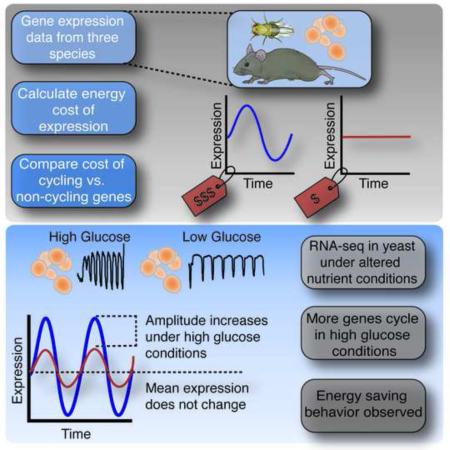
SUMMARY
Genes expressing circadian RNA rhythms are enriched for metabolic pathways, however, the adaptive significance of cyclic gene expression remains unclear. We estimated the genome-wide synthetic and degradative cost of transcription and translation in three organisms and found that the cost of cycling genes is strikingly higher compared to non-cycling genes. Cycling genes are expressed at high levels and constitute the most costly proteins to synthesize in the genome. We demonstrate that metabolic cycling is accelerated in yeast grown under higher nutrient flux and the number of cycling genes increases ~40% - achieved by increasing the amplitude and not the mean level of gene expression. These results suggest that rhythmic gene expression optimizes the metabolic cost of global gene expression and that highly expressed genes have been selected to be down-regulated in a cyclic manner for energy conservation.
INTRODUCTION
Circadian rhythms are an evolutionary adaptation of living systems to coordinate behavioral, physiologic and metabolic functions to the 24-hour cyclic environment (Bass and Takahashi, 2010; Dibner et al., 2010; Green et al., 2008; Mohawk et al., 2012). They are widely observed across members of prokaryotes and multiple eukaryotic kingdoms, including cyanobacteria, fungi, insects, mice, and humans (Bell-Pedersen et al., 2005; Dunlap, 1999). Significant advances have been made in the identification of the molecular mechanisms and genes driving these rhythms (Lowrey and Takahashi, 2011; Partch et al., 2014; Zhang and Kay, 2010). In eukaryotes, circadian rhythms are generated by cell-autonomous transcriptional feedback loops composed of positive transcriptional activators that drive the expression of negative feedback elements that repress their own transcription (Dunlap, 1999; Lowrey and Takahashi, 2004).
While the core circadian regulatory pathway includes genes such as Clock, Bmal1, Cry1/Cry2 and Per1/Per2 (Bass and Takahashi, 2010; Lowrey and Takahashi, 2011), thousands of transcripts have recently been identified as exhibiting circadian or cycling expression profiles using genome-wide approaches (Koike et al., 2012; Menet et al., 2012; Rey et al., 2011; Vollmers et al., 2012; Zhang et al., 2014). For example, about 800 transcripts have been detected during normal diurnal conditions and about 1200 transcripts have been detected during continual darkness in the brains of the wild type fruit fly, Drosophila (Hughes et al., 2012). In mouse liver, over 1,300 cycling pre-mRNA transcripts and 2,000 mRNA transcripts have been detected during 48 hours of continuous darkness (Koike et al., 2012). Additionally, more than half of the genes (~3500) in the yeast genome have been observed as showing periodic expression during metabolic cycling (Tu et al., 2005).
It has been hypothesized that circadian rhythm/periodic genes are closely related to metabolic pathways of the cell (Green et al., 2008; Rutter et al., 2002). Recently, ChIP-seq data suggest that genes that are enriched in metabolic pathways are preferentially bound by the mouse core transcriptional factors, including BMAL1, CLOCK, CRY1, CRY2, PER1, and PER2 (Koike et al., 2012; Menet et al., 2012; Rey et al., 2011; Vollmers et al., 2012). Moreover, genes that are involved in biosynthetic pathways also tend to be regulated in a periodic fashion, including glycolysis and gluconeogenesis pathways (Green et al., 2008). Thus, there are a number of essential cellular features that are driven by periodic gene expression; however, the underlying basis for whether a particular gene cycles or not is unclear.
Here we assess the role of energy needed to synthesize and degrade mRNAs and proteins in three species (yeast, Drosophila and mouse) and find that the expression of cycling genes costs as much as two times higher than other genes. We further show that the cycling expression of these expensive genes likely plays an important evolutionary function. For example, in genome-wide simulation experiments, we find that the periodic expression of empirically observed cycling gene sets leads to the least amount of energy consumed. Importantly, in yeast we find that increasing nutrient flux leads to an increase in the number and amplitude of cycling genes. Because the amplitude increase of cycling genes was achieved without an overall increase in the average expression level, these results reveal a previously unappreciated and efficient mode for increasing peak gene expression levels without an overall increase in energy expenditure. Thus, these results demonstrate that cyclic gene expression is an efficient strategy for optimizing metabolic cost.
RESULTS
Cycling Genes Are More Expensive Than Other Genes in Mouse
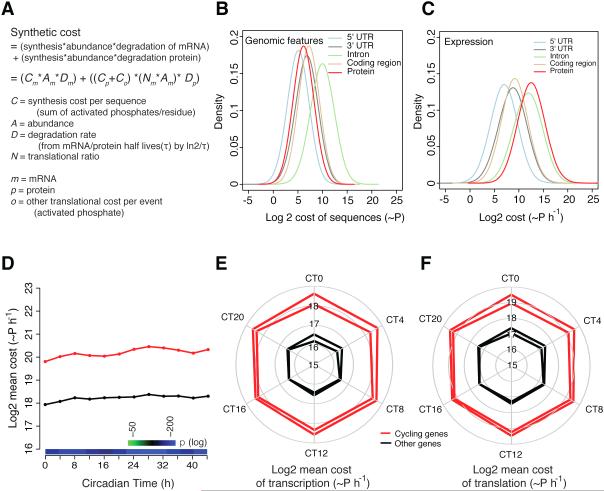
To identify potential mechanisms driving the expression of genes to be expressed in a cyclic manner, we evaluated the cost during mRNA and protein synthesis and degradation of whole transcriptome data from mouse liver (Koike et al., 2012). The synthetic cost of each mRNA and protein was calculated first based on the synthetic cost of each nucleotide or amino acid, which is determined from the number of activated phosphate bonds (~P) required for synthesizing each precursor (Wagner, 2005). Then the mRNA and protein cost per unit time were calculated by taking into account genome-wide mRNA abundance, protein abundance, mRNA and protein degradation rates and other costs such as amino acid charging of tRNA, translation initiation, translocation of ribosomes along the mRNA during elongation and termination (Wagner, 2005) (Figure 1A). This total “cost” for each gene, gene feature and protein sequence that takes into account all of the synthetic and degradative parameters listed above was then calculated for each circadian time point (Wagner, 2005). Similar to that described previously in yeast (Wagner, 2005), we find that translation rather than transcription of genes requires the greatest cost in mouse liver (Figures 1B-C).
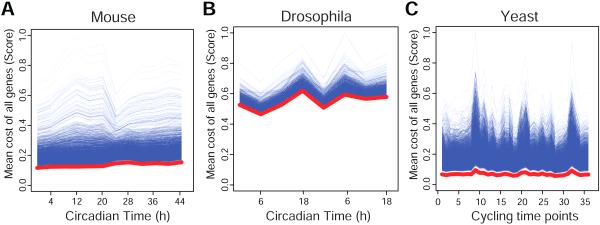
Figure 1. Cycling genes have an increased total cost.
(A) Schematic formula demonstrating that the cost of transcription and translation contribute to the overall cost of a gene. (B) The distribution of the cost for each genomic feature. (C) The distribution of the cost for each genomic feature after incorporation of gene expression data (only one time point from the transcriptomic experiment was plotted, however, all other time points are similar). Protein synthesis costs more energy than gene synthesis after taking into account the expression data. (D) Cycling genes have approximately a four times higher total mean cost than other expressed genes. The cost of cycling is increased at both (E) the transcriptional and (F) translational level (p < 2.2E-16 for all comparisons). Red line, cycling genes; black line, other genes. Two lines for red and black represent the two circadian cycles analyzed. In (D), significance levels are shown in the heatmap. See also Figure S1.
We next asked whether circadian RNA cycling genes in the mouse liver (2037 exon RNA cycling genes) require more energy for synthesis than other expressed genes in the liver (12,680 expressed genes minus 2037 cycling genes based on RNA-seq experiments) (Koike et al., 2012). We first examined the cost for a single mRNA and protein generation independently. The cost of protein generation of cycling genes has a small but significant decrease compared to other genes (0.41%; p = 9.00E-03) and there is no difference in the cost of mRNA generation between cycling genes and non-cycling genes (p = 3.53E-01). However, since the range of gene expression can vary by several orders of magnitude and contribute to energy costs, calculating the cost of mRNA sequences alone is not sufficient for estimating the total cost of genes. Thus, we calculated the total cost of expression using the magnitude of mRNA levels for both cycling and non-cycling genes. Strikingly, at each time point we examined, the total cost of cycling genes is ~4 times higher than other genes (Figure 1D; and see Table S1 for detailed information). Analysis using 1371 intron RNA cycling genes from the liver (Koike et al., 2012) also showed that cycling genes were more costly than non-cycling genes.
We next wanted to understand whether this increase in the total cost of cycling genes was being driven by either transcriptional or translational cost. We observed a ~4 fold increase in the cost of cycling genes at both the transcriptional and translational levels compared to the non-cycling genes (Figures 1E-F, Table S1). Thus, the increased cost of cycling genes is derived from an increase in cost at both the transcriptional and translational levels.
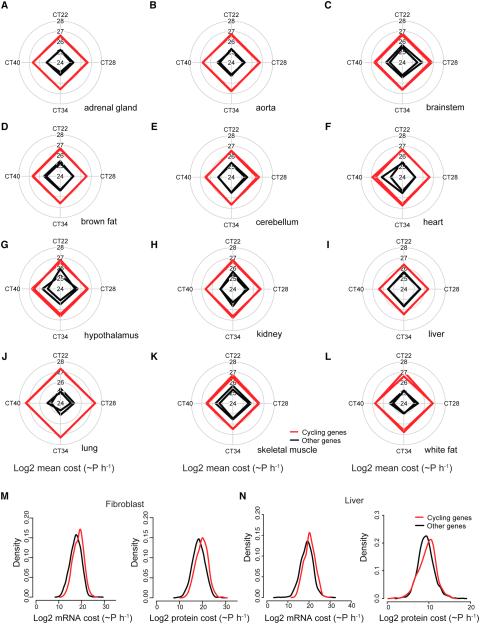
To explore whether the results found in the mouse liver apply generally to other tissues in the body, we analyzed recent circadian RNA-seq data from 12 different mouse tissues (Zhang et al., 2014). We find that cycling genes have increased cost in all 12 tissues implying that this feature is conserved (Figure 2A-L). Due to the lack of empirical protein measurement for every potential protein in our dataset, we estimated the protein abundance of genes lacking these data based on the mRNA expression data (see Experimental Procedures). To validate these estimates, we used the abundance of proteins from a mouse fibroblast proteomic dataset (Figure 2M) or from a mouse liver proteomic dataset (Figure 2N) that overlapped with the mouse liver cycling genes to calculate the transcriptional and translational costs for these two subsets of proteins (Schwanhausser et al., 2011; Shi et al., 2007). (Quantitative data from two recent circadian proteomics datasets (Mauvoisin et al., 2014; Robles et al., 2014) were not available for this analysis.) We found that cycling genes have an increased cost of 42% or 10% using both empirical datasets (p = 1.88E-38 and 2.09E-03, respectively, Table S1) consistent with our estimates based on mRNA expression levels alone. The difference in increased cost between the two empirical dataset is likely due to the different subset of proteins measured, as just 39% of the liver proteomic data and 10% of the fibroblast proteomic data overlap (764 genes).
Figure 2. Validation of increased cost in 12 mouse tissues and using proteomics datasets.
(A-L) Cost was calculated from all expressed genes in the mouse 12 tissue RNA-seq dataset from (Zhang et al. 2014; PNAS 111(45):16219-24). Eight time points (two cycles) from adrenal gland, aorta, brainstem, brown fat, cerebellum, heart, hypothalamus, kidney, liver, lung, skeletal muscle, and white fat were plotted and only the first time unit is shown for the two cycles (CT22, CT28, CT34, CT40). Red lines indicate cycling genes and black lines indicate other genes. The two lines for red and black represent the two cycling cycles analyzed. In all cases, the cycling genes exhibit significantly greater cost than non-cycling genes (p < 1E-04 in all comparisons). (M) Using only the overlap of the cycling gene dataset with empirical proteomic data from mouse fibroblasts, both the transcriptional (left panel) and translational (right panel) cost of cycling genes is increased in mouse. (N) Again, using only the overlap of the cycling gene dataset with empirical proteomic data from mouse liver, both the transcriptional and translational cost of cycling genes is increased in mouse. See also Figure S2.
The Increased Cost of Cycling Genes Can Be Extended to Drosophila and Yeast
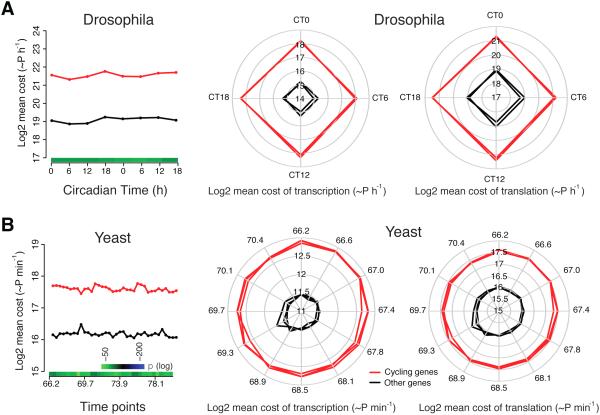
To address whether our observation in mouse is a conserved feature of circadian and metabolic cycling gene networks in other organisms, we performed cost analysis in Drosophila (Hughes et al., 2012) and yeast (Tu et al., 2005). As seen in mouse, we observed an increase in the total cost of cycling genes both for circadian genes in Drosophila (~5.5-fold increase) as well as for metabolic cycling genes in yeast (~2.5-fold increase) (Figures 3A-B, left panels). The increase in cost of cycling genes was seen at both the transcriptional and translational level in these organisms (Figures 3A-B, middle and right panels, Table S1 for more details). Thus, the increased energy requirement for cycling genes is conserved across both circadian and metabolic cycles as well as widely divergent species.
Figure 3. Cycling genes have an increased cost in Drosophila and yeast.
The increased cost of cycling genes in both (A) Drosophila and (B) yeast. The red lines indicate the cycling genes, whereas the black lines are non-cycling genes. Two and three lines for red and black in fruit fly and yeast represent the two circadian cycles and three yeast metabolic cycles analyzed. Only the time unit for the first cycle is shown for the radar plot of yeast.
The Increased Cost of Cycling Genes Can Not Be Explained by a Detection Artifact in Lowly Expressed Genes
Because it is possible that the detection of cycling genes may be influenced by expression level – where low levels of gene expression may compromise detection of cycling genes, we performed our cost analysis on subsets of the data that were partitioned by the level of gene expression. We first examined whether cycling genes have higher cost among the most highly expressed genes in all of the datasets. Figure S1 shows that cycling genes have significantly higher cost than non-cycling genes in the mouse and yeast datasets (p < 0.05) when only the genes with the highest expression are included (Figure S1A). In fact, regardless of whether the subset of genes have high, medium or low expression, cycling genes have a significantly higher cost than other genes in most of the datasets (Figure S1A). In addition, the mean cost of cycling genes remains higher than other genes even after removing the 20% lowest expressed genes (2583 genes in mouse liver, 3925 genes in Drosophila, 1355 genes in the yeast microarray dataset; Figure S1B; 3480 genes in 12 tissues in mouse, Figure S2; and Experimental Procedures).
Cycling of Expensive Genes Minimizes Genome-Wide Cost
To explore the potential benefit of generating energetically expensive genes in a cycling manner, we evaluated how perturbations in the composition of cycling gene sets would affect the overall cost of the system. We randomized which genes were cycling compared to other genes in a series of 10,000 simulations for each circadian time point, and calculated the resultant mean cost of all of the genes. As shown in Figure 4A, the experimentally-defined transcriptional and translational system results in one of the lowest energy cost usage combinations compared to the simulated genomes. In fact, none of the 120,000 simulations performed in total for the 12 circadian time points in mouse have a lower energy usage than the experimentally observed transcriptome (Table S1). Similar findings were observed in both Drosophila and yeast (Figure 4B, 4C and Table S1). Thus, the cycling expression of the more expensive genes is a conserved strategy for minimizing overall cellular energy usage.
Figure 4. Cycling gene expression yields an optimized overall cost.
Simulation experiments were performed by randomly switching the cycling genes with other genes and measuring the mean cost of all the genes in (A) mouse, (B), Drosophila or (C) yeast. The red lines indicate the experimentally observed results, whereas the blue lines are the simulations.
Cycling Gene Paralogs Exhibit Increased Cost
As an independent test of the utility of cycling genes, we leveraged whole genome duplication information, which has been shown to be integral for protein interaction networks and metabolic functions in yeast (DeLuna et al., 2008; Presser et al., 2008), to compare the cost of paralogous genes. We observed that for duplicated genes that originated from whole genome duplication in yeast, the mean cost of the cycling copies is higher than the cost of the non-cycling copies (p = 2.4E-02). Moreover, there is an enrichment of cases where the copy with the higher cost is regulated by the metabolic cycle (108 vs. 94, p = 4.1E-02). This effect is stronger in mouse as there were two rounds of whole genome duplication: 240 cases with a higher cost for the cycling copy and only 116 cases for the reverse (p = 4.97E-11). The mean cost of the cycling copies is higher than that of the non-cycling copies as well (p = 1.24E-20). These results further support evolutionary mechanisms for cyclical regulation of genes with higher cost.
Increased Expression Level is a Conserved Feature of Cyclical Gene Expression
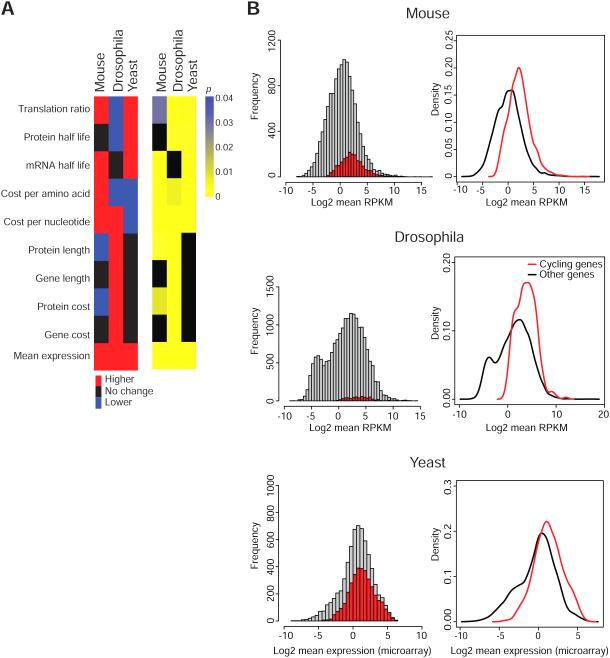
We next examined the contribution of specific molecular factors driving synthetic and degradative costs that could be responsible for the increased cost of cycling genes. We examined gene cost, protein cost, gene length, protein length, cost per nucleotide, cost per amino acid, mRNA half-life, protein half-life, and translation ratio across the three species. The only factor that we find consistently positively contributes to the high cost of the cycling genes in the three species examined is expression level (Figure 5A and Figure S3). For all of the 36 metabolic time points in yeast, 8 circadian time points in Drosophila and 12 circadian times points in mouse, the expression level of cycling genes is significantly higher than other genes (Figure 5A and 5B and see Table S1 for details), which is consistent with recent findings (Wu et al., 2012). To summarize, expression levels always contribute positively to an increased cost for cycling genes, however, other genomic features such as the use of more expensive building blocks or the length of sequences can also contribute to the increased cost of cycling genes, in a species dependent manner.
Figure 5. Expression levels positively contribute to the increased cost of cycling genes.
(A) Factors that contribute to the increased cost of cycling genes. Among the factors examined, only expression level positively contributes to the increased cost of the cycling genes in all three genomes. Red: positive contributions; blue: negative contributions; black: no significant changes. Significance levels are shown in the blue-yellow heatmap with p > 0.05 indicated by black (Wilcoxon rank-sum test). (B) Cycling genes have increased gene expression in all three species. Cycling genes are indicated by red bars in histograms (left panels) and red lines in density plots (right panels). See also Figure S3.
High Glucose Results in Increased Numbers of Cycling Genes in Yeast
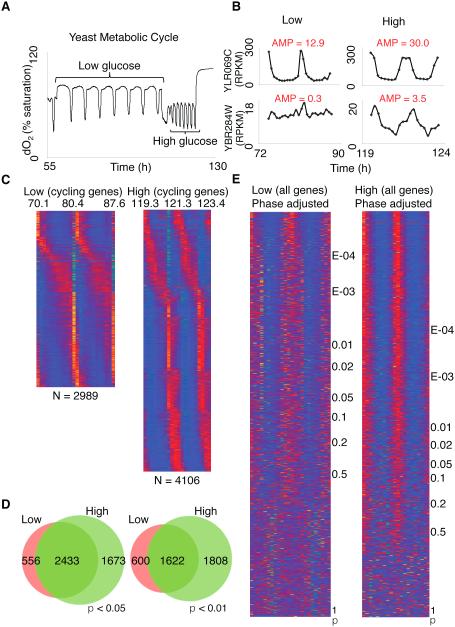
To test the hypothesis that differential energy requirements lead to alterations in cyclical gene expression, we designed a genome-scale experiment to manipulate the yeast metabolic cycle. We compared the effects of low and high steady-state glucose infusion rates on cyclic gene expression by changing the chemostat dilution rate of glucose-limited cells (Figure 6A and Experimental Procedures for more details). Although both conditions are energy-restricted, high glucose accelerated the speed of the metabolic cycle from ~6 h (slow cycling in low glucose) to ~2 h (fast cycling in high glucose) and led to an overall increase in oxygen consumption, as reflected in significantly lower mean dissolved oxygen (dO2) levels (Figure 6A; p < 5.0E-04, Table S1). We then conducted RNA sequencing over two consecutive cycles from equally spaced samples from each group [24 samples from the low glucose and 20 samples from the high glucose conditions, respectively (Figure S4A)]. We found genes that are periodically expressed in both low and high glucose conditions (e.g. YLR069C, Figure 6B, adjusted p = 1.93E-08 for the low and adjusted p = 8.97E-07 for the high condition) and genes that only showed periodic expression in one condition (e.g. YBR284W, Figure 6B, adjusted p = 1 for the low and adjusted p = 4.56E-07 for the high condition). Surprisingly, there are over 1,000 more genes with cyclical expression under high glucose conditions compared to low (Figures 6C, D, adjusted p < 0.05 and p < 0.01). In total, we detected more than 4,500 genes with periodic gene expression, which accounts for greater than 70% of the transcribed yeast genome (Table S1). To control for the sensitivity of detection of cycling genes between the low and high glucose conditions, we also display heatmaps for expression of all ~6000 genes in yeast (Figure 6E). At all levels of significance, the number of cycling genes is much greater in the high glucose condition (note p values to the right of each heatmap in Figure 6E).
Figure 6. Speed of metabolic cycling in yeast is linked to cyclical gene expression.
(A) Differential nutrient content leads to either slow (low glucose) or fast (high glucose) metabolic cycling in yeast. (B) Individual genes show differential periodic expression patterns within the low and high glucose conditions. Expression of YBR284W is only cyclical in the high glucose condition while YLR069C is cyclically expressed under both conditions. AMP indicates amplitude values. (C) Expression pattern of cycling genes in the low and high glucose conditions across the time points sampled. Red, high expression; blue, low expression. (D) More cycling genes are observed in the high compared to the low glucose condition. Left, p < 0.05; right, p < 0.01. At both significance levels, more than 3,000 genes have periodic expression in the high glucose condition. (E) Expression pattern of all genes in the low and high glucose conditions. Red, high expression; blue, low expression. Genes were phase adjusted and ranked by P values. See also Figure S4 and S7.
To assess whether the higher number of cycling genes in the high glucose condition could be due to misclassification of cycling genes by the algorithm (JTK_CYCLE), we evaluated the performance of JTK_CYCLE on the highly and lowly expressed genes using permutation tests. We asked whether JTK_CYCLE preferentially calls highly expressed genes as cycling. To determine this, we randomly shuffled the order of the time points while maintaining the mean gene expression values. We then determined the frequency of occurrence of cycling genes at six different levels of gene expression from high to low expression in either glucose condition. As shown in Figure S4B, there is a significant increase of “artificial cycling genes” only in the lowest 1,000 expressed genes in both high and low glucose conditions (approximately the bottom 20%; p < 1E-04 for both datasets), which goes against the expectation that JTK_CYCLE would detect fewer cycling genes at low expression levels. Finally, after removing the bottom 20% of all expressed genes (1339 genes), we found that there are still more cycling genes in the high glucose than in low glucose condition (Figure S4C). In addition, cycling genes have higher cost than other genes regardless of whether the bottom 20% expressed genes are removed or not (Figure S1B and S2). Thus, counter to expectation, the detection of cycling genes by JTK_CYCLE does not decrease but rather increases at low expression levels, providing additional evidence that cycling gene expression is not biased towards highly expressed genes.
Increased Glucose Leads to Increased Cyclical Amplitude without Increasing Expression
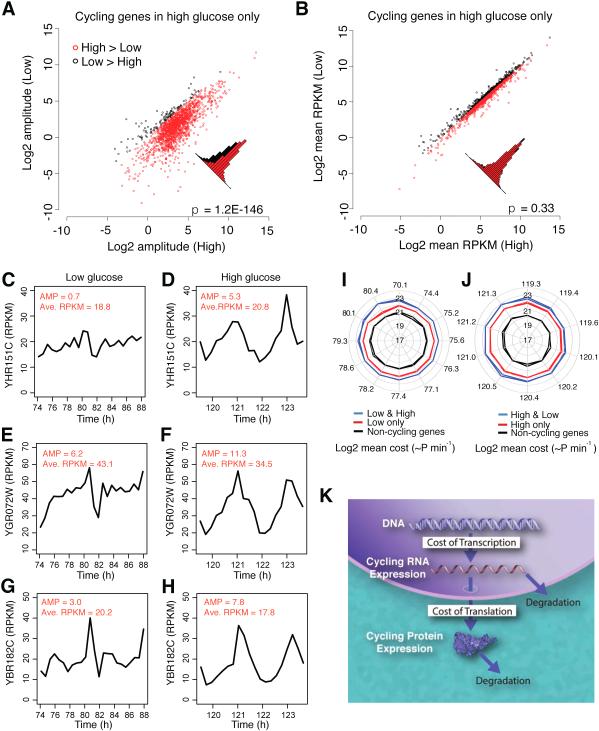
In addition to an increase in the number of cycling genes, the amplitude of the expression of the cycling genes in high glucose was higher than in low glucose even though their mean expression levels were similar (p < 1E-50, Figures 7A, 7B, and S5A-B). This is exemplified in Figure 6B, where we show the amplitude (calculated from JTK_CYCLE, see Experimental Procedures) of the YBR284W gene is 3.5 in high glucose compared to 0.3 in low glucose. Additional examples are presented in Figures 7C-7H. Thus under higher metabolic conditions, the number of cycling genes increases as well as the amplitude of these oscillations. Because the mean expression level of cycling genes does not increase, increasing the amplitude of cycling genes is an extremely efficient mode for increasing the peak expression levels.
Figure 7. Differential Cycling Amplitude and Cost of Genes with Differential Metabolic Cycling in Yeast.
(A) Amplitude and (B) RPKM comparisons of cycling genes in low and high glucose conditions. (A) Red, the genes whose amplitude is greater in high glucose compared to low glucose; black, the genes whose amplitude is smaller in high glucose than in low glucose. (B) Red points, the genes with mean RPKM greater in high glucose compared to low glucose; black points, the genes with mean RPKM smaller in high glucose compared to low glucose. (C-H) Examples of genes with higher cycling amplitude but not significantly higher RPKM in high compared to low glucose conditions. Amplitude strength and average RPKM in the two conditions are indicated in red. (I-J) Differential cost of genes with differential metabolic cycling in yeast. The cost of cycling genes and non-cycling genes in low (I) compared to high (J) glucose conditions. Blue, cycling genes; black, non-cycling genes; red, genes with periodic expression in only one condition. Two lines for red and black represent the two metabolic cycles analyzed. Only the time unit for the first cycle is shown for the radar plot (K) Schematic representation of cost of cycling genes during protein synthesis. See also Figure S5 and S6.
Surprisingly, our data suggest that increasing amplitude without increasing mean expression is an energy saving behavior in high glucose. When more nutrients/resources are available to yeast in high glucose, more molecules are expressed to "consume" those nutrients, thus the expression levels are increased. However, this increase in expression is offset by a concomitant downregulation in expression during another time period of the cycle. As such, the median of the peak (75th percentile) of gene expression in high glucose is significantly higher than in low glucose (p = 4.49E-05), which leads to an increased cost (p = 0.011). In line with this, we found that the median of the trough (25th percentile) of gene expression in high glucose is significantly lower than in low glucose (p = 0.003), and the cost is lower as well (p = 0.019) (Figure S5C). Therefore, it is cost-effective to reduce the expression of these cycling genes during the metabolic cycle when they are not needed. Interestingly, we observed 1162 out of 1673 genes following this pattern at the peak of the cycle and 1151 out of 1673 genes follow this pattern at the trough of the cycle, which is greater than expected by chance (p = 7.83E-57 and p =2.3E-53, respectively). These data suggest a mechanism for why more cycling genes are observed under the high glucose condition as more genes are using this strategy to reduce cost. Finally, a strong positive correlation is observed between the amplitude and the expression of genes (r = 0.838 for high glucose and r = 0.784 for low glucose condition, p < 2E-16 for both, Figure S5D). This indicates that for genes that are increasing their amplitude (experimental conditions - from low to high glucose), overall expression levels are upregulated. However, we have observed unchanged mean expression levels for those cycling genes in the experimental data, which additionally indicates that this is an energy saving behavior.
Increased Glucose Alters Yeast Metabolic Pathway Costs
Previous work has shown that there are three major phases of the yeast metabolic cycle (Oxidative, Ox; Reductive, Building, R/B; Reductive, Charging, R/C) (Tu et al., 2005). We also find the same three major clusters of genes in these new analyses (Table S1). We also find an enrichment of genes in different cellular pathways among the cycling genes in the two conditions, and the genes periodically expressed in both glucose conditions are strongly enriched in mitochondrial and ribosomal functions (Table S1).
Because cycling genes can express higher peak levels without an increase in the overall mean level of expression, we asked whether this was also the case for the three metabolic phases of the cycle. We calculated the total amount of gene expression (RPKM) for proteins that are involved in the metabolic cycle in each condition. We found that there is less gene expression of mitochondrial ribosomal genes (e.g. MRPL10 and related genes, Figure S6A, 28.5% less, p = 2.2E-09), the large (60S) ribosomal subunit and related genes (e.g. RPL17B, Figure S6A, 15.1% less, p =6.2E-07), or genes encoding nuclear-encoded mitochondrial ribosomal proteins (Figure S6A, 24.9% less, p = 6.8E-13) under high versus low glucose conditions. We next investigated whether each of the yeast metabolic phases have different molecular requirements. As expected, in each of the three phases (Ox, R/B, and R/C), there is less of a change in gene expression (from highly expressed to low expressed) observed in each of these phases in the high glucose condition compared to the low (7.3%, 2.3% and 13.0% less, p = 4.4E-06, 4.5E-11 and 6.8E-18, respectively, Figure S6A). We also observed that the Ox and R/B phase contain more genes with conserved cycling across the two cycling conditions compared to genes in the R/C phases (65% and 62% vs. 33%), and the overall phase distribution of the cycling genes is likely determined by the Ox and R/B phases (Figure S6B and S6C). As the Ox phase is enriched for genes involved in ribosome synthesis and the R/B phase is enriched for genes involved in mitochondria biogenesis (Cai and Tu, 2012; Tu et al., 2005), these results indicate that the costs related to gene expression in yeast metabolic pathways are reduced in the high compared to the low glucose condition.
Cycling Genes in Both Nutrient Conditions Have the Highest Cost
To further distinguish the genes with periodic expression in each condition, we estimated the cost at each time point (Figures 7I and 7J, Table S1). We found that the periodically expressed genes have a higher cost on average in both high and low glucose conditions (Figures 7I and 7J). More importantly, the genes with periodic expression in both the high and low glucose conditions have a higher cost than the genes with periodic expression in only one condition (Figures 7I and 7J). These results strongly suggest that the yeast metabolic cycle promotes the periodic expression of genes with a higher cost, consistent with the prediction of our hypothesis. Figure 7K illustrates the model of our calculations, demonstrating that the cost of a gene from DNA to protein determines the cyclical expression of that gene.
DISCUSSION
Cycling genes, whether circadian or metabolic, define an evolutionarily conserved mechanism for cellular energy conservation in three divergent eukaryotic organisms. Empirically we find that cycling genes are expressed at high levels and constitute the most costly proteins to transcribe and translate in the genome. The essence of the strategy for utilizing cycling genes is that peak cycling gene expression (amplitude) can be elevated relative to constitutive expression without an increase in overall mean levels of expression. The peak is offset by the trough. Thus, abundant proteins that are required at one time can be down-regulated at other times to economize on overall production. In testing this hypothesis using the yeast metabolic cycle, we discover the non-intuitive result that when metabolic rate increases under higher glucose conditions, the number of cycling genes increases dramatically. This result is in line with previous work demonstrating that yeast grown under higher glucose conditions exhibit altered gene expression patterns that correspond to growth rates (Slavov and Botstein, 2011). However, our sampling time period under high glucose was within a 2h period (Figure S4A) making effects of cell cycling less likely to be involved. This is supported by studies demonstrating that metabolic cycling in yeast occurs in the absence of cell division (Slavov et al., 2011). Paradoxically, in our study, the mean levels of the cycling genes do not increase and in some cases actually decreases. Thus, in yeast and in cells, cyclic gene expression is a potent mechanism for energy conservation. If a protein is not needed at a particular time, its production is shut down. In turn, if higher expression is needed, cyclic expression is efficient and thus increases in metabolic demand would be expected to lead to additional cycling genes under this scenario, as we have observed in yeast.
As translation requires a greater cost compared to transcription, we speculate that the cycling of proteins might be greater than for transcripts. However, such comparisons require comparable quantitative proteomic datasets to what is currently available for mRNA. It is possible that cost is not as relevant for directing cyclical expression of genes that are core components of the regulatory network of cycling behavior and biosynthetic pathways, such as Bmal1, Clock, Cry, Dbp, Per, and Nampt (Green et al., 2008). In fact, when we specifically examine the cost of core circadian genes, we do not observe that these genes are typically among the most expensive genes because these transcriptional regulatory genes are expressed at low levels. Among 16 of the canonical circadian genes, only one gene (Nampt) is among the top 10% of expensive cycling genes, and only 3 other genes (Atf6, Clock, and Creb1) are among the top 25%.
One might ask why it is necessary for a cell to synthesize new proteins in a cyclic manner instead of utilizing stable, long-lived proteins? We can offer at least two explanations. First, it is known that many cellular processes are incompatible, such as oxidative vs. reductive metabolic pathways. This has led to two different solutions in cells: subcellular compartmentalization and temporal partitioning of metabolic pathways. In cases in which subcellular compartmentalization is not efficient, then temporal partitioning (time sharing) may be the only solution. Indeed in many primordial photosynthetic organisms, temporal partitioning is the major strategy for separating processes such as photosynthesis during the day, which involves oxygen, and nitrogen fixation during the night, which must occur in an oxygen-poor environment (Fay, 1992; Schneegurt et al., 1994; Stockel et al., 2008). Furthermore, there is considerable evidence in plants and animals that mis-expression of genes in the cell can cause unexpected deleterious effects (Fernandez et al., 2013; Lai et al., 2012; Manansala et al., 2013; Montgomery et al., 2013), again discounting the long-term maintenance of global protein expression.
Second, in addition to partitioning of cellular processes, in yeast (Bristow et al., 2014) and parasites (Bozdech et al., 2003; Suvorova and White, 2014) there is considerable gene expression turnover and gene expression occurs “as needed” in these organisms. In the yeast metabolic cycle, the three phases (Oxidative; Reductive, Building; Reductive, Charging) follow the strategy of “just-in-time” delivery of components (Kuang et al., 2014). That is, at each of these phases, the basic building blocks of the cell are synthesized at the time that they are needed, in order to flexibly adapt with the environment. Cells do not store these components throughout the metabolic cycle. An example showing the importance of this “just-in-time” strategy is that cyanobacteria show higher reproductive fitness if the patterns of their the internal circadian oscillator and environmental cycles are similar, while fitness is decreased if the internal circadian system does not match the environment well (e.g. in constant light) (Woelfle et al., 2004). Why might this occur? Such “just-in-time” strategies have been successfully implemented in manufacturing as the cost of maintaining storage and completing regular inventory exceeds the cost of manufacturing and delivery of goods in real time (Gonzalez et al., 2006; Qureshi et al., 2013). Perhaps inventory storage is either not efficient in cells, or as in the first example, components might be incompatible to be stored together. Thus, we see in this temporal view of gene expression, a surprisingly efficient strategy for both the partitioning and deliver of cellular metabolic components on a genome scale. The hypothesis for a “just-in-time” strategy in transcriptional networks has been previously proposed (Zaslaver et al., 2004). However, our results not only provide evidence in support of this strategy in metabolic processes occurring in more simple organisms such as yeast, but our work expands and supports this hypothesis to mammals such as mouse. Our findings are also in line with the hypothesis that ultradian or time-keeping strategies are employed at the molecular level (such as in gene expression) to integrate cellular functions in yeast as well as mammalian systems (Lloyd and Murray, 2005)
Although the cost during synthesis and degradation of the transcription and translation of cycling genes has been evaluated here, there are other cellular processes that consume energy in the cell that may play a part in cycling gene expression, such as the transport of mRNA and protein outside of the nucleus (Gorlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Vargas et al., 2005), protein folding and misfolding (Beissinger and Buchner, 1998; Goldberg, 2003), alternative splicing (Staley and Guthrie, 1998; Wahl et al., 2009) and DNA repair (Lindahl and Wood, 1999; Sancar et al., 2004). Because there is little difference among the cost of nucleotides (Table S1), it is unlikely that codon bias is a major contributor to changes in energetic cost; however, this needs to be investigated further especially with regards to translational efficiency (Quax et al., 2015) once quantitative proteomic datasets across cycling time points are available. The evaluation of noncoding RNAs also needs to be considered as these transcripts may have rapid turnover but also contribute to the regulation of whether coding transcripts are ultimately expressed as proteins. Future studies that determine targets and functions of these noncoding RNAs on a genome scale will need to be incorporated. Also energy generation and consumption are linked to the temporal compartmentalization of metabolic functions, which allows for increased efficiency of metabolism especially under depleted nutrient conditions (Tu et al., 2005; Tu and McKnight, 2007). The organization of the genome may also play a role in energy usage as genes physically near each other on chromosomes have similar expression profiles (Wang et al., 2011), and such relationships may lead to similar cost and energy usage during expression. How the constraints on the cost of gene expression constrain other biological circuits such as feedback loops, enzymatic activities, proportional regulation of promoter activities in coexpression networks, or transcriptional networks that are involved in cyclical gene expression still needs to be investigated (Alon, 2007; Hart and Alon, 2013; Keren et al., 2013; Koike et al., 2012; Milo and Last, 2012; Wagner, 2007). In addition, overall energy utilization in cells includes processes other than the ones leading to protein expression. These include, but are not limited to, lipid, carbohydrate, and triglyceride production and turnover (Palinkas et al., 2015), and ion transport across plasma membranes. Future studies that empirically calculate these parameters over time can ultimately be incorporated into this model to determine total energy use and whether cycling genes contribute to an energy saving mechanism.
Future experiments that empirically measure the energetic properties of all of these processes on a genome-wide basis will contribute to our overall knowledge of cycling gene energy usage. In all, the data presented here highlight the importance of investigating energy usage, and how such fundamental processes can deeply influence cellular and organismal physiology.
EXPERIMENTAL PROCEDURES
Transcriptome data
Three large-scale transcriptomic profiling data sets were used to characterize the cycling behavior of the three species (Table S1): a ~300 min metabolic cycling data set for the diploid yeast strain CEN.PK (Tu et al., 2005), a 12 hour light/dark transcriptomic data set for wild type Drosophila brain (Hughes et al., 2012), and a 48-hour constant darkness transcriptomic profiling of mouse liver (Koike et al., 2012). For each of these datasets, only the expressed genes were used for further analysis. For each gene, the average expression values were used if multiple expression signals were detected.
Mouse liver proteome data
The mouse liver proteome data were obtained from a public mouse liver proteome database (Shi et al., 2007).
Calculation of cost of each mRNA and protein
The cost of each gene/protein that met our criteria described above was calculated. The energy usage of synthesizing each amino acid and nucleotide was based on a previous analysis of the yeast metabolic system, which is calculated by the activated phosphate bonds (~P) (Wagner, 2005) (Table S1). On average, the synthetic cost per nucleotide residue (mean cost: 49.5 ~P) is greater than that of an amino acid (mean cost: 29.1 ~P).
Cycling genes
The cycling genes from these three studies (Hughes et al., 2012; Koike et al., 2012; Tu et al., 2005) were used for the primary analyses. Cycling genes in 12 tissues from mouse (Zhang et al., 2014) were also used for confirmation. For the mouse liver data, only exon based cycling genes are included, although we found that intronic cycling genes have higher cost (transcriptional) than other genes as well (data not shown). For the genes that do not have an annotated Ensembl ID, the transcript names were first mapped to an Ensembl ID by the BioMart data mining tool.
The cost during mRNA and protein synthesis and degradation of the transcriptome
The expression cost of a gene was calculated based on two parts, as previously proposed (Wagner, 2005). Please note that the degradation rate of a particular mRNA and protein are assumed constant in the given environment as the genome-wide measurement of this parameter across each cycling time point is not available. That assumption should have limited influence on our calculation of the cost of peak and trough as the transcriptional burst is a major mode of gene expression regulation (Cai et al., 2006; Dar et al., 2012).
Yeast metabolic cycle experiments
Yeast strains and methods
Yeast manipulations were performed using standard methods (Sherman, 2002).
Continuous culture conditions
Yeast cultures were grown as previously described (Tu et al., 2005). Samples were collected over two metabolic cycles. For the low glucose condition, samples were taken every 36 minutes for ~14.5 hours. For the high glucose condition samples were taken every 13 minutes for ~4.25 hours.
Library Preparation
RNA-seq libraries were prepared as described in detail previously (Takahashi et al., 2015).
Bioinformatic analysis of metabolic cycles under low and higher glucose
TopHat v2.0.10 was used as the mapping program (Trapnell et al., 2009); the unmapped reads and the reads with mapping quality score less than 10 were filtered out after mapping (Table S1). The mapped and filtered reads were used to calculate the RPKM values with HOMER (Heinz et al., 2010). Mapping the reads to the less well-annotated CEN.PK genome (Nijkamp et al., 2012; Otero et al., 2010) or normalizing the RPKM between samples did not change the major findings of this study (Figure S7). JTK_CYCLE was used to determine the circadian behaviors of the genes (Hughes et al., 2010). Genes with an adjusted P value < 0.05 were further regarded as cycling genes.
Full experimental procedures are available in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Hughes, and Michael Nitabach for providing additional information about Drosophila datasets, John B. Hogenesch for providing the 12 tissue datasets in mouse, and Laurence D. Hurst, Tae-Kyung Kim and Yonghao Yu for helpful discussions. This work was supported by NIH grants (R00MH090238 and R21MH107672) and the Jon Heighten Scholar in Autism Research award at UT Southwestern (G.K.), and NIH grant (R01AG045795) and the Howard Hughes Medical Institute (J.S.T). J.S.T. is a cofounder of, a Scientific Advisory Board member of, and a paid consultant for Reset Therapeutics, Incorporated, a biotechnology company aimed at discovering small-molecule therapies that modulate circadian activity for a variety of disease indications.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The NCBI Gene Expression Omnibus (GEO) accession number for the RNA-seq data reported in this manuscript is GSE57683.
AUTHOR CONTRIBUTIONS
G.-Z. W. initiated the project, designed and conducted most of the analyses; S.L., L.S., P.K., B.T. performed yeast experiments; H.-C. H. mapped the yeast reads and calculated the yeast cycling genes; N.K. prepared the mouse cycling expression data; J.S.T. and G.K. supervised the project; G.-Z. W., S.L., H.-C. H., J.S.T. and G.K. wrote the manuscript.
REFERENCES
- Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissinger M, Buchner J. How chaperones fold proteins. Biol. Chem. 1998;379:245–259. [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristow SL, Leman AR, Haase SB. Cell cycle-regulated transcription: effectively using a genomics toolbox. Methods Mol. Biol. 2014;1170:3–27. doi: 10.1007/978-1-4939-0888-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Tu BP. Driving the cell cycle through metabolism. Annu. Rev. Cell Dev. Biol. 2012;28:59–87. doi: 10.1146/annurev-cellbio-092910-154010. [DOI] [PubMed] [Google Scholar]
- Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- Dar RD, Razooky BS, Singh A, Trimeloni TV, McCollum JM, Cox CD, Simpson ML, Weinberger LS. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc. Natl. Acad. Sci. USA. 2012;109:17454–17459. doi: 10.1073/pnas.1213530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuna A, Vetsigian K, Shoresh N, Hegreness M, Colon-Gonzalez M, Chao S, Kishony R. Exposing the fitness contribution of duplicated genes. Nat. Genet. 2008;40:676–681. doi: 10.1038/ng.123. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Fay P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 1992;56:340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L, Chaib J, Martinez-Zapater JM, Thomas MR, Torregrosa L. Mis-expression of a PISTILLATA-like MADS box gene prevents fruit development in grapevine. Plant J. 2013;73:918–928. doi: 10.1111/tpj.12083. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Gonzalez CM, Jang T, Raines M, Lys TZ, Schaeffer AJ. Significant clinical practice cost savings through downsizing office supply inventory and just in time ordering. J. Urol. 2006;176:267–269. doi: 10.1016/S0022-5347(06)00501-5. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart Y, Alon U. The utility of paradoxical components in biological circuits. Mol. Cell. 2013;49:213–221. doi: 10.1016/j.molcel.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Grant GR, Paquin C, Qian J, Nitabach MN. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012;22:1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren L, Zackay O, Lotan-Pompan M, Barenholz U, Dekel E, Sasson V, Aidelberg G, Bren A, Zeevi D, Weinberger A, et al. Promoters maintain their relative activity levels under different growth conditions. Mol. Syst. Biol. 2013;9:701. doi: 10.1038/msb.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z, Cai L, Zhang X, Ji H, Tu BP, Boeke JD. High-temporal-resolution view of transcription and chromatin states across distinct metabolic states in budding yeast. Nat. Struct. Mol. Biol. 2014;21:854–863. doi: 10.1038/nsmb.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JH, Dijkwel PP. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA. 2012;109:17129–17134. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- Lloyd D, Murray DB. Ultradian metronome: timekeeper for orchestration of cellular coherence. Trends Biochem. Sci. 2005;30:373–377. doi: 10.1016/j.tibs.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genom. Hum. Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv. Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manansala MC, Min S, Cleary MD. The Drosophila SERTAD protein Taranis determines lineage-specific neural progenitor proliferation patterns. Dev. Biol. 2013;376:150–162. doi: 10.1016/j.ydbio.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, Waridel P, Quadroni M, Gachon F, Naef F. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. USA. 2014;111:167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Last RL. Achieving diversity in the face of constraints: lessons from metabolism. Science. 2012;336:1663–1667. doi: 10.1126/science.1217665. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. A nnu. Rev. Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery JR, Whitt JP, Wright BN, Lai MH, Meredith AL. Mis-expression of the BK K(+) channel disrupts suprachiasmatic nucleus circuit rhythmicity and alters clock-controlled behavior. Am. J. Physiol. Cell Physiol. 2013;304:C299–311. doi: 10.1152/ajpcell.00302.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Nijkamp JF, van den Broek M, Datema E, de Kok S, Bosman L, Luttik MA, Daran-Lapujade P, Vongsangnak W, Nielsen J, Heijne WH, et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell Fact. 2012;11:36. doi: 10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero JM, Vongsangnak W, Asadollahi MA, Olivares-Hernandes R, Maury J, Farinelli L, Barlocher L, Osteras M, Schalk M, Clark A, et al. Whole genome sequencing of Saccharomyces cerevisiae: from genotype to phenotype for improved metabolic engineering applications. BMC Genomics. 2010;11:723. doi: 10.1186/1471-2164-11-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinkas A, Bulik S, Bockmayr A, Holzhutter HG. Sequential metabolic phases as a means to optimize cellular output in a constant environment. PLoS One. 2015;10:e0118347. doi: 10.1371/journal.pone.0118347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presser A, Elowitz MB, Kellis M, Kishony R. The evolutionary dynamics of the Saccharomyces cerevisiae protein interaction network after duplication. Proc. Natl. Acad. Sci. USA. 2008;105:950–954. doi: 10.1073/pnas.0707293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TE, Claassens NJ, Soll D, van der Oost J. Codon Bias as a Means to Fine-Tune Gene Expression. Mol. Cell. 2015;59:149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi MI, Iftikhar M, Bhatti MN, Shams T, Zaman K. Critical elements in implementations of just-in-time management: empirical study of cement industry in Pakistan. Springerplus. 2013;2:645. doi: 10.1186/2193-1801-2-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10:e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu. Rev. Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Schneegurt MA, Sherman DM, Nayar S, Sherman LA. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 1994;176:1586–1597. doi: 10.1128/jb.176.6.1586-1597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Shi R, Kumar C, Zougman A, Zhang Y, Podtelejnikov A, Cox J, Wisniewski JR, Mann M. Analysis of the mouse liver proteome using advanced mass spectrometry. J. Proteome Res. 2007;6:2963–2972. doi: 10.1021/pr0605668. [DOI] [PubMed] [Google Scholar]
- Slavov N, Botstein D. Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol. Biol. Cell. 2011;22:1997–2009. doi: 10.1091/mbc.E11-02-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Macinskas J, Caudy A, Botstein D. Metabolic cycling without cell division cycling in respiring yeast. Proc. Natl. Acad. Sci. USA. 2011;108:19090–19095. doi: 10.1073/pnas.1116998108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stockel J, Welsh EA, Liberton M, Kunnvakkam R, Aurora R, Pakrasi HB. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc. Natl. Acad. Sci. USA. 2008;105:6156–6161. doi: 10.1073/pnas.0711068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova ES, White MW. Transcript maturation in apicomplexan parasites. Curr. Opin. Microbiol. 2014;20C:82–87. doi: 10.1016/j.mib.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Kumar V, Nakashe P, Koike N, Huang HC, Green CB, Kim TK. ChIP-seq and RNA-seq Methods to Study Circadian Control of Transcription in Mammals. Methods Enzymol. 2015;551:285–321. doi: 10.1016/bs.mie.2014.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. The yeast metabolic cycle: insights into the life of a eukaryotic cell. Cold Spring Harb. Symp. Quant. Biol. 2007;72:339–343. doi: 10.1101/sqb.2007.72.019. [DOI] [PubMed] [Google Scholar]
- Vargas DY, Raj A, Marras SA, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proc. Natl. Acad. Sci. USA. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S. Circadian Oscillations of Protein-Coding and Regulatory RNAs in a Highly Dynamic Mammalian Liver Epigenome. Cell Metabolism. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Energy constraints on the evolution of gene expression. Mol. Biol. Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- Wagner A. Energy costs constrain the evolution of gene expression. J. Exp. Zool. B. Mol. Dev. Evol. 2007;308:322–324. doi: 10.1002/jez.b.21152. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang GZ, Chen WH, Lercher MJ. Coexpression of linked gene pairs persists long after their separation. Genome Biol. Evol. 2011;3:565–570. doi: 10.1093/gbe/evr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Wu G, Zhu J, He F, Wang W, Hu S, Yu J. Gene and Genome Parameters of Mammalian Liver Circadian Genes (LCGs) PLoS One. 2012;7:e46961. doi: 10.1371/journal.pone.0046961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U. Just-in-time transcription program in metabolic pathways. Nature Genet. 2004;36:486–491. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat. Rev. Mol. Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.