Abstract
CLIP proteases are non-digestive serine proteases present in hemolymph of insects and other arthropods. They are composed of one or more amino-terminal clip domains followed by a linker sequence and a carboxyl-terminal S1A family serine protease domain. The genes for CLIP proteases have evolved as four clades (CLIPA, CLIPB, CLIPC, CLIPD), each present as multigene families in insect genomes. CLIP proteases in hemolymph function in innate immune responses. These include proteolytic activation of the cytokine Spätzle, to form an active Toll ligand leading to synthesis of antimicrobial peptides, and specific activation of prophenoloxidase, required for the melanization response. CLIP proteases act in cascade pathways. In the immune pathways that have been characterized, microbial surface molecules stimulate activation of an initiating modular serine protease, which then activates a CLIPC, which in turn activates a CLIPB. The active CLIPB then cleaves and activates an effector molecule (proSpätzle or prophenoloxidase). CLIPA proteins are pseudoproteases, lacking proteolytic activity, but some can function as regulators of the activity of other CLIP proteases and form high molecular weight immune complexes. A few three dimensional structures for CLIP proteases are now available for structure-function analysis of these immune factors, revealing structural features that may act in specific activation or in formation of immune complexes. The functions of most CLIP proteases are unknown, even in well studied insect species. It is very likely that additional proteins activated by CLIP proteases and acting in immunity remain to be discovered.
Introduction
Specific proteolysis is the most prominent posttranslational modification of extracellular proteins and a mechanism for regulating their activity [1]. Extracellular serine protease pathways have evolved in animals to stimulate rapid responses to tissue damage and pathogen invasion [2]. Protease cascade pathways offer mechanisms for rapid, local amplification of a small initial signal, with regulation at multiple levels [3]. The proteases circulate as inactive zymogens and become sequentially activated upon recognition of aberrant tissues or microbial polysaccharides. Through specific molecular interactions and limited proteolysis, a localized reaction is rapidly initiated to stop bleeding, dismantle clots, or attack invading microorganisms. After accomplishing their functions, the active enzymes are inactivated by serine protease inhibitors, especially members of the serpin superfamily [4,5].
Protease cascade systems have evolved in innate immune systems of insects [6,7]. Biochemical, genetic, and molecular biological approaches have led to varying degrees of understanding of these pathways in a few insect species. Serine proteases (SPs) containing one or more amino-terminal clip domains [8] function in extracellular pathways that regulate some immune responses of insects (Fig. 1, 2). Clip domains were named because a diagram of the disulfide bond pattern in the N-terminal domain of horseshoe crab clotting enzyme [9] resembled a paper clip (Shun-ichiro Kawabata, personal communication). CLIP proteases (Snake and Easter) also participate in a protease cascade that regulates dorsal-ventral pattern in Drosophila melanogaster embryos [10]. The CLIP proteases represent a protein architecture apparently unique to invertebrates. They have been identified in arthropods and molluscs, and they form large gene families in the insect genomes studied so far (ranging from 15 CLIP protease genes in Bombyx mori to 42 in Manduca sexta and 63 in Aedes aegypti) [11-16]. Infections can stimulate activation of CLIP protease zymogens in hemolymph, with specific cleavage at a site at the amino-terminus of the protease domain, creating a two-chain form active form of the enzyme, in which the clip domain and protease domain remain connected by a disulfide bond (Fig. 1). Immune cascade pathways containing CLIP proteases lead to activation of prophenoloxidase (proPO) [17-26] or the Toll-ligand Spätzle [19,27-30] (Fig. 2). Once CLIP proteases are activated, they are regulated by serpin inhibitors in hemolymph plasma [31-34]. Some members of the CLIP superfamily contain a protease domain with mutations of one or more of the catalytic triad residues, such that they lack proteolytic activity. Such serine protease homologs (SPH) can function as cofactors required for proPO activation by an active CLIP protease [35], and they can also negatively regulate the melanization response [36].
Figure 1. Domain architecture of CLIP serine proteases.
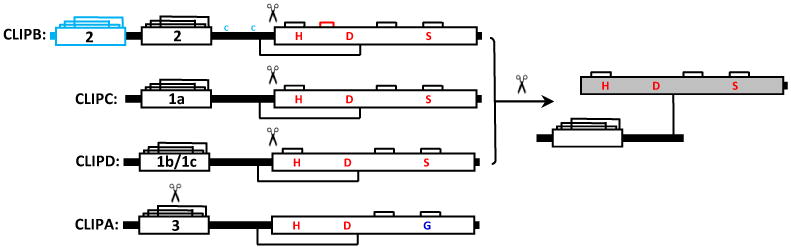
CLIP proteases contain one or more amino-terminal clip domains connected by a linker sequence to a carboxyl-terminal serine protease domain. The protease zymogen is activated by specific cleavage at the beginning of the catalytic domain. After this cleavage, the clip domain and protease domain remain connected by an interchain disulfide bond. CLIP proteases that have active sites with an intact catalytic triad (H, D, S) fall into three groups based on sequence alignments, known as clades B, C, and D. CLIPB proteases contain one or two amino-terminal clip domains from sequence type 2. CLIPB proteases include Manduca PAP1, PAP2, PAP3, HP8, Bombyx PPAE and BAEEase, Holotrichia PPAF1 and PPAF3, Tenebrio SPE, Drosophila SPE, MP1, MP2, easter, and Grass, Aedes IMP1, IMP2, TMP, B5, B29, B35, and Anopheles B4, B8, B9, B14. Manduca PAP2, PAP3, and Bombyx PPAE have two clip domains and two extra Cys residues in the linker (shown in light blue). CLIPC proteases, containing a single clip domain from group 1a include Manduca HP6 and Drosophila Persephone and Spirit. CLIPD proteases contain one clip domain from type 1b or 1c. At this time, there are no members of the CLIPD family with a known function. CLIPA pseudoproteases, known as serine protease homologs, have an amino terminal clip domain from type 3 and a protease-like domain in which the active site serine residue is changed to glycine, and therefore these proteins lack protease activity. CLIPA proteins are apparently activated by a specific cleavage in the clip domain. Manduca SPH1a, SPH2, and Anopheles CLIPA8 are examples of CLIPAs. (For simplicity, additional Cys residues in some of the linkers and protease domains are not indicated.)
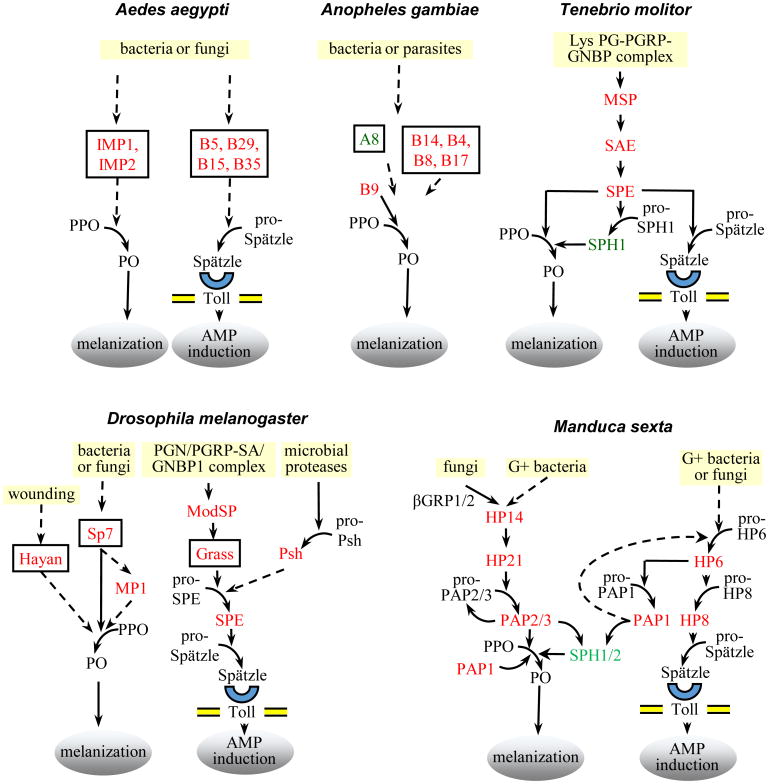
Figure 2. Protease cascades in insect immune responses.
Microbial stimuli lead to activation of CLIP proteases, organized in pathways that result in activation of proPO or proSpätzle. For protease names shown in boxes, genetic evidence indicates participation in an immune pathway, but the activating protease and the protease's substrate are not yet known. Dashed arrows indicate putative steps that have not been verified experimentally. The diagrams summarize data from the following: Aedes aegypti [55,56], Anopheles gambiae [36,45,57-59], Tenebrio molitor [24,29,60,61], Drosophila melanogaster [25-28,62-68], Manduca sexta [19,21-23,30,35,42,43,54,69-75] and unpublished results from the authors' laboratories.
In D. melanogaster, Ae. aegypti, and Anopheles gambiae, genetic and RNAi analyses have demonstrated that certain CLIP proteases have a role in regulation of melanization or the Toll pathway (Fig. 2), but substrates of the proteases and the identity of their activating enzymes are generally not yet known. Exceptions are D. melanogaster SPE, which activates proSpätzle [27,29], D. melanogaster SP7 (MP2), which can activate proPO [20], and An. gambiae CLIPB9, which can activate M. sexta proPO [37]. In two beetles, Tenebrio molitor and Holotrichia diomphalia [33], and in a moth, M. sexta [34], biochemical studies have led to more detailed understanding of cascade pathways including CLIP proteases, which activate proPO and proSpätzle. In these systems, direct activation of downstream proteins by specific proteases has been achieved using purified proteins (Fig. 2). However, it is clear that much remains to be learned about the functions of hemolymph CLIP proteases, with biochemical functions known for only a few members of this family.
Structure of CLIP proteases
Phylogenetic analyses of CLIP proteases has revealed four distinct clades in this family named A, B, C, and D [13,16]. Detailed understanding of structure and function of CLIP proteases is currently limited by a lack of three-dimensional structural data for most members of this family. Available structures include the crystal structures of the D. melanogaster Grass zymogen, a CLIPB (Protein Data Bank accession 2XXL) [38], the protease domain of H. diomphalia proPO-activating factor-1 (PPAF1), also a CLIPB (2OLG) [39], clip domain pseudoprotease H. diomphalia PPAF2, a CLIPA (2B9L) [40], and the two clip domains from M. sexta proPO-activating protease-2 (PAP2, a CLIPB), solved by NMR spectroscopy (2IKD, 2IKE) [41].
The carboxyl-terminal catalytic or protease-like domains of CLIP proteases adopt a chymotrypsin fold consisting of two adjacent β-barrel-like structures arranged perpendicularly to each other (Fig. 3). Each unit contains six antiparallel β-strands, with hydrophobic residues holding the barrels together at their interface. The catalytic residues (His, Asp, Ser) are in the cleft formed between the two barrels. Following chymotrypsin nomenclature, surface loops 30, 60, and 140 connect the secondary structure elements and control the access of protein substrates. CLIPBs possess an additional loop closed by a disulfide bond that does not exist in the other groups of CLIP proteases (Figs. 1 and 3). This protruding structure may block access to the activation cleavage site, enhancing the specificity of zymogen activation [38,39].
Figure 3. Three dimensional representations of insect CLIP proteases.
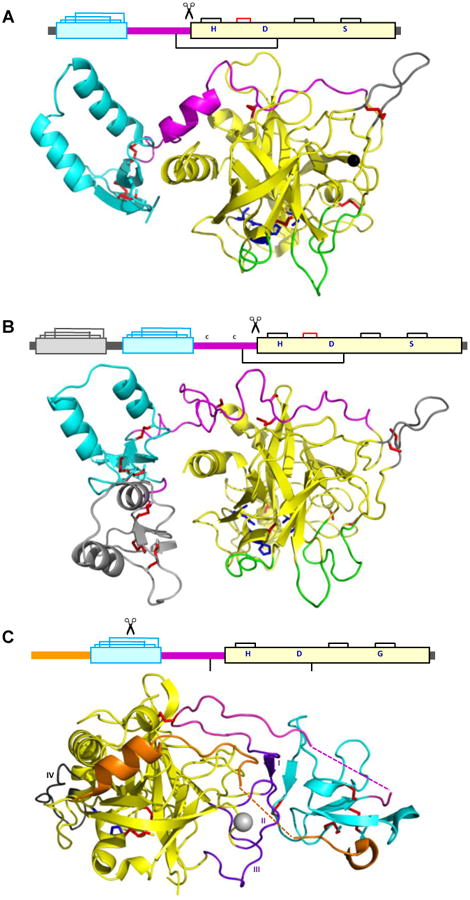
(A) Drosophila melanogaster Grass zymogen structure solved by x-ray crystallography (2XXL) [38]. The ribbon diagram shows the clip domain (cyan), linker (magenta), and the catalytic domain (yellow). Loops 30, 60, and 140 in the catalytic domain are shown in green; the signature loop 75 of CLIPBs is in gray; a calcium ion is shown as a gray sphere. Side chains of the Cys and catalytic residues (S, H and D) are colored red and blue, respectively. (B) Manduca sexta PAP2 structural model based on the NMR structure of its clip domains [41] and a homology model of the PAP2 catalytic domain. The first clip domain is in gray, the second clip domain is in cyan, and the rest of the molecule is colored using the same scheme described in (A). (C) Holotrichia diomphalis PPAF2 crystal structure (2B9L) [39]. In the PPAF2 structure, regions I, II and III (purple) of the protease-like domain interact with the clip domain (cyan). Region IV (gray) may interact with another protein. The amino-terminal extension (orange) and the linker (magenta), including the unstructured parts (dashed line) and the “interchain” disulfide bond, further stabilize the association of the clip domain with the protease-like domain.
Clip domains are ∼35–55 residue sequences with a conserved pattern of three disulfide bonds (Pfam accession number PF12032). Based on the CLIP proteases identified so far, this domain can be broadly defined as CX1-14CX3-10CX8-36CX4-18CC, with substantial sequence variation in the regions between Cys residues. Clip domain sequences fall into three major groups based on sequence similarity and the distance between certain Cys residues. These clip domain groups are labeled types 1, 2, and 3, with three subgroups identified in type 1 (1a, 1b, 1c) [16]. There is a conserved association of certain clip domain types with protease domain types, indicating a co-evolution of distinct combinations of the clip domain and protease domain types. CLIPA proteins have type 3 clip domains, CLIPB proteins have type 2 clip domains, CLIPC proteins have clip domain type 1a, and CLIPD proteins have type 1b or 1c clip domains. The disulfide linkage patterns of most clip domains have not been experimentally determined, but the absolutely conserved Cys1 and Cys5, Cys2 and Cys4, and Cys3 and Cys6 are predicted to form three disulfide bonds, based on the disulfide pattern known from a few CLIP proteases [9,38,40]. Cys1, Cys2, and Cys5-Cys6 are located in three β-strands that form an antiparallel β-sheet, and the region between Cys3 and Cys4 provides unique sequence features that help to define in the different types of clip domains. Group-2 clip domains in Drosophila Grass [38] and Manduca PAP2 (27) contain a helix-turn-helix structure in this region. This feature is predicted in 21 domain models of all 13 of the CLIPBs in M sexta, and may also exist in group-1a and 1b clip domains [16], although the length between Cys3 and Cys4 is shorter in group-1, and one or both helices may be shorter or deformed. A hydrophobic cavity present in the known clip domain structures and models [16,38,41] may function in binding interactions with other proteins, helping to regulate or localize components of CLIP protease cascades.
An evolutionary conserved protease cascade module: CLIPC proteases activate CLIPB proteases, which in turn activate immune effector proteins or cytokines
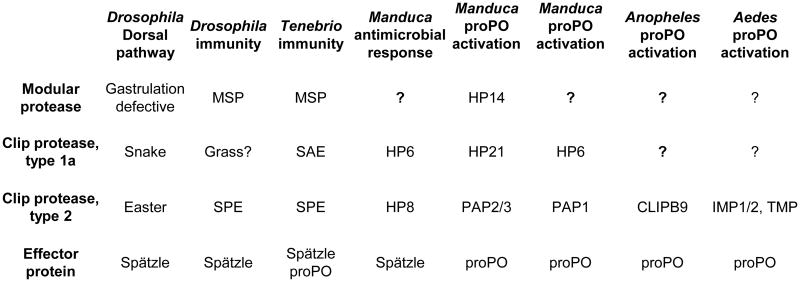
As described above, insect CLIP proteases with known functions fall into two main groups, CLIPB and CLIPC, based on overall sequence comparison and on structural features of their clip domains [8,12,19,38]. For CLIP proteases of known function, these two groups of proteases can also be distinguished based on their position in cascade pathways [19] (Figs. 4, 5). Phylogenetic analysis based on alignment of the protease domains places the CLIP proteases of known function in two clades. One clade contains the enzymes known to activate proSpätzle or proPO (terminal proteases, all from the CLIPB group). The other clade, the CLIPC group, contains enzymes upstream in the pathways; those with known substrates are penultimate proteases in the pathways, cleaving and activating the terminal proteases. Within the terminal protease clade, the enzymes that activate proSpätzle (Easter, SPEs, HP8) cluster together, as do those that activate proPO (Manduca PAPs, Bombyx PPAE, Holotrichia PPAF1, Tenebrio SPE, Drosophila SP7). The terminal proteases all have a basic residue, Arg or Lys, at their activation site, whereas the penultimate proteases instead have Leu (Manduca HP21, Tenebrio SAE, Drosophila Snake), His (Drosophila Persephone,, Manduca HP6), or Ser (Drosophila Spirit) at this position. The terminal proteases all contain the distinctive 75-loop in the protease domain, whereas penultimate proteases do not [38]. These two groups of proteases also differ consistently in the length of the sequence between the third and fourth Cys residues of their clip domains, a feature previously used to define two groups of CLIP proteases [8]. Type 2 clip domains have 22-24 residues between Cys 3-4, whereas type 1 clip domains typically have 15-17 residues at the same position. This region, forming two antiparallel alpha-helices in the clip domains of M. sexta PAP2, has been proposed as a recognition/binding site [41]. A conserved structural difference in this region between the CLIP proteases that occupy different positions in cascade pathways may contribute to binding interactions required for their function to either activate another protease or to activate proPO or proSpätzle. These relationships also hold for the clip domain and protease domain sequences of Ae. aegypti, and An. gambiae [13]. There is a very good correlation between branches of the CLIP tree based on alignment of only the protease domain or the type of associated clip domain.
Figure 4. Conservation of a protease cascade module.
In pathways in which clip domain substrates have been determined experimentally, a conserved pattern is evident. A non-CLIP modular serine protease activates a penultimate CLIP protease from clade C with a type 1 clip domain, which activates a terminal CLIP protease from clade B with a type 2 clip domain, which then activates an effector protein.
Figure 5. Comparison of insect CLIP proteases known to participate in specific pathways.

The phylogenetic tree is based on an alignment of the protease domain sequences, with horseshoe crab proclotting enzyme (Limulus PCE) as an outgroup. Numbers at branches indicate bootstrap value, as a percent of 1000 repetitions. There are two clades that correspond with groups of proteases that are either the terminal protease in a known pathway (CLIPB proteases) or the penultimate proteolytic step of a pathway (CLIPC protease, labeled in red). The tree derived from the protease domain sequences correlates with the type of associated N-terminal clip domain, with terminal proteases having type 2 clip domains and penultimate proteases having type 1a clip domains. The activation site P1 residue is the amino acid residue (determined experimentally or predicted based on sequence alignment) on the amino-terminal side of the peptide bond that is cleaved to activate the protease zymogen.
Some exceptions to this general scheme may occur. For example, active M. sexta PAP3 can cleave and activate proPAP3 zymogen, providing a feed-forward regulation of the melanization cascade, and activation of a CLIPB by another CLIPB [42]. Another is D. melanogaster Hayan, a CLIPC with strong genetic evidence for its requirement in wound-activated melanization [25]. A recombinant form of Hayan, lacking its clip domain, can cleave PPO1 with requirement for the natural sequence at the PPO activation site, although production of an active PO resulting from this reaction has not yet been demonstrated. Further investigation of the substrate (s) for Hayan in vivo will be important for resolving the wound-activated melanization pathway.
Roles of CLIP pseudoproteases: serine protease homologs
Insect genomes contain multiple genes that encode proteins with one or more aminoterminal clip domains joined to a serine protease-like domain that lacks a complete catalytic triad needed for proteolysis. Most often the active site serine residue is changed to glycine in these serine protease homologs (SPHs). Most such pseudoproteases are in the CLIPA lineage. Some CLIP-SPH proteins function as regulators of immune pathways and in some cases are known to modify the function of CLIPBs. The most studied example of this phenomenon is the observation that CLIP-SPHs can greatly increase the efficiency of proteolytic activation of proPOs by CLIPB proteases in M. sexta and T. molitor [24,35,43,44]. These SPHs functioning as cofactors must themselves be activated by a specific proteolytic cleavage, adding another level of regulation to the pathway. CLIPA proteins can also act as negative regulators of immune pathways [45,46]. It is likely that the domains of CLIPA SPHs function by forming protein interactions between active proteases and their substrates or by helping to localize the members of protease cascades at the surface of a pathogen or parasite.
As shown in the Holotrichia PPAF2 structure [40], the protease-like domain adopts a chymotrypsin-like fold and interacts with the nearby clip domain through three loops (Fig. 3C). Sequences of these regions are conserved among CLIPAs but not CLIPBs, and the close association of clip domain and protease-like domain may be a distinguishing feature of CLIPAs. This orientation of the two domains in PPAF2 may be enhanced by interactions of an aminoterminal extension preceding the clip domain with the protease-like domain. Similar extensions also exist in other CLIPAs and may have a similar role in bringing the two domains into close association. The clip domain of PPAF2 is oriented toward the protease domain very differently from the clip domain in Grass (Fig. 3A), near a different surface of the protease domain and forming significant contacts between the two domains. This orientation and other structural features may participate in the formation of a 600 kDa PPAF2 dodecamer, after its clip domain is cleaved by PPAF3 at Arg99 between Cys3 and Cys4. The two stacked hexameric rings serve as a scaffold for anchoring PO, activated by PPAF1 [40]. In M. sexta, generation of high Mr active POs by PAPs also requires a complex of clip-domain SPHs [43]. Remarkably, proPOs cleaved by PAP-1 (a CLIPB) did not display PO activity; active POs were produced only in the presence of an active PAP and the SPH1 and SPH2 complex [43]. Perhaps the interaction of CLIPA proteins alters the conformation of CLIPB substrates, promoting efficient cleavage and activation by CLIPB proteases. There are many insect CLIPA pseudoproteases of unknown function, even in well studied model systems. Understanding how these serine protease homologs regulate immune pathways is an important topic for future research.
Conclusions
We hypothesize that many new immune proteins that are activated by protease cleavage in hemolymph remain to be discovered. There are 35 CLIP proteases in the M. sexta genome, but only four immune effector proteins activated by proteases are known: proPO-1 and proPO-2 [47], proSpätzle [30], and pro-plasmatocyte spreading peptide [48], which promotes plasmatocyte attachment, spreading, and decreases bleeding. It seems unlikely that 35 proteases function to activate only four immune proteins, and we predict that there may be a significantly larger number of protease-activated cytokines, enzymes, or antimicrobial molecules in plasma. CLIP proteases might have additional non-immune functions in insect development, as they do in Drosophila embryonic dorsal/ventral pattern formation, or in other physiological systems remaining to be discovered.
Researchers in protease biochemistry have come to realize that “proteases do not act in isolation, but form cascades, biochemical pathways, and regulatory circuits” [49] and that protease pathways intersect to form a protease web [50]. Most of the regulatory action in such systems occurs as post-translational modifications (zymogen activation, protease cleavage of substrates, protease interaction with inhibitors) rather than at the level of gene expression. Even for most human plasma proteases, the in vivo substrate profiles have not been elucidated [51], and those which have display astonishing complexity. Thrombin, for example, has at least nine important substrates, with pro-clotting and anti-clotting activities depending on physiological context and interaction with cofactors [52]. Proteomics offers one of the only experimental means to investigate protease functions in complex in vivo systems [49,51,53], but there has been practically no use of this technology for the study of the activity of insect hemolymph proteases (other than studies of protease-serpin interactions in Manduca [54]. With such a large number of CLIP proteases in insect hemolymph, it seems probable that they function in immune pathways beyond activation of proPO and proSpätzle. Progress in this field requires assays to identify natural substrates of these proteases, some of which may represent undiscovered immune effector molecules and reveal new defense mechanisms in insect immune systems.
Highlights.
CLIP proteases contain one or more amino-terminal clip domains and a carboxyl-terminal serine protease domain.
CLIP proteases in insect hemolymph are rapidly activated by specific proteolysis after immune stimulation and participate in cascade pathways to trigger activation of prophenoloxidase or the Toll ligand spätzle.
Some members of the CLIP protease superfamily are pseudoproteases known as serine protease homologs, which can function to regulate immune pathways.
The function of most insect CLIP proteases is unknown, even in well-studied model systems.
Acknowledgments
We thank Yingxia Hu for modeling M. sexta proPAP2 structure and Xiaolong Cao for constructing the phylogenetic tree. This is contribution 16-035-J from the Kansas Agricultural Experiment Station. Research carried out in the authors' laboratories was supported by NIH grants GM041247 and GM058634.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lange PF, Overall CM. Protein TAILS: when termini tell tales of proteolysis and function. Curr Opin Chem Biol. 2013;17:73–82. doi: 10.1016/j.cbpa.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 2.Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27:67–74. doi: 10.1016/s0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- 3.Salvesen GS. New perspectives on proteases. In: Nature Publishing Group, editor. Horizon Symposia. 2004. [Google Scholar]
- 4.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 5.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 6.Kanost M, Clem RJ. Insect Proteases. In: Gilbert L, editor. Insect Molecular Biology and Biochemistry. Elsevier; 2012. pp. 346–364. [Google Scholar]
- 7.Cerenius L, Kawabata SI, Lee BL, Nonaka M, Söderhäll K. Proteolytic cascades and their involvement in invertebrate immunity. Trends Biochem Sci. 2010;35:575–583. doi: 10.1016/j.tibs.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 9.Muta T, Hashimoto R, Miyata T, Nishimura H, Toh Y, Iwanaga S. Proclotting enzyme from horseshoe crab hemocytes. cDNA cloning, disulfide locations, and subcellular localization. J Biol Chem. 1990;265:22426–22433. [PubMed] [Google Scholar]
- 10.Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo--shaping and transducing a morphogen gradient. Curr Biol. 2005;15:R887–899. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 12.Ross J, Jiang H, Kanost MR, Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene. 2003;304:117–131. doi: 10.1016/s0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- 13.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou Z, Lopez DL, Kanost MR, Evans JD, Jiang HB. Comparative analysis of serine protease-related genes in the honey bee genome: possible involvement in embryonic development and innate immunity. Insect Molecular Biology. 2006;15:603–614. doi: 10.1111/j.1365-2583.2006.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **16.Cao X, He Y, Hu Y, Zhang X, Wang Y, Zou Z, Chen Y, Blissard GW, Kanost MR, Jiang H. Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem Mol Biol. 2015;62:51–63. doi: 10.1016/j.ibmb.2014.10.006. RNA-Seq based identification of 125 expressed genes for serine proteases and pseudoproteases predicted to have regulatory rather than digestive function, with analysis of evolutionary relationships, structural features, and expression patterns of 42 CLIP proteases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanost MR, Gorman MG. Phenoloxidases in insect immunity. In: Beckage N, editor. Insect Immunology. Academic Press/Elsevier; 2008. pp. 69–96. [Google Scholar]
- 18.Cerenius L, Lee BL, Söderhäll K. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–271. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- *19.An C, Ishibashi J, Ragan EJ, Jiang H, Kanost MR. Functions of Manduca sexta hemolymph proteinases HP6 and HP8 in two innate immune pathways. J Biol Chem. 2009;284:19716–19726. doi: 10.1074/jbc.M109.007112. Biochemical experiments with purified recombinant proteins showed that hemolymph protease 6, a CLIPC orthologous with Drosophila Persephone, activates hemolymph protease 8, a CLIPB that activates proSpätzle and also activates proPAP1, a CLIPB that activates prophenoloxidase, demonstrating a branched immune protease pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **20.An C, Zhang M, Chu Y, Zhao Z. Serine protease MP2 activates prophenoloxidase in the melanization immune response of Drosophila melanogaster. PLoS One. 2013;8:e79533. doi: 10.1371/journal.pone.0079533. The authors demonstrated by genetic and biochemical experiments that CLIPB protease MP2 from Drosophila can activate prophenoloxidase and that it is inhibited by serpin27a, known to regulate the melanization response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang H, Wang Y, Kanost MR. Pro-phenol oxidase activating proteinase from an insect, Manduca sexta: a bacteria-inducible protein similar to Drosophila easter. Proc Natl Acad Sci U S A. 1998;95:12220–12225. doi: 10.1073/pnas.95.21.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang H, Wang Y, Yu XQ, Kanost MR. Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta. A bacteria-inducible serine proteinase containing two clip domains. J Biol Chem. 2003;278:3552–3561. doi: 10.1074/jbc.M205743200. [DOI] [PubMed] [Google Scholar]
- 23.Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol. 2003;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- **24.Kan H, Kim CH, Kwon HM, Park JW, Roh KB, Lee H, Park BJ, Zhang R, Zhang J, Söderhäll K, et al. Molecular control of phenoloxidase-induced melanin synthesis in an insect. J Biol Chem. 2008;283:25316–25323. doi: 10.1074/jbc.M804364200. This paper presents results of biochemical experiments showing that Tenebrio molitor Spätzle-processing enzyme has two substrates in the melanization pathway, activating prophenoloxisase and the CLIP SPH1 cofactor, which together participate in forming a melanization complex. [DOI] [PubMed] [Google Scholar]
- 25.Nam HJ, Jang IH, You H, Lee KA, Lee WJ. Genetic evidence of a redox-dependent systemic wound response via Hayan protease-phenoloxidase system in Drosophila. EMBO J. 2012;31:1253–1265. doi: 10.1038/emboj.2011.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang H, Kambris Z, Lemaitre B, Hashimoto C. Two proteases defining a melanization cascade in the immune system of Drosophila. J Biol Chem. 2006;281:28097–28104. doi: 10.1074/jbc.M601642200. [DOI] [PubMed] [Google Scholar]
- 27.Jang IH, Chosa N, Kim SH, Nam HJ, Lemaitre B, Ochiai M, Kambris Z, Brun S, Hashimoto C, Ashida M, et al. A spatzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Developmental Cell. 2006;10:45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Roh KB, Kim CH, Lee H, Kwon HM, Park JW, Ryu JH, Kurokawa K, Ha NC, Lee WJ, Lemaitre B, et al. Proteolytic cascade for the activation of the insect toll pathway induced by the fungal cell wall component. J Biol Chem. 2009;284:19474–19481. doi: 10.1074/jbc.M109.007419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An C, Jiang H, Kanost MR. Proteolytic activation and function of the cytokine Spätzle in the innate immune response of a lepidopteran insect, Manduca sexta. Febs J. 2010;277:148–162. doi: 10.1111/j.1742-4658.2009.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gulley MM, Zhang X, Michel K. The roles of serpins in mosquito immunology and physiology. J Insect Physiol. 2013;59:138–147. doi: 10.1016/j.jinsphys.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gubb D, Sanz-Parra A, Barcena L, Troxler L, Fullaondo A. Protease inhibitors and proteolytic signalling cascades in insects. Biochimie. 2010;92:1749–1759. doi: 10.1016/j.biochi.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Park JW, Kim CH, Rui J, Park KH, Ryu KH, Chai JH, Hwang HO, Kurokawa K, Ha NC, Söderhäll I, et al. Beetle immunity. Adv Exp Med Biol. 2010;708:163–180. doi: 10.1007/978-1-4419-8059-5_9. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu XQ, Jiang H, Wang Y, Kanost MR. Nonproteolytic serine proteinase homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 2003;33:197–208. doi: 10.1016/s0965-1748(02)00191-1. [DOI] [PubMed] [Google Scholar]
- 36.Volz J, Muller HM, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 2006;8:1392–1405. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 37.An C, Budd A, Kanost MR, Michel K. Characterization of a regulatory unit that controls melanization and affects longevity of mosquitoes. Cell Mol Life Sci. 2011;68:1929–1939. doi: 10.1007/s00018-010-0543-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **38.Kellenberger C, Leone P, Coquet L, Jouenne T, Reichhart JM, Roussel A. Structure-function analysis of grass clip serine protease involved in Drosophila Toll pathway activation. J Biol Chem. 2011;286:12300–12307. doi: 10.1074/jbc.M110.182741. Crystal structure of a clip-domain serine protease zymogen with detailed analysis of structure and zymogen activation. The structure shows a distinctive loop in CLIPB proteases potentially important in specificity of activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piao S, Kim S, Kim JH, Park JW, Lee BL, Ha NC. Crystal structure of the serine protease domain of prophenoloxidase activating factor-I. J Biol Chem. 2007;282:10783–10791. doi: 10.1074/jbc.M611556200. [DOI] [PubMed] [Google Scholar]
- **40.Piao S, Song YL, Kim JH, Park SY, Park JW, Lee BL, Oh BH, Ha NC. Crystal structure of a clip-domain serine protease and functional roles of the clip domains. EMBO J. 2005;24:4404–4414. doi: 10.1038/sj.emboj.7600891. Structure of a clip-domain serine protease homolog reveals features that may stabilize domain association, induce oligomer formation, and localize proPO activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang R, Lu Z, Dai H, Velde DV, Prakash O, Jiang H. The solution structure of clip domains from Manduca sexta prophenoloxidase activating proteinase-2. Biochemistry. 2007;46:11431–11439. doi: 10.1021/bi7010724. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Lu Z, Jiang H. Manduca sexta proprophenoloxidase activating proteinase-3 (PAP3) stimulates melanization by activating proPAP3, proSPHs, and proPOs. Insect Biochem Mol Biol. 2014;50:82–91. doi: 10.1016/j.ibmb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **43.Gupta S, Wang Y, Jiang H. Manduca sexta prophenoloxidase (proPO) activation requires proPO-activating proteinase (PAP) and serine proteinase homologs (SPHs) simultaneously. Insect Biochem Mol Biol. 2005;35:241–248. doi: 10.1016/j.ibmb.2004.12.003. Active phenoloxidase was generated only when proPO was treated with the actvie CLIPB PAP1 and cofactor SPHs at the same time. ProPOs cleaved by PAP1 alone, or cleaved by PAP1 followed by addition of SPHs did not become active. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Jiang H. Prophenoloxidase (proPO) activation in Manduca sexta: an analysis of molecular interactions among proPO, proPO-activating proteinase-3, and a cofactor. Insect Biochem Mol Biol. 2004;34:731–742. doi: 10.1016/j.ibmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Barillas-Mury C. CLIP proteases and Plasmodium melanization in Anopheles gambiae. Trends Parasitol. 2007;23:297–299. doi: 10.1016/j.pt.2007.05.001. [DOI] [PubMed] [Google Scholar]
- *46.Yassine H, Kamareddine L, Chamat S, Christophides GK, Osta MA. A serine protease homolog negatively regulates TEP1 consumption in systemic infections of the malaria vector Anopheles gambiae. J Innate Immun. 2014;6:806–818. doi: 10.1159/000363296. The authors present evidence indicating that the the serine protease homolog CLIPA2 localizes to microbial surfaces and negatively regulates a putative thioester protein convertase, thereby decreasing intensity of an immune response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang H, Wang Y, Ma C, Kanost MR. Subunit composition of pro-phenol oxidase from Manduca sexta: molecular cloning of subunit ProPO-P1. Insect Biochem Mol Biol. 1997;27:835–850. doi: 10.1016/s0965-1748(97)00066-0. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Jiang HB, Kanost MR. Biological activity of Manduca sexta paralytic and plasmatocyte spreading peptide and primary structure of its hemolymph precursor. Insect Biochemistry And Molecular Biology. 1999;29:1075–1086. doi: 10.1016/s0965-1748(99)00086-7. [DOI] [PubMed] [Google Scholar]
- 49.Doucet A, Overall CM. Protease proteomics: revealing protease in vivo functions using systems biology approaches. Mol Aspects Med. 2008;29:339–358. doi: 10.1016/j.mam.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Doucet A, Butler GS, Rodriguez D, Prudova A, Overall CM. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol Cell Proteomics. 2008;7:1925–1951. doi: 10.1074/mcp.R800012-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Auf dem Keller U, Schilling O. Proteomic Techniques and Activity-based Probes for the System-wide Study of Proteolysis. Biochimie. 2010;92:1705–1714. doi: 10.1016/j.biochi.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 52.Lane DA, Philippou H, Huntington JA. Directing thrombin. Blood. 2005;106:2605–2612. doi: 10.1182/blood-2005-04-1710. [DOI] [PubMed] [Google Scholar]
- 53.Agard NJ, Wells JA. Methods for the proteomic identification of protease substrates. Curr Opin Chem Biol. 2009;13:503–509. doi: 10.1016/j.cbpa.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tong YR, Jiang HB, Kanost MR. Identification of plasma proteases inhibited by Manduca sexta serpin-4 and serpin-5 and their association with components of the prophenol oxidase activation pathway. Journal Of Biological Chemistry. 2005;280:14932–14942. doi: 10.1074/jbc.M500532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **55.Zou Z, Shin SW, Alvarez KS, Kokoza V, Raikhel AS. Distinct melanization pathways in the mosquito Aedes aegypti. Immunity. 2010;32:41–53. doi: 10.1016/j.immuni.2009.11.011. This investigation demonstrated immune and tissue melanization pathways functionally differentiated by different modules of CLIP proteases in an important vector species. [DOI] [PubMed] [Google Scholar]
- 56.Shin SW, Bian G, Raikhel AS. A toll receptor and a cytokine, Toll5A and Spz1C, are involved in toll antifungal immune signaling in the mosquito Aedes aegypti. J Biol Chem. 2006;281:39388–39395. doi: 10.1074/jbc.M608912200. [DOI] [PubMed] [Google Scholar]
- 57.Volz J, Osta MA, Kafatos FC, Muller HM. The roles of two clip domain serine proteases in innate immune responses of the malaria vector Anopheles gambiae. Journal Of Biological Chemistry. 2005;280:40161–40168. doi: 10.1074/jbc.M506191200. [DOI] [PubMed] [Google Scholar]
- 58.Paskewitz SM, Andreev O, Shi L. Gene silencing of serine proteases affects melanization of Sephadex beads in Anopheles gambiae. Insect Biochem Mol Biol. 2006;36:701–711. doi: 10.1016/j.ibmb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 59.Michel K, Suwanchaichinda C, Morlais I, Lambrechts L, Cohuet A, Awono-Ambene PH, Simard F, Fontenille D, Kanost MR, Kafatos FC. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum. Proc Natl Acad Sci U S A. 2006;103:16858–16863. doi: 10.1073/pnas.0608033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang R, Kim EH, Gong JH, Kwon HM, Kim CH, Ryu KH, Park JW, Kurokawa K, Zhang J, Gubb D, et al. Three pairs of protease-serpin complexes cooperatively regulate the insect innate immune responses. J Biol Chem. 2009;284:35652–35658. doi: 10.1074/jbc.M109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim CH, Kim SJ, Kan H, Kwon HM, Roh KB, Jiang R, Yang Y, Park JW, Lee HH, Ha NC, et al. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J Biol Chem. 2008;283:7599–7607. doi: 10.1074/jbc.M710216200. [DOI] [PubMed] [Google Scholar]
- 62.El Chamy L, Leclerc V, Caldelari I, Reichhart JM. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat Immunol. 2008;9:1165–1170. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang H, Kambris Z, Lemaitre B, Hashimoto C. A serpin that regulates immune melanization in the respiratory system of Drosophila. Dev Cell. 2008;15:617–626. doi: 10.1016/j.devcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, Jiang H, Hoffmann JA, Reichhart JM. A serpin mutant links Toll activation to melanization in the host defence of Drosophila. Embo J. 2002;21:6330–6337. doi: 10.1093/emboj/cdf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Gregorio E, Han SJ, Lee WJ, Baek MJ, Osaki T, Kawabata S, Lee BL, Iwanaga S, Lemaitre B, Brey PT. An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev Cell. 2002;3:581–592. doi: 10.1016/s1534-5807(02)00267-8. [DOI] [PubMed] [Google Scholar]
- 66.Scherfer C, Tang H, Kambris Z, Lhocine N, Hashimoto C, Lemaitre B. Drosophila Serpin-28D regulates hemolymph phenoloxidase activity and adult pigmentation. Dev Biol. 2008;323:189–196. doi: 10.1016/j.ydbio.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 67.Ahmad ST, Sweeney ST, Lee JA, Sweeney NT, Gao FB. Genetic screen identifies serpin5 as a regulator of the toll pathway and CHMP2B toxicity associated with frontotemporal dementia. Proc Natl Acad Sci U S A. 2009;106:12168–12173. doi: 10.1073/pnas.0903134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **68.Buchon N, Poidevin M, Kwon HM, Guillou A, Sottas V, Lee BL, Lemaitre B. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc Natl Acad Sci U S A. 2009;106:12442–12447. doi: 10.1073/pnas.0901924106. Genetic analysis demonstrated that two pattern recognition receptors, a modular serine protease, Grass, and SPE constitute an extracellular pathway to generate spätzle, which triggers Toll-mediated immune signaling in Drosophila. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *69.Gorman MJ, Wang Y, Jiang H, Kanost MR. Manduca sexta hemolymph proteinase 21 activates prophenoloxidase-activating proteinase 3 in an insect innate immune response proteinase cascade. J Biol Chem. 2007;282:11742–11749. doi: 10.1074/jbc.M611243200. The authors demonstrated by biochemical experiments that interaction with the modular serine protease HP14 can lead to activation of CLIPC protease HP21, without cleavage of the putative HP21 zymogen activation site, and that this HP21 can cleave and activate prophenoloxidase-activating protease-3, a CLIPB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang H, Ma C, Lu ZQ, Kanost MR. Beta-1,3-glucan recognition protein-2 (betaGRP- 2) from Manduca sexta; an acute-phase protein that binds beta-1,3-glucan and lipoteichoic acid to aggregate fungi and bacteria and stimulate prophenoloxidase activation. Insect Biochem Mol Biol. 2004;34:89–100. doi: 10.1016/j.ibmb.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 71.Tong Y, Kanost MR. Manduca sexta serpin-4 and serpin-5 inhibit the prophenol oxidase activation pathway: cDNA cloning, protein expression, and characterization. J Biol Chem. 2005;280:14923–14931. doi: 10.1074/jbc.M500531200. [DOI] [PubMed] [Google Scholar]
- 72.Wang Y, Jiang H. Interaction of beta-1,3-glucan with its recognition protein activates hemolymph proteinase 14, an initiation enzyme of the prophenoloxidase activation system in Manduca sexta. J Biol Chem. 2006;281:9271–9278. doi: 10.1074/jbc.M513797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **73.Wang Y, Jiang H. Reconstitution of a branch of the Manduca sexta prophenoloxidase activation cascade in vitro: snake-like hemolymph proteinase 21 (HP21) cleaved by HP14 activates prophenoloxidase-activating proteinase-2 precursor. Insect Biochem Mol Biol. 2007;37:1015–1025. doi: 10.1016/j.ibmb.2007.05.013. Biochemical analysis elucidated a branch of the immune protease system in Manduca sexta, including βGRP2, HP14, HP21, PAP2, and proPOs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang GM, Lu ZQ, Jiang HB, Asgari S. Negative regulation of prophenoloxidase (proPO) activation by a clip-domain serine proteinase homolog (SPH) from endoparasitoid venom. Insect Biochemistry And Molecular Biology. 2004;34:477–483. doi: 10.1016/j.ibmb.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 75.Zhu YF, Wang Y, Gorman MJ, Jiang HB, Kanost MR. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. Journal Of Biological Chemistry. 2003;278:46556–46564. doi: 10.1074/jbc.M309682200. [DOI] [PubMed] [Google Scholar]