Abstract
It is striking that, while central to sexual reproduction, the genomic regions determining sex or mating-types are often characterized by suppressed recombination that leads to a decrease in the efficiency of selection, shelters genetic load, and inevitably contributes to their genic degeneration. Research on model and lesser-explored fungi has revealed similarities in recombination suppression of the genomic regions involved in mating compatibility across eukaryotes, but fungi also provide opposite examples of enhanced recombination in the genomic regions that determine their mating types. These contrasted patterns of genetic recombination (sensu lato, including gene conversion and ectopic recombination) in regions of the genome involved in mating compatibility point to important yet complex processes occurring in their evolution. A number of pieces in this puzzle remain to be solved, in particular on the unclear selective forces that may cause the patterns of recombination, prompting theoretical developments and experimental studies. This review thus points to fungi as a fascinating group for studying the various evolutionary forces at play in the genomic regions involved in mating compatibility.
Keywords: bipolar, homothallism, heterothallism, MAT, Muller’s ratchet, tetrapolar, Microbotryum violaceum, Cryptococcus neoformans, Neurospora tetrasperma, Podospora anserina
1. Introduction
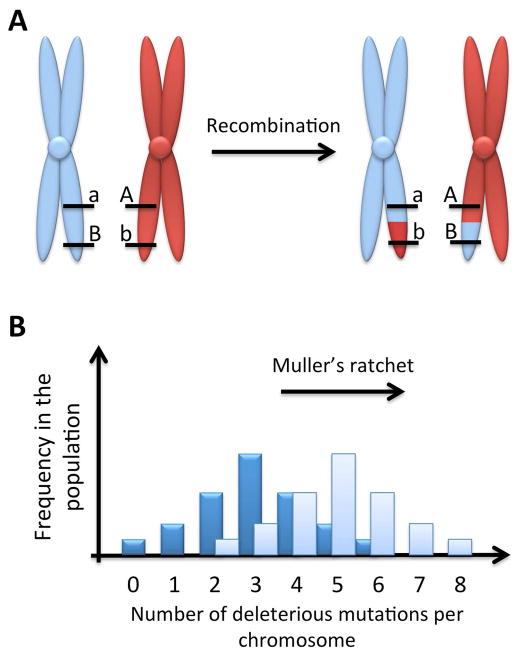
A decade ago an alarming hypothesis captured public imagination: men were going to become extinct within the next 5–10 million years (Marshall Graves, 2002). Behind this prediction was the degeneration of the male-specific Y chromosome. The Y chromosome, which was once an autosome carrying a sex-determining gene, has indeed experienced repeated and expanding stages in its accumulation of mutations relative to its homologous X chromosome. Degeneration of gene content and mutation accumulation are consequences of the lack of crossing-over during meiosis, because it is through such recombination that chromosomes with fewer deleterious mutations than the minimum number in the population can be produced (Graves, 2006; Bergero and Charlesworth, 2009) (Fig. 1a). It was thus first reasoned by Hermann Muller that, in the absence of recombination, chromosomes containing the fewest deleterious mutations would be regularly lost from the population by chance, which would lead to an irreversible and increasing loss in coding capability over time (Muller, 1964; Bergero and Charlesworth, 2009) (Fig. 1b). Although extensions of this model, known as “Muller’s ratchet,” have been formulated [e.g., (Kondrashov, 1982)], they all predict a connection between the suppression of recombination and the accumulation of mutations. Furthermore, large regions united by recombination suppression prevent selection from acting independently upon variation in separate loci, causing hitchhiking of deleterious mutations together with positive selection of a beneficial allele [i.e., Hill-Robertson interference (Gillespie, 2000)].
Fig 1.
(A) Illustration of recombination as a mechanism for purging deleterious mutations. A crossing-over event between two chromosomes carrying different deleterious mutations (a and b) produces a chromosome free of deleterious mutations (A and B). (B) Illustration of the concept of Muller’s ratchet. Without recombination, the number of deleterious mutations per chromosome increases over time in the population, and the chromosomes free of deleterious mutations will be lost by chance. The dark blue shading shows a theoretical original state, with a given mean and variance for the number of deleterious alleles per chromosome in a population. The light blue shading shows the resultant state after many generations without recombination, illustrating the Muller’s ratchet effect of an increase in the mean number of deleterious mutations.
Suppression of recombination and the consequent molecular degeneration around genes controlling sexual compatibility is not unique to humans as it is found in other animals, plants, and in the fungi (Hood, 2002; Fraser and Heitman, 2004a, b; Whittle et al., 2011; Hood et al., 2013; Fontanillas et al., 2015). An apparent irony is found in that the very elements regulating genetic exchange between individuals are themselves largely excluded from the benefits of this recombination (Idnurm, 2011), especially because sex does not have to be determined genetically but can be through, as examples, environmental sex determination or sequential hermaphroditism (Bachtrog et al., 2014; Beukeboom and Perrin, 2014).
There may be processes that counter the trend toward degeneration and the recombinationally-inert nature of mating-type or sex chromosome regions. In one view, the nonreciprocal transfer of DNA sequence achieved through gene conversion may counter-act some of the deleterious effects of suppressed recombination (Marais et al., 2010; Trombetta et al., 2010), but this phenomenon remains rare in the regions where crossing-over frequencies are low (Bachtrog, 2013). A process of cyclical renewal of sex-determining regions has also been suggested, where new regions evolve to control the process of mating when old sex chromosomes become too degraded (Mank and Avise, 2009; Blaser et al., 2014). However, as recent studies expand both the diversity of species and the genetic tools for documenting recombination, there are some rare exceptions in which presumed dead spots of genetic exchange permit recombination.
Regions controlling mating compatibility are in some cases associated with suppressed recombination while in other cases they harbor crossing-over hotspots (Burgoyne, 1982; Yi and Li, 2005; Hsueh et al., 2006; Brick et al., 2012; Sarbajna et al., 2012; Jakočiūnas et al., 2013; Bolton et al., 2014). The evolutionary drivers for these contrasted relationships between recombination and mating-type determination are still unclear. That is, the regions controlling sex determination or mating compatibility take a broad range of values in their association with either recombination suppression or enhancement.
This article outlines three contrasted situations regarding recombination associated with regions that control mating compatibility; suppressed recombination, enhanced recombination, and non-homologous or non-reciprocal exchange that may counteract molecular degeneration. We outline some of the puzzles about these relationships between mating-type determination and recombination, and we suggest possible explanations. Recent discoveries in fungi provide valuable insights into the evolutionary genomics of sexual compatibility. While previous articles have highlighted the similarities between fungal mating-type chromosomes and the sex chromosomes in other eukaryotes (Fraser and Heitman, 2004a, b; Menkis et al., 2008; Whittle and Johannesson, 2011), here we focus on the evolutionary causes for these similarities, point out the evolutionary differences, and highlight questions that remain to be answered.
2. Fungi are key model organisms for understanding the evolutionary genomics of sexual compatibility systems
The fungi are a large group of eukaryotes, with estimates of the total number of species in the millions (Blackwell, 2011). Mating has long been studied in fungi because they are tractable model organisms (Kniep, 1919; Gäumann, 1952), and the spores produced from sexual processes in the pathogen fungal species can be infectious propagules. Mating compatibility is determined by chromosomal regions that are referred to as the mating-type loci. Many species have a single mating-type locus that encodes alternate transcription factors that regulate expression of pheromone and pheromone receptor genes as well as genes involved in post-mating compatibility (Debuchy et al., 2010). Pheromones are used for partner recognition and attraction. Some basidiomycete species have incorporated the genes that encode the pheromone and pheromone receptor proteins into the mating-type locus (Raudaskoski and Kothe, 2010). With the exceptions discussed below, crossing-over is not known to occur within the mating-type loci, where the intercompatibility of alternate alleles maintains the divergent forms and even ancient trans-specific polymorphisms (Devier et al., 2009; van Diepen et al., 2013), to the extent that homology is sometimes difficult to recognize or has been questioned (Metzenberg and Glass, 1990; Debuchy et al., 2010). This feature resembles the mammalian sex chromosomes, yet in fungi the mating compatibility of the cell type can be defined by as little as 1 kb of DNA rather than the majority of the chromosome (Butler, 2007).
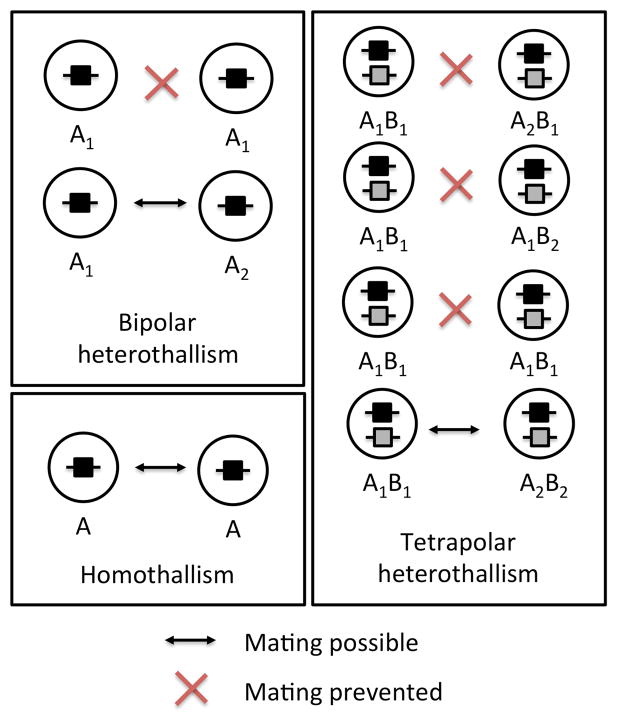
The fungal system in which mating can only occur between individuals of different mating types is called heterothallism (Fig. 2). A bipolar system of mating where a single genetic locus governs mating type (Fig. 2) is found in heterothallic ascomycetes, heterothallic Mucoromycotina species and a few heterothallic basidiomycetes. Most often such bipolar systems harbor only two alleles, and thus at the population level yield individuals of two mating types (Billiard et al., 2011). Most basidiomycete fungi exhibit a tetrapolar mating system; mating is controlled by two loci, with mating being possible only between haploid cells carrying different alleles of both loci (Fig. 2). In these species tightly-linked genes that encode pheromones and pheromone receptors regulate cell fusion. The second locus encodes homeodomain proteins that function in non-self recognition, controlling post-fusion compatibility (Feldbrügge et al., 2004). These two loci, encoding pheromones/receptors and homeodomain proteins, can be linked to form a single locus (a system called bipolarity, because only two haploid mating types are produced) or in crosses they may segregate independently (a system called tetrapolarity because the diploid, being heterozygous at both loci, can produce four mating type combinations after meiosis). The independent segregation of the two mating type loci in tetrapolar systems is associated with high levels of allelic polymorphism, yielding potentially thousands of mating types (Casselton and Olesnicky, 1998; Brown and Casselton, 2001; Nieuwenhuis et al., 2013). Multiple mating types being determined by one locus are found in other eukaryotes, such as self-incompatibility in angiosperms, ciliates and tunicates (Hurst et al., 1992). Likewise, the existence of a two-locus system to determine mating types is not just an oddity of mushrooms, as it is also found in the self-incompatibility systems of some angiosperms (Barrett and Shore, 2008; Klaas et al., 2011) and the mating-type systems of tunicates and social amoebae (Harada et al., 2008; Bloomfield et al., 2010).
Fig 2.
Different breeding systems in the fungi. Homothallism means that each haploid is compatible with all other haploids and heterothallism is when only cells of different mating types can mate. Bipolarity qualifies heterothallism where the mating type is determined by a single locus while under tetrapolarity mating can only occur when the two mating-type loci carry different alleles. Letters A and B indicate mating-type loci and numbered subscripts indicate alternate alleles, where they occur.
The opposite condition of heterothallism is called homothallism, under which mating is possible between mitotic descendants of the same haploid genotype (Fig. 2). Homothallism can be achieved by different mechanisms: 1) switching such as in ascomyete yeasts, in which non-allelic recombination regularly replaces the active cassette in the mating-type locus by a copy of the alternative transcription factor gene(s) that is present elsewhere in the genome (Haber, 2012); 2) the presence of both mating-type alleles in a haploid genome (Beatty et al., 1994; Paoletti et al., 2007; Gioti et al., 2012); and 3) the possibility, in the absence of mating, for endoduplication to restore the diploid condition or fusion between two haploid nuclei carrying the same mating-type allele (Glass and Smith, 1994; Lin et al., 2005; Wik et al., 2008). Some fungi exhibit a particular system where post-meiotic nuclei of opposite mating types are packaged into spores for dispersal so that restitution of the diploid condition can always occur, with no requirement of finding another compatible mate (Billiard et al., 2011, 2012). This situation has long been called pseudo- or secondary-homothallism based on observation of mating compatibility in vitro, but actually corresponds to a heterothallic breeding system of molecular non-self recognition, with automixis being favored as a mating system.
Taken together, the fungi display a variety of systems for mating-type determination and mating compatibility. These are associated with different, and even opposite, scenarios regarding the extent of recombination around the genes essential for mating compatibility. We outline below the three most contrasting and puzzling situations.
3. Three impacts of recombination associated with mating type
3.1 Recombination suppressed around mating-type loci – driving forces and evolutionary consequences
In plants and animals, suppression of recombination on sex chromosomes is considered to have evolved due to the benefit of linking together multiple genes involved in sex determination, with the further suppression of recombination linking genes that display antagonistic effects in the opposite sex (Bergero and Charlesworth, 2009). The linking of sexually-antagonistic genes is often viewed as occurring in successive steps, creating “evolutionary strata” of ages of linkage and thus degrees of divergence that are usually observed between sex chromosomes (Bergero and Charlesworth, 2009). However, this adaptive explanation for the stepwise increase in the region of recombination cessation is more difficult to apply to fungi because mating types are, with few exceptions (Dettman et al., 2003), not associated with male/female function. In addition, the fungal mating type has generally been considered to have little influence on the fitness of individuals in any part of the life cycle other than mate recognition (Brasier, 1999), with some exceptions that are described later. Selection for linking dozens or hundreds of additional genes to the mating-type loci due to antagonistic selection on mating-type is therefore difficult to imagine.
Yet, recombination is suppressed over substantial distances of 100s of kbp to several mbp around the mating-type loci of several fungi, e.g., Neurospora tetrasperma (Menkis et al., 2008; Ellison et al., 2011), Microbotryum spp. (Hood, 2002; Hood et al., 2013; Badouin et al., 2015; Fontanillas et al., 2015; Whittle et al., 2015), Ustilago hordei (Bakkeren and Kronstad, 1994), Cryptococcus spp. (Fraser and Heitman, 2004b), and Podospora anserina (Grognet et al., 2014). The evolutionary forces resulting in the cessation of recombination appear to differ among fungi. Recombination suppression in some basidiomycetes may have evolved due to the benefit of linking the two mating loci, as the pheromone/receptor genes and the homeodomain genes are found at the edges of the region of suppressed recombination, e.g., in U. hordei and Cryptococcus spp. (Billiard et al., 2012; Nieuwenhuis et al., 2013), and within the region of suppressed recombination in Microbotryum spp. (Petit et al., 2012; Badouin et al., 2015; Fontanillas et al., 2015). Such a linkage of the pre- and post-mating compatibility genes may be advantageous under certain conditions (Nieuwenhuis et al., 2013; Vuilleumier et al., 2013). First, when diploid selfing (i.e., mating between products of meiosis of a given diploid individual) is favored, linkage of compatibility factors may be beneficial because it results in the production of a larger proportion of inter-compatible haploid mating-types. Noting that compatibility requires differences at both pheromone/receptor genes and homeodomain genes, any given haploid in a linked bipolar system is compatible with 50% of the other meiotic products from its diploid parent, versus compatibility with 25% in an unlinked tetrapolar system. Second, a similar force may select for linkage between the pre- and post-mating-type loci in outcrossing basidiomycete species when there are few mating-type alleles in the population (Billiard et al., 2011; Nieuwenhuis et al., 2013).
In N. tetrasperma and Microbotryum spp., recombination is suppressed along most of the mating-type chromosomes (Hood, 2002; Hood et al., 2013; Badouin et al., 2015; Fontanillas et al., 2015; Whittle et al., 2015), and selection for linkage of the mating-type locus to the centromere may have been a contributing factor (Menkis et al., 2008; Hood et al., 2013). In N. tetrasperma, suppressed recombination between the mating-type locus and the centromere ensures that mat A and mat a will segregate at the first division of meiosis, to package meiotic products of different mating type into a single ascospore for dispersal (Raju and Perkins, 1994). This is achieved through the cytological feature of nuclear movement following meiosis in N. tetrasperma that brings together non-sister daughter nuclei of meiosis II. Thus, recombination suppression provides the species with the ability to readily self, since compatible nuclei coexist in the vegetative mycelia emerging from germinated ascospores (Menkis et al., 2008). In addition, the linkage of the mating-type to the centromere may be beneficial in these species under automixis (i.e., intra-tetrad mating or meiotic parthenogenesis), as it allows the restoration of heterozygosity linked to all centromeres and to mating type, which may shelter deleterious mutations or provide conditions for maintaining heterosis (Hood and Antonovics, 2004).
In the face of suppressed recombination causing reduced efficiency of selection and degeneration, the question remains why mating-type loci in some fungi have evolved to encompass so many genes (over 20 in Cryptococcus, more than 200 in P. anserina and over a thousand in N. tetrasperma and Microbotryum spp.), instead of just the essential compatibility genes and/or the centromere. The mechanism to suppress recombination is often the formation of chromosomal inversions (Ellison et al., 2011), ironically caused precisely by unequal crossing-overs, although in some cases the non-recombining regions have remained collinear such as in P. anserina (Grognet et al., 2014). Because few of the genes within these large non-recombining regions seem to be involved in mating-type determination (Bakkeren and Kronstad, 1994; Lengeler et al., 2002; Fraser et al., 2004; Menkis et al., 2008; Badouin et al., 2015; Fontanillas et al., 2015), one may hypothesize that selection would favor their escape into recombining regions where they would not suffer from Muller’s ratchet and Hill-Robertson interference that reduce the power of selection. These cases of non-collinear mating-type chromosomes may represent transient conditions, and selection against genic degeneration may favor the escape of these genes trapped within mating-type loci perhaps by selection for further rearrangements or retrotransposition, as occurred on animal sex chromosomes (Bachtrog, 2013).
The most straightforward hypothesis for explaining large regions of suppressed recombination would seem to be that there is antagonistic selection on mating types for other traits than just mating-type determinism. In fact, some studies have reported mating-type specific traits in fungi, such as gene expression profiles (Samils et al., 2013; Grognet et al., 2014), virulence (Kwon-Chung et al., 1992; Nielsen et al., 2005), or mitochondrial inheritance (Billiard et al., 2011, 2012). In N. tetrasperma, data on mating-type biased gene expression indicate patterns of expression divergence associated with growth on different nutrient regimes, suggesting that individuals of different mating types differ in their fitness optima under different growth conditions (Samils et al., 2013). The latter finding is also supported by a rapid diversification of the mating-type biased expression, leading to the speculation that the rapid divergence of expression is due to selection for a diversification of strains of different mating types (Whittle et al., 2014). In the human pathogen Cryptococcus neoformans alternate mating types also can exhibit different life history trait optima, such as differences in hyphal growth rates and virulence in animal models (Kwon-Chung et al., 1992; Nielsen et al., 2005; Lin et al., 2006, 2008). However, these mating-type specific differences in gene expression or traits may alternatively result from degeneration in some alleles linked to one mating type rather than from antagonistic selection (Fontanillas et al., 2015).
In fact, as haploids of different mating-types need to co-occur in the same ecological niche for mating, it would seem suboptimal that mating types would be selected for different virulence or fitness optima. In addition, the main model of antagonistic pleiotropy between sexes does not seem to apply in fungi for explaining that regions of suppressed recombination encompass several genes around the mating type loci. However, there are alternative hypotheses (Ironside, 2010). For example, in inbred populations, as is strikingly the case for the automictic N. tetrasperma, P. anserina and Microbotryum spp., suppression of recombination can help to shelter deleterious, recessive alleles (Oudemans et al., 1998; Hood and Antonovics, 2000; Thomas et al., 2003), which has been predicted to occur in successive steps (Antonovics and Abrams, 2004; Johnson et al., 2005). Ironically, in the long term this expansion of recombination suppression will lead to further accumulation in the load of deleterious substitutions and insertions.
Alternatively, such large regions of recombination suppression may be hypothesized to protect co-adapted gene complexes, i.e., that suppressed recombination is favored for linking alleles with antagonistic effects, but not in direct association with mating types, instead with the adaptation to different selection regimes (Navarro and Barton, 2003). Linkages would be selected for reasons independent of mating types, and different mating-types would bear the same co-adapted complexes, but linkage would be maintained over longer evolutionary times when they include mating types, due to balancing selection on mating types (Ironside, 2010). In the highly specialized and sympatric plant pathogenic Microbotryum species for instance, one hypothesis is that a suite of genes having co-adapted alleles for host plant specialization are located within the mating-type chromosome. Combinations allowing higher fitness on a given host plant could be selected to be linked altogether by avoiding breaking beneficial allelic combinations in the face of interspecific gene flow (Navarro and Barton, 2003). Both mating-type chromosomes would have the same genes involved in adaptation on a given host within a Microbotryum species, but the linkage of such genes to regions determining mating compatibility in each host-specialized species would allow these genes to display suppressed recombination over longer time frames compared to if they were on autosomes, because negative frequency-dependent selection maintain mating types in balanced frequencies (Ironside, 2010).
3.2 Recombination enhanced adjacent to mating-type loci and independent assortment
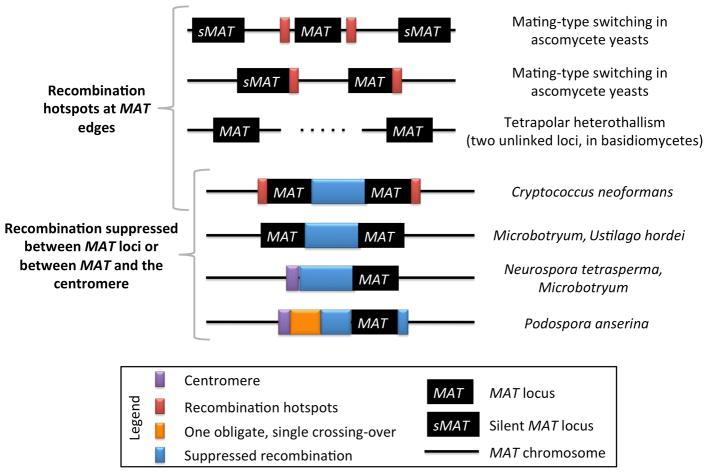
In contrast to the situations outlined in section 3.1, recombination frequencies are extremely high in association with mating-type regions in some fungi, including independent assortment of the component factors on different chromosomes or recombination enhancers in regions that flank mating-type loci (Fig. 3).
Fig 3.
Patterns of recombination suppression or enhancement associated with mating-type loci (MAT) of fungi. Recombination may be enhanced in association with the mating-type loci of fungi, through non-allelic mating-type switching or by allelic recombination hotspots. A “silent” MAT indicates a potentially coding region placed within a transcriptionally quiescent part of the genome. If these regions are recombined into the active parts they are functional. Recombination may be suppressed, in examples in the basidiomycetes in which the two sex-determination regions are linked and/or by linkage to the centromere.
In the tetrapolar basidiomycetes the two mating-type loci, containing the pheromone/receptor and homeodomain protein genes, can be located on different chromosomes and thus experience independent assortment (Kües et al., 2011; Nieuwenhuis et al., 2013). Such independent assortment of two mating-type loci has been interpreted as evidence of selection for discrimination against inbreeding because mating compatibility is being determined by a larger number of independently-segregating, haploid self-incompatibility loci (Billiard et al., 2011, 2012). However, as noted above, whether such a selective process would function to promote outcrossing over diploid-selfing would depend on the level of polymorphism for mating-type alleles in the population. For example, if there is no mating-type diversity among diploid genotypes, the transition from bipolarity to tetrapolarity serves only to decrease the compatibility of random gamete pairs from 50% to 25% regardless of whether they are derived from the same or different diploid parents. When there is polymorphism for intercompatible mating-type alleles, then random gamete compatibility under tetrapolarity will rise above 25%; however, the interdependent drivers of outcrossing (e.g., benefits such as Fisher-Muller effects) and of multiallelism at mating types (e.g., negative frequency-dependent selection) remain difficult to disentangle (Billiard et al., 2011, 2012; Nieuwenhuis et al., 2013).
In ascomycetous yeasts like Saccharomyces cerevisiae and Schizosaccharomyces pombe, a particular mode of mitotic recombination and associated transposition enhancers has evolved around the active mating-type locus to allow mating-type switching. Mating-type switching is a process by which a strain of one mating-type can change to the opposite mating-type, with the replacement of active mating-type transcription factor genes by one of the two alternative quiescent mating-type loci also present in the genome. At least in the case of the yeasts in the Saccharomycotina, it appears that the systems of one active and two silent loci likely evolved from a simpler situation that used chromosome inversions brought about by identical DNA sequence in two regions, as observed in Hansenula polymorpha and Pichia pastoris (Hanson et al., 2014; Maekawa and Kaneko, 2014). Independent domestications of transposable elements have occurred in yeast lineages that allow more complex forms of non-allelic recombination events (Butler et al., 2004; Barsoum et al., 2010; Rajaei et al., 2014). Mating-type switching is one form of homothallism, which may be advantageous because it allows the haploid cell lineage to be compatible for the sexual cycle with any mitotic cell lineage in the population, it allows recombination within a clonal lineage, or can generate karyotypic variants that may be favored by selection (Billiard et al., 2011, 2012).
Aside from the ectopic recombination associated with mating-type switching, in some species the regions flanking the mating-type loci can exhibit greatly enhanced homologuous recombination mediated through crossing-over hotspots. For example, in the basidiomycete C. neoformans recombination is suppressed over a large (~120 kb) mating type-determining region (Lengeler et al., 2002), flanked on either side by hot spots for recombination 10–50 times the genome-wide average frequency (Hsueh et al., 2006). This enhanced recombination rate is not just a physical consequence of a large region with suppressed recombination, because it is also observed in experimental diploids that are homozygous for mating type, and is instead due to the presence of GC-rich motifs (Hsueh et al., 2006; Sun et al., 2014). The evolutionary explanation for these GC-rich recombination hotspots remains undetermined.
An interesting parallel to C. neoformans are the recombining regions at both edges of the mammalian sex chromosomes, called the “pseudo-autosomal regions.” These also constitute hotspots of recombination, being the only defined intra-chromosomal region where recombination occurs in 100% of the cells (Burgoyne, 1982; Yi and Li, 2005; Brick et al., 2012; Sarbajna et al., 2012). The occurrence of at least one crossing-over event per chromosome is thought to facilitate chromosome segregation at meiosis (Whitby, 2005), but it may also be selected for by the benefits that favor recombination (Otto et al., 2011). In P. anserina an obligate and single crossing-over between the mating-type region and the centromere allows heterokaryotic ascospores to be produced that carry sister nuclei of meiosis II that are of opposite mating types. Upon ascospore germination, the resulting mycelia will therefore be able to undergo sex without waiting for a mate (Grognet et al., 2014); this is automixis as the two nuclei enclosed in ascospores result from a single meiosis event (Lewis and John, 1963). It is striking that automixis functions in concert with mating-type determination in N. tetrasperma and M. violaceum by preventing recombination between the mating-type locus and the centromere, while in P. anserina this is achieved by an obligate crossing-over.
3.3 Recombination and gene conversion within MAT – mechanisms of mating-system transition and counteracting molecular degeneration
The accumulation of deleterious mutations in regions of suppressed recombination may be counteracted by gene conversion. Gene conversion is a mechanism of non-reciprocal transfer of DNA sequences between homologous or non-homologous sites of high sequence similarity, such that both copies are the same after conversion. Although this does not constitute recombination per se, it yields similar results of information transfer between homologous chromosomes. Gene conversion between mating-type alleles has been suggested in fungi with large regions of suppressed recombination linked to mating-type loci, such as in N. tetrasperma (Menkis et al., 2010), M. violaceum (Fontanillas et al. 2015) and C. neoformans (Sun et al., 2012a).
In N. tetrasperma, introgression from a related species, freely recombining around its mating-type locus, has also been observed on the mating-type chromosomes with suppressed recombination. The correlation between introgression and a weaker signal of molecular degeneration suggests that introgression is adaptive in reinvigorating the genome from the negative effects of recombination suppressions (Sun et al., 2012b).
An unusual recombination feature is present within the mating-type locus of Sclerotinia sclerotiorum. In this ascomycete, a region of the mating-type locus is inverted every meiotic generation (Chitrampalam et al., 2013). There is no apparent benefit to this process; however, this is one example in which a mating-type locus is capable of high frequency recombination.
Further evidence of recombination associated with mating-type regions exists in homothallic species that appear to have originated from heterothallic ancestors by non-homologous crossing-over event between mating-type alleles. These non-homologous recombination events brought together both mating-type alleles in the same haploid genome, which conferred both mating-type functions and thus universal mating compatibility in most ascomycetes (Gioti et al., 2012). Such a rearrangement to the genomic structures affecting mating compatibility may confer an advantage when waiting for a compatible haploid mate is costly (Billiard et al., 2011), as it confers a universal compatibility for mating, and therefore increases the chance to find a compatible mate (Billiard et al., 2011, 2012). Such events of unusual recombination can cause changes in breeding systems from heterothallism to homothallism as observed in Aspergillus nidulans (Paoletti et al., 2007), Neurospora spp. (Ellison et al., 2011; Gioti et al., 2012), S. sclerotiorum (Chitrampalam et al., 2013), Grosmannia clavigera (Tsui et al., 2013), and Zygosaccharomyces rouxii (Watanabe et al., 2013). Meiotic crossing-over events that reorganize the mating-type locus as a step toward homothallism likely occurred between the homologous chromosomes bearing the mating-type regions.
Mitotic recombination within mating-type loci has been demonstrated, albeit under laboratory conditions, by targeted mutation by replacement with selectable markers of genes within mating-type loci in fungi, with examples from the Mucoromycotina, Ascomycota and Basidiomycota lineages, such as reported in Shen et al., 2002; El-Khoury et al., 2008; Li et al., 2011; Grognet et al., 2014. These experiments provide additional lines of evidence that mating-type loci are not by default impaired for crossing over, although with a caveat that meiotic recombination may use pathways that are similar to mitotic crossing over but also those that are specific to meiosis (Kohl and Sekelsky, 2013).
4. Concluding remarks and future directions
The genes involved with mating compatibility are structurally arranged in a diversity of ways in fungi, which has resulted in a range of effects on recombination. There may be highly localized regions of meiotic recombination or conversely a great expansion of linkage that incorporates the majority of the chromosome. Fungi thus represent a fascinating group to study the evolutionary causes and consequences of recombination suppression and enhancement, being tractable systems, with their relative simplicity of mating compatibility mechanisms and their small genomes.
Many interesting questions remain to be investigated about the evolution of mating-type chromosomes in fungi. In particular, the question of why so many genes not involved in mating-type determination are sometimes trapped within non-recombining mating-type loci in fungi deserves further investigations. The contrasted patterns of cross-over and mating-type are based on examples from a small number of species. We thus also need a broader survey of the diversity of fungal systems that have been little studied to be able to begin to draw generalities. Further experimental work should address the adaptive significance of traits (e.g., tetrapolarity and outcrossing, recombination suppression or hotspots) versus non-adaptive explanations (Ironside, 2010). The discoveries from such studies promise to enlighten understanding of the evolution of sex-determination regions of genomes from a new perspective.
Mating type loci and sex chromosomes are associated with suppressed recombination.
Exceptions to recombination suppression are known in the fungi.
Some fungal mating systems rely on or include increased recombination frequencies.
Acknowledgments
Our research has been supported by the US National Science Foundation grants DEB-1115765 and DEB-0747222, National Institutes of Health grant R21 AI094364, the Australian Research Council, the FungiSex ANR-09-0064-01 grant, and the ERC starting grant GenomeFun 309403.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonovics J, Abrams JY. Intratetrad mating and the evolution of linkage relationships. Evolution. 2004;58:702–709. doi: 10.1111/j.0014-3820.2004.tb00403.x. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman T-L, Hahn MW, Kitano J, Mayrose I, Ming R, Perrin N, Ross L, Valenzuela N, Vamosi JC. Sex determination: why so many ways of doing it? PLoS Biol. 2014;12:e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouin H, Hood ME, Gouzy J, Aguileta G, Siguenza S, Perlin MH, Cuomo CA, Fairhead C, Branca A, Giraud T. Chaos of rearrangements in in the mating-type chromosomes of the anther-smut fungus Microbotryum lychnidis-dioicae. Genetics. 2015 doi: 10.1534/genetics.115.177709. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkeren G, Kronstad JW. Linkage of mating type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc Natl Acad Sci USA. 1994;91:7085–7089. doi: 10.1073/pnas.91.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SCH, Shore JS. New insights on heterostyly: Comparative biology, ecology and genetics. In: Franklin-Tong V, editor. Self-Incompatibility in Flowering Plants: Evolution, Diversity and Mechanisms. Springer-Verlag; Berlin: 2008. pp. 3–32. [Google Scholar]
- Barsoum E, Martinez P, Åström SU. α3, a transposable element that promotes host sexual reproduction. Genes Dev. 2010;24:33–44. doi: 10.1101/gad.557310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty NP, Smith ML, Glass NL. Molecular characterization of mating-type loci in selected homothallic species of Neurospora, Gelasinospora and Anixiella. Mycol Res. 1994;98:1309–1316. [Google Scholar]
- Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Beukeboom L, Perrin N. The Evolution of Sex Determination. Oxford University Press; Oxford, UK: 2014. [Google Scholar]
- Billiard S, López-Villavicencio M, Hood ME, Giraud T. Sex, outcrossing and mating types: unsolved questions in fungi and beyond. J Evol Biol. 2012;25:1020–1038. doi: 10.1111/j.1420-9101.2012.02495.x. [DOI] [PubMed] [Google Scholar]
- Billiard S, López-Villavicencio M, Devier B, Hood ME, Fairhead C, Giraud T. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev. 2011;86:421–442. doi: 10.1111/j.1469-185X.2010.00153.x. [DOI] [PubMed] [Google Scholar]
- Blackwell M. The fungi: 1, 2, 3 … 5.1 million species? Am. J Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- Blaser O, Neuenschwander S, Perrin N. Sex-chromosome turnovers: the hot-potato model. Am Nat. 2014;183:140–146. doi: 10.1086/674026. [DOI] [PubMed] [Google Scholar]
- Bloomfield G, Skelton J, Ivens A, Tanaka Y, Kay RR. Sex determination in the social amoeba Dictyostelium discoideum. Science. 2010;330:1533–1536. doi: 10.1126/science.1197423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton MD, de Jonge R, Inderbitzin P, Liu Z, Birla K, Van de Peer Y, Subbarao KV, Thomma BPHJ, Secor GA. The heterothallic sugarbeet pathogen Cercospora beticola contains exon fragments of both MAT genes that are homogenized by concerted evolution. Fungal Genet Biol. 2014;62:43–54. doi: 10.1016/j.fgb.2013.10.011. [DOI] [PubMed] [Google Scholar]
- Brasier CM. Fitness, continuous variation and selection in fungal populations: an ecological perspective. In: Worrall JJ, editor. Structure and dynamics of fungal populations. Springer; 1999. pp. 307–339. [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Casselton LA. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet. 2001;17:393–400. doi: 10.1016/s0168-9525(01)02343-5. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum Genet. 1982;61:85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- Butler G. The evolution of MAT: the Ascomycetes. In: Heitman J, et al., editors. Sex in Fungi: molecular determination and evolutionary implications. ASM Press; Washington, D.C: 2007. pp. 3–18. [Google Scholar]
- Butler G, Kenny C, Fagan A, Kurischko C, Gaillardin C, Wolfe KH. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc Natl Acad Sci USA. 2004;101:1632–1637. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitrampalam P, Inderbitzin P, Maruthachalam K, Wu BM, Subbarao KV. The Sclerotinia sclerotiorum mating type locus (MAT) contains a 3.6-kb region that is inverted in every meiotic generation. PLoS One. 2013;8:e56895. doi: 10.1371/journal.pone.0056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R, Berteaux-Lecellier V, Silar P. Mating systems and sexual morphogenesis in ascomycetes. In: Borkovich KA, et al., editors. Cellular and Molecular Biology of Filamentous Fungi. ASM Press; Washington, DC: 2010. pp. 501–535. [Google Scholar]
- Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW. Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution. 2003;57:2721–2741. doi: 10.1111/j.0014-3820.2003.tb01515.x. [DOI] [PubMed] [Google Scholar]
- Devier B, Aguileta G, Hood ME, Giraud T. Ancient trans-specific polymorphism at pheromone receptor genes in basidiomycetes. Genetics. 2009;181:209–223. doi: 10.1534/genetics.108.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khoury R, Sellem CH, Coppin E, Boivin A, Maas MFPM, Debuchy R, Sainsard-Chanet A. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr Genet. 2008;53:249–258. doi: 10.1007/s00294-008-0180-3. [DOI] [PubMed] [Google Scholar]
- Ellison CE, Stajich JE, Jacobson DJ, Natvig DO, Lapidus A, Foster B, Aerts A, Riley R, Lindquist EA, Grigoriev IV, Taylor JW. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics. 2011;189:55–69. doi: 10.1534/genetics.111.130690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrügge M, Kämper J, Steinberg G, Kahmann R. Regulation of mating and pathogenic development in Ustilago maydis. Curr Opin Microbiol. 2004;7:666–672. doi: 10.1016/j.mib.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Fontanillas E, Hood ME, Petit E, Barbe V, Gouzy J, Badouin H, de Vienne DM, Aguileta G, Poulain J, Wincker P, Chen Z, Toh SS, Cuomo C, Perlin MH, Giraud T. Degeneration of the non-recombining regions in the mating type chromosomes of the anther smut fungi. Mol Biol Evol. 2015;32:928–943. doi: 10.1093/molbev/msu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Heitman J. Evolution of fungal sex chromosomes. Mol Microbiol. 2004a;51:299–306. doi: 10.1046/j.1365-2958.2003.03874.x. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Heitman J. Fungal mating-type loci. Curr Biol. 2004b;31:R792–R795. doi: 10.1016/j.cub.2003.09.046. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:2243–2255. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gäumann EA. The Fungi. Hafner Publishing Company; New York, London: 1952. [Google Scholar]
- Gillespie JH. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics. 2000;155:909–919. doi: 10.1093/genetics/155.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioti A, Mushegian AA, Strandberg R, Stajich JE, Johannesson H. Unidirectional evolutionary transitions in fungal mating systems and the role of transposable elements. Mol Biol Evol. 2012;29:3215–3226. doi: 10.1093/molbev/mss132. [DOI] [PubMed] [Google Scholar]
- Glass NL, Smith ML. Structure and function of a mating-type gene from the homothallic species Neurospora africana. Mol Gen Genet. 1994;244:401–409. doi: 10.1007/BF00286692. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Grognet P, Bidard F, Kuchly C, Tong LCH, Coppin E, Benkhali JA, Couloux A, Wincker P, Debuchy R, Silar P. Maintaining two mating types: structure of the mating type locus and its role in heterokaryosis in Podospora anserina. Genetics. 2014;197:421–432. doi: 10.1534/genetics.113.159988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SJ, Byrne KP, Wolfe KH. Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. Proc Natl Acad Sci USA. 2014;111:E4851–8. doi: 10.1073/pnas.1416014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Takagaki Y, Sunagawa M, Saito T, Yamada L, Taniguchi H, Shoguchi E, Sawada H. Mechanism of self-sterility in a hermaphroditic chordate. Science. 2008;320:548–550. doi: 10.1126/science.1152488. [DOI] [PubMed] [Google Scholar]
- Hood ME. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics. 2002;160:457–461. doi: 10.1093/genetics/160.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Antonovics J. Intratetrad mating, heterozygosity, and the maintenance of deleterious alleles in Microbotryum violaceum (=Ustilago violacea) Heredity. 2000;85:231–241. doi: 10.1046/j.1365-2540.2000.00748.x. [DOI] [PubMed] [Google Scholar]
- Hood ME, Antonovics J. Mating within the meiotic tetrad and the maintenance of genomic heterozygosity. Genetics. 2004;166:1751–1759. doi: 10.1534/genetics.166.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Petit E, Giraud T. Extensive divergence between mating-type chromosomes of the anther-smut fungus. Genetics. 2013;193:309–315. doi: 10.1534/genetics.112.146266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y-P, Idnurm A, Heitman J. Recombination hotspots flank the Cryptococcus mating-type locus: Implications for the evolution of a fungal sex chromosome. PLoS Genet. 2006;2:e184. doi: 10.1371/journal.pgen.0020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Hamilton WD, Ladle RJ. Covert sex. Trends Ecol Evol. 1992;7:144–5. doi: 10.1016/0169-5347(92)90205-P. [DOI] [PubMed] [Google Scholar]
- Idnurm A. Sex and speciation: The paradox that non-recombining DNA promotes recombination. Fungal Biol Rev. 2011;25:121–127. doi: 10.1016/j.fbr.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ironside JE. No amicable divorce? Challenging the notion that sexual antagonism drives sex chromosome evolution. BioEssays. 2010;32:718–726. doi: 10.1002/bies.200900124. [DOI] [PubMed] [Google Scholar]
- Jakočiūnas T, Holm LR, Verhein-Hansen J, Trusina A, Thon G. Two portable recombination enhancers direct donor choice in fission yeast heterochromatin. PLoS Genet. 2013;9:e1003762. doi: 10.1371/journal.pgen.1003762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LJ, Antonovics J, Hood ME. The evolution of intratetrad mating rates. Evolution. 2005;59:2525–2532. [PubMed] [Google Scholar]
- Klaas M, Yang B, Bosch M, Thorogood D, Manzanares C, Armstead IP, Franklin FCH, Barth S. Progress towards elucidating the mechanisms of self-incompatibility in the grasses: further insights from studies in Lolium. Ann Bot. 2011;108:677–685. doi: 10.1093/aob/mcr186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniep H. Untersuchungen über den Antherenbrand (Ustilago violacea Pers.). Ein Beitrag zum Sexualitätproblem. Z Bot. 1919;11:257–284. [Google Scholar]
- Kohl KP, Sekelsky J. Meiotic and mitotic recombination in meiosis. Genetics. 2013;194:327–334. doi: 10.1534/genetics.113.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS. Selection against harmful mutations in large sexual and asexual populations. Genet Res. 1982;40:325–332. doi: 10.1017/s0016672300019194. [DOI] [PubMed] [Google Scholar]
- Kües U, James TY, Heitman J. Mating type in basidiomycetes: Unipolar, bipolar, and tetrapolar paterns of sexuality. In: Wöstemeyer J, Pöggeler S, editors. The Mycota XIV. Evolution of fungi and fungal-like organisms. Springer-Verlag; Berlin, Heidelberg: 2011. [Google Scholar]
- Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, Heitman J. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell. 2002;1:704–718. doi: 10.1128/EC.1.5.704-718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KR, John B. Chromosome Marker. Little, Brown & Co; Boston: 1963. [Google Scholar]
- Li CH, Cervantes M, Springer DJ, Boekhout T, Ruiz-Vazquez RM, Torres-Martinez SR, Heitman J, Lee SC. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 2011;7:e1002086. doi: 10.1371/journal.ppat.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- Lin X, Huang JC, Mitchell TG, Heitman J. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATα allele enhances filamentation. PLoS Genet. 2006;2:e187. doi: 10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Nielsen K, Patel S, Heitman J. Impact of mating type, serotype, and ploidy on the virulence of Cryptococcus neoformans. Infect Immun. 2008;76:2923–2938. doi: 10.1128/IAI.00168-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H, Kaneko Y. Inversion of the chromosomal region between two mating type loci switches the mating type in Hansenula polymorpha. PLoS Genet. 2014;10:e1004796. doi: 10.1371/journal.pgen.1004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Avise JC. Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex Dev. 2009;3:60–67. doi: 10.1159/000223071. [DOI] [PubMed] [Google Scholar]
- Marais GAB, Campos PRA, Gordo I. Can intra-Y gene conversion oppose the degeneration of the human Y chromosome? A simulation study. Genome Biol Evol. 2010;2:347–357. doi: 10.1093/gbe/evq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Graves JA. The rise and fall of SRY. Trends Genet. 2002;18:259–264. doi: 10.1016/s0168-9525(02)02666-5. [DOI] [PubMed] [Google Scholar]
- Menkis A, Whittle CA, Johannesson H. Gene genealogies indicates abundant gene conversions and independent evolutionary histories of the mating-type chromosomes in the evolutionary history of Neurospora tetrasperma. BMC Evol Biol. 2010;10:234. doi: 10.1186/1471-2148-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkis A, Jacobson DJ, Gustafsson T, Johannesson H. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 2008;4:e1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg RL, Glass NL. Mating type and mating strategies in Neurospora. Bioessays. 1990;12:53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;1:1–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton NH. Chromosomal speciation and molecular divergence--accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Marra RE, Hagen F, Boekhout T, Mitchell TG, Cox GM, Heitman J. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics. 2005;171:975–983. doi: 10.1534/genetics.105.045039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis BPS, Billiard S, Vuilleumier S, Petit E, Hood ME, Giraud T. Evolution of uni- and bifactorial sexual compatibility systems in fungi. Heredity. 2013;111:445–455. doi: 10.1038/hdy.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Pannell JR, Peichel CL, Ashman T-L, Charlesworth D, Chippindale AK, Delph LF, Guerrero RF, Scarpino SV, McAllister BF. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 2011;27:358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Oudemans PV, Alexander HM, Antonovics J, Altizer S, Thrall PH, Rose L. The distribution of mating-type bias in natural populations of the anther-smut Ustilago violacea on Silene alba in Virginia. Mycologia. 1998;90:372–381. [Google Scholar]
- Paoletti M, Seymour FA, Alcocer MJC, Kaur N, Calvo AM, Archer DB, Dyer PS. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol. 2007;17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Petit E, Giraud T, de Vienne DM, Coelho MA, Aguileta G, Amselem J, Kreplak J, Poulain J, Gavory F, Wincker P, Young SK, Cuomo C, Perlin MH, Hood ME. Linkage to the mating-type locus across the genus Microbotryum: insights into nonrecombining chromosomes. Evolution. 2012;66:3519–3533. doi: 10.1111/j.1558-5646.2012.01703.x. [DOI] [PubMed] [Google Scholar]
- Rajaei N, Chiruvella KK, Lin F, Åström SU. Domesticated transposase Kat1 and its fossil imprints induce sexual differentiation in yeast. Proc Natl Acad Sci USA. 2014;111:15491–15496. doi: 10.1073/pnas.1406027111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju NB, Perkins DD. Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora, and Podospora. Dev Genet. 1994;15:104–118. doi: 10.1002/dvg.1020150111. [DOI] [PubMed] [Google Scholar]
- Raudaskoski M, Kothe E. Basidiomycete mating type genes and pheromone signaling. Eukaryot Cell. 2010;9:847–859. doi: 10.1128/EC.00319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samils N, Gioti A, Karlsson M, Sun Y, Kasuga T, Bastiaans E, Wang Z, Li N, Townsend JP, Johannesson H. Sex-linked transcriptional divergence in the hermaphrodite fungus Neurospora tetrasperma. Proc R Soc B. 2013;280 doi: 10.1098/rspb.2013.0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbajna S, Denniff M, Jeffreys AJ, Neumann R, Soler Artigas M, Veselis A, May CA. A major recombination hotspot in the XqYq pseudoautosomal region gives new insight into processing of human gene conversion events. Hum Mol Genet. 2012;21:2029–2038. doi: 10.1093/hmg/dds019. [DOI] [PubMed] [Google Scholar]
- Shen WC, Davidson RC, Cox GM, Heitman J. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot. Cell. 2002;1:366–377. doi: 10.1128/EC.1.3.366-377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Hsueh Y-P, Heitman J. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 2012a;8:e1002810. doi: 10.1371/journal.pgen.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Billmyre RB, Mieczkowski PA, Heitman J. Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans. PLoS Genet. 2014;10:e1004849. doi: 10.1371/journal.pgen.1004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Corcoran P, Menkis A, Whittle CA, Andersson SGE, Johannesson H. Large-scale introgression shapes the evolution of the mating-type chromosomes of the filamentous ascomycete Neurospora tetrasperma. PLoS Genet. 2012b;8:e1002820. doi: 10.1371/journal.pgen.1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Shykoff J, Jonot O, Giraud T. Mating-type ratio bias in populations of the phytopathogenic fungus Microbotryum violaceum from several host species. Int J Plant Sci. 2003;164:641–647. [Google Scholar]
- Trombetta B, Cruciani F, Underhill PA, Sellitto D, Scozzari R. Footprints of X-to-Y gene conversion in recent human evolution. Mol Biol Evol. 2010;27:714–725. doi: 10.1093/molbev/msp231. [DOI] [PubMed] [Google Scholar]
- Tsui CKM, DiGuistini S, Wang Y, Feau N, Dhillon B, Bohlmann J, Hamelin RC. Unequal recombination and evolution of the mating-type (MAT) loci in the pathogenic fungus Grosmannia clavigera and relatives. G3. 2013;3:465–480. doi: 10.1534/g3.112.004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Diepen LTA, Olson Å, Ihrmark K, Stenlid J, James TY. Extensive trans-specific polymorphism at the mating type locus of the root decay fungus Heterobasidion. Mol Biol Evol. 2013;30:2286–2301. doi: 10.1093/molbev/mst126. [DOI] [PubMed] [Google Scholar]
- Vuilleumier S, Alcala N, Niculita-Hirzel H. Transitions from reproductive systems governed by two self-incompatible loci to one in fungi. Evolution. 2013;67:501–516. doi: 10.1111/j.1558-5646.2012.01783.x. [DOI] [PubMed] [Google Scholar]
- Watanabe J, Uehara K, Mogi Y. Diversity of mating-type chromosome structures in the yeast Zygosaccharomyces rouxii caused by ectopic exchanges between MAT-like loci. PLoS One. 2013;8:e62121. doi: 10.1371/journal.pone.0062121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC. Making crossovers during meiosis. Biochem Soc Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Johannesson H. Evolution of mating-type loci and mating-type chromosomes in model species of filamentous ascomycetes. In: Wöstemeyer J, Pöggeler S, editors. The Mycota XIV. Evolution of fungi and fungal-like organisms. Springer-Verlag; Berlin, Heidelberg: 2011. pp. 277–292. [Google Scholar]
- Whittle CA, Sun Y, Johannesson H. Degeneration in codon usage within the region of suppressed recombination in the mating-type chromosomes of Neurospora tetrasperma. Eukaryot. Cell. 2011;10:594–603. doi: 10.1128/EC.00284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CA, Sun Y, Johannesson H. Dynamics of transcriptome evolution in the model eukaryote Neurospora. J Evol Biol. 2014;27:1125–1135. doi: 10.1111/jeb.12386. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Votintseva A, Ridout K, Filatov DA. Recent and massive expansion of the mating-type-specific region in the smut fungus Microbotryum. Genetics. 2015;199:809–816. doi: 10.1534/genetics.114.171702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wik L, Karlsson M, Johannesson H. The evolutionary trajectory of the mating-type (mat) genes in Neurospora relates to reproductive behavior of taxa. BMC Evol Biol. 2008;8:109. doi: 10.1186/1471-2148-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Li W-H. Molecular evolution of recombination hotspots and highly recombining pseudoautosomal regions in hominoids. Mol Biol Evol. 2005;22:1223–1230. doi: 10.1093/molbev/msi106. [DOI] [PubMed] [Google Scholar]