. sabaeus) from Senegal, while in Env, SIVagmTAN-CM545 clustered with SIVagmTAN1, identified in a tantalus monkey (C. tantalus) from Uganda. To analyze more in detail the mosaic structure of SIVagm-TAN-CM545, we performed a bootscan analysis on the full-length genome sequence using the software SimPlot (Lole et al., 1999). Results from this analysis, shown in Fig. 5, confirmed the mosaic genomic structure of SIVagmTAN-CM545 (Fig. 5, panel A), with portions closely related to SIVagmSAB in gag, pol, accessory genes (Fig. 5 panel B) and nef (panel D) and a portion closely related to SIVagmTAN in env. (panel C). However, at the 3′ end of GP41 where env and nef genes overlap (#300 bp), SIVagmTNACM545 clusters with SIVagmSAB. These results obtained by bootscanning using SimPlot software were confirmed by using RDP software, version 4.35 (Martin et al., 2010) (not shown). The recombinant nature of this novel SIVagm isolate was further confirmed through nucleotide similarity calculation between SIVagmTAN-CM545 and previously characterized SIVagm in the 5 main viral genes (Table 1). In gag, pol, vif and nef, SIVagmTAN-CM545 is more closely related to SIVagmSAB from C. sabaeus with similarities higher that 70% in gag, pol and vif and around 60% in nef. In env, SIVagmTAN-CM545 is more closely related to C. tantalus with similarity above 70%.
Fig. 4.
Phylogenetic relationships of the new SIVagm strain (SIVagmTAN-CM545) to previously characterized SIVs and HIV-2 strains in Gag (371 amino acids) (A), Pol (826 amino acids) (B) and Env (627 amino acids) (C) genes. The trees show SIVagm and other SIV/HIV-2 phylogeny based on maximum likelihood (ML) methods using PHYML (Guindon and Gascuel, 2003). SIVagmTAN-CM545 is highlighted in green or blue and boxed, other SIVagm are color-coded as on Fig. 1. The remaining SIV/HIV-2 are in light gray. Numbers on branches represent bootstrap values >70% from 1000 pseudo-replicates. Scale bars are in substitutions per site. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Recombination analysis on the full-length genome sequence of the new SIVagmTAN-CM545 strain. Bootscan analysis of nucleotide sequences of the new SIVagmTAN-CM545 strain versus a selection of SIVagm strains (A). The y-axis indicates the % of permutated trees (bootstrap support) between the query sequence (SIVagmTAN-CM545) and the other sequences. Bootscan analysis was performed on the nucleotide alignment using the SIMPLOT package version 3.5.1 (Lole et al., 1999) using a sliding window of 400 nucleotides (nt) moved in steps of 30 nt. Regions where the new SIVagmTAN-CM545 strain is more closely related to a particular SIV strain are highlighted with letters (b, c, d) and the corresponding phylogenetic tree analysis are shown (panels B, C, D). The same code of colors for AGM distribution range was used on the phylogenetic tree. SIVagmTAN-CM545 is boxed. The trees were inferred by maximum likelihood (ML) methods using PHYML (Guindon and Gascuel, 2003). Appropriate nucleotide substitution models were selected for each data set using MEGA5. Numbers on branches represent bootstrap values (1000 pseudo-replicates) equal and above 70%. Scale bars are in substitutions per site. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Nucleotides similarities between the novel SIVagmTAN-CM545 and previously characterized SIVagm in the 5 main genes.
| % Nucleotide identity of different SIVagm with SIVagmTAN-CM545 in: |
|||||
|---|---|---|---|---|---|
| gag | pol | vif | env | nef | |
| SIVagmTAN-CM545 | 100 | 100 | 100 | 100 | 100 |
| SIVagmTAN-TANl | 64 | 67 | 60 | 76 | 56 |
| SIVagmGRI-GRI677 | 67 | 66 | 62 | 67 | 55 |
| SIVagmSAB-SABl | 78 | 78 | 77 | 72 | 63 |
| SIVagmSAB-Sab92018 | 79 | 78 | 74 | 72 | 62 |
| SIVagmVER-AGM3 | 67 | 66 | 56 | 67 | 56 |
| SIVagmVER-TYOl | 65 | 66 | 59 | 67 | 54 |
| SIVagmVER-9063 | 66 | 66 | 59 | 66 | 53 |
| SIVagmVER-AGM155 | 65 | 66 | 60 | 68 | 55 |
Shaded area represent positions with the similarity with novel SIVagmTAN-CM545 isolate.
3.3. Genetic diversity of the new SIVagmTAN sequence compared to other partial SIVagm sequences from different geographic areas
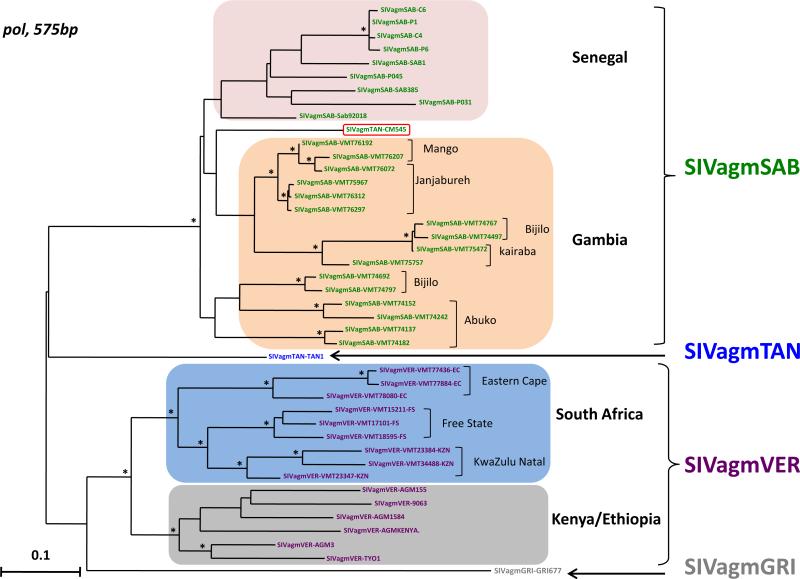
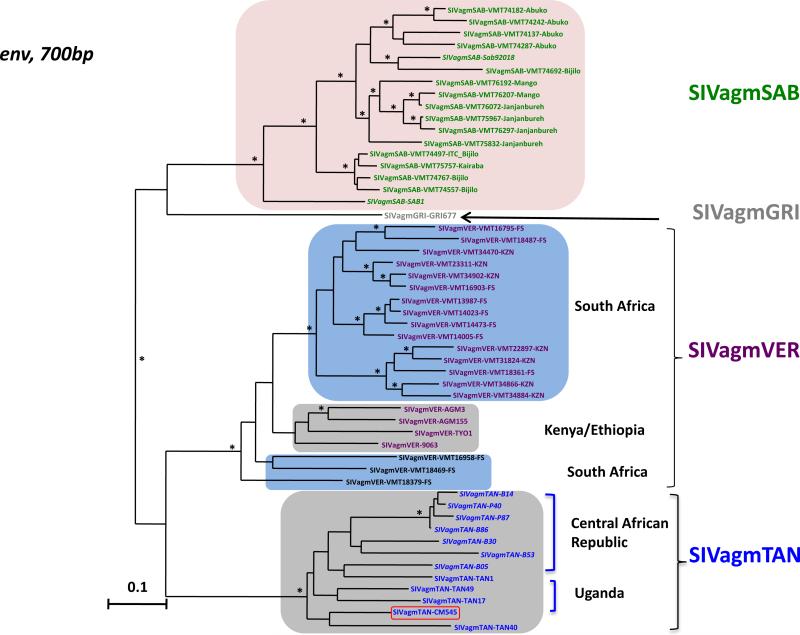
Recent studies on wild vervet monkeys (C. pygerythrus) showed a high genetic diversity among SIVagmVER strains and showed phylogeographic clusters of SIVagmVER among east and southern African strains, but also among south African SIVagmVER strains populations (Ma et al., 2013). Less markedly, a clustering by geographic origin was also observed between green monkeys (C. sabaeus) from Senegal and the Gambia infected by SIVagmSAB (Ma et al., 2014). Therefore we compared the new SIVagmTAN-CM545 sequence with previously reported SIVagmSAB and SIVagmTAN strains in pol and env partial fragments. Results from these analyses confirmed the phylogeographic clustering of SIVagm sequences, especially in the integrase region of the polymerase gene (Fig. 6). In this phylogeny, the novel SIVagmTAN-CM545 is nested between SIVagmSAB sequences from the Gambia and Senegal, basal to the former (Fig. 6). In the envelope gene (Fig. 7), SIVagmTAN-CM545 clustered with other SIVagmTAN sequences from Uganda and the Central African Republic, without marked phylogeography.
Fig. 6.
Phylogenetic relationships of the new SIVagm strain (SIVagmTAN-CM545) to previously characterized SIVagm strains in the integrase region of pol gene. A 575 bp alignment in the integrase region of the polymerase gene of SIVagmTAN-CM545 together with other SIVagm was phylogenetically analyzed with maximum likelihood (ML) methods using PHYML (Guindon and Gascuel, 2003). Highlighted group of sequences indicate geographic clusters of different SIVagm strains. SIVagmTAN-CM545 is boxed in red, between the clusters of SIVagmSAB from Senegal and Gambia. SIVagm are color-coded as on Fig. 1. Asterisks on branches represent bootstrap values >70% from 1000 pseudo-replicates. Scale bars are in substitutions per site. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Phylogenetic relationships of the new SIVagm strain (SIVagmTAN-CM545) to previously characterized SIVagm strains in the env gene. A 700 bp alignment of the env gene of SIVagmTAN-CM545 together with other SIVagm was phylogenetically analyzed with maximum likelihood (ML) methods using PHYML (Guindon and Gascuel, 2003). Highlighted group of sequences indicate geographic clusters of different SIVagm strains. SIVagmTAN-CM545 is boxed in red intermixed with SIVagmTAN from Uganda and the Central African Republic. SIVagm are color-coded as on Fig. 1. SIVagmSAB from Senegal are in italics font in the cluster of SIVagmSAB from the Gambia. Asterisks on branches represent bootstrap values >70% from 1000 pseudo-replicates. Scale bars are in substitutions per site. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Our present findings show that the genetic diversity among the different SIVagm lineages is more complex than initially observed. In a large part of the genome, SIVagmTAN-CM545 is more closely related to viruses infecting C. sabaeus monkeys from west Africa than the ones infecting C. tantalus monkeys from Central Africa. This new SIVagm sequence challenges thus the initial observations on co-evolution between SIV and the host in the different African green monkey species.
The major question is how this new recombinant SIVagmTAN virus originated and how widespread this variant is among the tantalus monkeys. This is the first SIVagmTAN complete sequence obtained from African green monkeys in the western part of the tantalus range and it is thus possible that the initial SIVagmTAN strain from a Ugandan tantalus monkey is not representative for the entire SIVagmTAN diversity. For the other SIVagmTAN strains obtained from Uganda, only partial env and gag sequences are available and they form in each genomic region a species-specific lineage. However, for the tantalus monkeys from the Central African Republic only partial env sequences are available and it cannot be excluded that they could have the same recombinant structure as the new strain from the Cameroonian monkey.
Recombination can only occur when simultaneous infection with SIVagmSAB and SIVagmTAN occurs in the same host, as a result of co- or superinfection. Hybridization among the different Chlorocebus species has been extensively reported at borders of the geographic ranges of the different species (Groves, 2001; Haus et al., 2013b), and could thus be at the origin of the emergence of this recombinant SIVagmSAB/TAN strain which became subsequently established in certain AGM populations. It is also interesting to note that SIVagmSAB infecting C. sabaeus monkeys has a mosaic structure (Jin et al., 1994) with Gag and Pol proteins more closely related to HIV-2/SIVsmm than to SIVagmGRI and SIVagmVER, while Env and Nef proteins, clustered with other SIVagm lineages illustrating that cross-species transmission and recombination played a role in SIV evolution in the Chlorocebus species. Finally, the new SIVagmTAN strain was characterized in a naturally infected but captive tantalus monkey. However, all molecular studies on SIV were done on the sample collected during the quarantine period and co-infection or superinfection with SIV from other SIV infected NHP in the sanctuary is thus excluded. Moreover, the range of Sabaeus monkeys ends in west Africa at the Volta River and is thus is unlikely that the animal encountered a SIVagmSAB infected monkey during his captivity in the village in Cameroon.
Our current findings are to some extent inline with the possibility also raised by Wertheim and Worobey (2007) on the dynamics of SIVagm and the evolution of AGMs. These authors postulated for a migration from west towards the east of Africa, with C. sabaeus being infected first by SIVagmSAB. Following this reasoning, the hypothesis of ancient AGMs and SIVagm co-divergence is seriously weakened. On the other hand, a more recent study on the genetic diversity of SIVagmVER in vervet monkeys from South-Africa revealed that geographically distinct vervet populations, physically separated by the Drakensberg Mountains, carry different viral variants. The old age of these mountains, and the genetic distance between the SIVagmVER strains of the vervet living in this isolated area suggests that SIVagmVER was present in the ancestral vervet populations before this physical barrier has effectively separated them from each other. These results reinforce the initial theory on a very old origin of SIVagm, and molecular clock analyses of the SIVagmVER env and pol gene sequences from South African vervets estimate the most recent common ancestor of South African SIVagm strains several hundred thousand to several million years ago (Jasinska et al., 2013; Ma et al., 2013).
In conclusion, all these observations together with the identification of this recombinant SIVagm strain, illustrate that our knowledge on SIVs in African green monkeys is still not resolved. Whether this novel SIVagmTAN-CM545 isolate is currently circulating in AGMs in Cameroon and other regions of the tantalus range or is a relic of historical times, needs further investigation and suggests that evolution of SIVagm is more complex than previously thought. In order to better understand SIVagm evolution in the different African green monkey species, a more comprehensive screening for SIV infection in a wider geographic range of each Chlorocebus species is necessary. It is also possible that the AGM phylogeny and species classification is more complex when molecular tools are used as suggested by recent studies. Therefore, future studies should simultaneously characterize SIVagm strains and host species. The fact that there is congruence between SIVagm and AGM phylogenies suggests also that SIVagm phylogenies based on partial genome sequences might be misleading.
Acknowledgments
This work was supported in part by grants from the Agence Nationale de Recherches sur le SIDA, France (ANRS 12255), from the National Institutes of Health (RO1 AI 50529), the Institut de Recherche pour le Développement (IRD), France and the Centre Pasteur du Cameroun, Yaoundé, Cameroon. We thank the Cameroonian Ministries of Health, Research, and Environment and Forestry and Wildlife, for the permission to perform this study.
Footnotes
Nucleotides accession numbers
Nucleotide sequences reported in the present work have been deposited at the European Nucleotide Archive (ENA) under the accession numbers LM999936 to LM999948.
References
- Accola MA, Bukovsky AA, Jones MS, Gottlinger HG. A conserved dileucine-containing motif in p6(gag) governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIV(mac) and SIV(agm). J. Virol. 1999;73:9992–9999. doi: 10.1128/jvi.73.12.9992-9999.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghokeng AF, Ayouba A, Ahuka S, Liegoies F, Mbala P, Muyembe JJ, Mpoudi-Ngole E, Delaporte E, Peeters M. Genetic diversity of simian lentivirus in wild De Brazza's monkeys (Cercopithecus neglectus) in Equatorial Africa. J. Gen. Virol. 2010;91:1810–1816. doi: 10.1099/vir.0.021048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuka-Mundeke S, Liegeois F, Ayouba A, Foupouapouognini Y, Nerrienet E, Delaporte E, Peeters M. Full-length genome sequence of a simian immunodeficiency virus (SIV) infecting a captive agile mangabey (Cercocebus agilis) is closely related to SIVrcm infecting wild red-capped mangabeys (Cercocebus torquatus) in Cameroon. J. Gen. Virol. 2010;91:2959–2964. doi: 10.1099/vir.0.025767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuka-Mundeke S, Ayouba A, Mbala-Kingebeni P, Liegeois F, Esteban A, Lunguya-Metila O, Demba D, Bilulu G, Mbenzo-Abokome V, Inogwabini BI, Muyembe-Tamfum JJ, Delaporte E, Peeters M. Novel multiplexed HIV/simian immunodeficiency virus antibody detection assay. Emerg. Infect. Dis. 2011;17:2277–2286. doi: 10.3201/eid1712.110783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan JS, Short M, Taylor ME, Su S, Hirsch VM, Johnson PR, Shaw GM, Hahn BH. Species-specific diversity among simian immunodeficiency viruses from African-green monkeys. J. Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewley JP, Lewis JC, Brown DW, Gadsby EL. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J. Virol. 1998;72:10305–10309. doi: 10.1128/jvi.72.12.10305-10309.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves C. Primate Taxonomy. Smithsonian Institution Press; Washington and London: 2001. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haus T, Akom E, Agwanda B, Hofreiter M, Roos C, Zinner D. Mitochondrial diversity and distribution of African green monkeys (Chlorocebus Gray, 1870). Am. J. Primatol. 2013a;75:350–360. doi: 10.1002/ajp.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haus T, Roos C, Zinner D. Discordance between spatial distributions of Y-chromosomal and mitochondrial haplotypes in African green monkeys (spp.): a result of introgressive hybridization or cryptic diversity? Int. J. Primatol. 2013b;34:986–999. doi: 10.1007/s10764-013-9717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry RM, Wells MA, Phelan MA, Schneider AL, Epstein JS, Quinnan GV. Antibodies to simian immunodeficiency virus in African green monkeys in Africa in 1957–62. Lancet. 1986;2:455. doi: 10.1016/s0140-6736(86)92156-2. [DOI] [PubMed] [Google Scholar]
- Hirsch V, Riedel N, Kornfeld H, Kanki PJ, Essex M, Mullins JI. Cross-reactivity to human T-lymphotropic virus type-III/lymphadenopathy-associated virus and molecular-cloning of simian T-cell lymphotropic virus type-III from African-green monkeys. Proc. Natl. Acad. Sci. U.S.A. 1986;83:9754–9758. doi: 10.1073/pnas.83.24.9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, Mcgann C, Dapolito G, Goldstein S, Ogenodoi A, Biryawaho B, Lakwo T, Johnson PR. Identification of a new subgroup of SIVagm in tantalus monkeys. Virology. 1993;197:426–430. doi: 10.1006/viro.1993.1606. [DOI] [PubMed] [Google Scholar]
- Jasinska AJ, Schmitt CA, Service SK, Cantor RM, Dewar K, Jentsch JD, Kaplan JR, Turner TR, Warren WC, Weinstock GM, Woods RP, Freimer NB. Systems biology of the vervet monkey. ILAR J. 2013;54:122–143. doi: 10.1093/ilar/ilt049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MJ, Hui HX, Robertson DL, Muller MC, Barresinoussi F, Hirsch VM, Allan JS, Shaw GM, Sharp PM, Hahn BH. Mosaic genome structure of simian immunodeficiency virus from West-African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. Chimpanzee reservoirs of pandemic and non-pandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegeois F, Lafay B, Formenty P, Locatelli S, Courgnaud V, Delaporte E, Peeters M. Full-length genome characterization of a novel simian immunodeficiency virus lineage (SIVolc) from olive colobus (Procolobus verus) and new SIVwrcPbb strains from Western red colobus (Piliocolobus badius badius) from the tai forest in Ivory Coast. J. Virol. 2009;83:428–439. doi: 10.1128/JVI.01725-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegeois F, Schmidt F, Boue V, Butel C, Mouacha F, Ngari P, Ondo BM, Leroy E, Heeney JL, Delaporte E, Peeters M, Rouet F. Full-length genome analyses of two new simian immunodeficiency virus (SIV) strains from mustached monkeys (C. Cephus) in Gabon illustrate a complex evolutionary history among the SIVmus/mon/gsn lineage. Viruses. 2014;6:2880–2898. doi: 10.3390/v6072880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli S, Peeters M. Cross-species transmission of simian retroviruses: how and why they could lead to the emergence of new diseases in the human population. AIDS. 2012;26:659–673. doi: 10.1097/QAD.0b013e328350fb68. [DOI] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstine LJ, Pedersen NC, Higgins J, Pallis KC, Uyeda A, Marx P, Lerche NW, Munn RJ, Gardner MB. Seroepidemiologic survey of captive old-world primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from sooty mangabeys (Cercocebus atys). Int. J. Cancer. 1986;38:563–574. doi: 10.1002/ijc.2910380417. [DOI] [PubMed] [Google Scholar]
- Ma D, Jasinska A, Kristoff J, Grobler JP, Turner T, Jung Y, Schmitt C, Raehtz K, Feyertag F, Martinez Sosa N, Wijewardana V, Burke DS, Robertson DL, Tracy R, Pandrea I, Freimer N, Apetrei C. SIVagm infection in wild African green monkeys from South Africa: epidemiology, natural history, and evolutionary considerations. PLoS Pathog. 2013;9:e1003011. doi: 10.1371/journal.ppat.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Jasinska AJ, Feyertag F, Wijewardana V, Kristoff J, He T, Raehtz K, Schmitt CA, Jung Y, Cramer JD, Dione M, Antonio M, Tracy R, Turner T, Robertson DL, Pandrea I, Freimer N, Apetrei C. Factors associated with simian immunodeficiency virus transmission in a natural African nonhuman primate host in the wild. J. Virol. 2014;88:5687–5705. doi: 10.1128/JVI.03606-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics (Oxford, England) 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Masuda T, Tsujimoto H, Ishikawa K, Kodama T, Morikawa S, Nakai M, Honjo S, Hayami M. Isolation of simian immunodeficiency virus from African-green monkeys and seroepidemiologic survey of the virus in various non-human primates. Int. J. Cancer. 1988;41:115–122. doi: 10.1002/ijc.2910410121. [DOI] [PubMed] [Google Scholar]
- Peeters M, Courgnaud V, Abela B, Auzel P, Pourrut X, Bibollet-Ruche F, Loul S, Liegeois F, Butel C, Koulagna D, Mpoudi-Ngole E, Shaw GM, Hahn BH, Delaporte E. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 2002;8:451–457. doi: 10.3201/eid0805.01-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin. Microbiol. Rev. 2006;19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Worobey M. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathog. 2007;3:866–873. doi: 10.1371/journal.ppat.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]