Abstract
Chlamydia are Gram-negative, obligate intracellular bacteria responsible for significant diseases in humans and economically important domestic animals. These pathogens undergo a unique biphasic developmental cycle transitioning between the environmentally stable elementary body (EB) and the replicative intracellular reticulate body (RB), a conversion that appears to require extensive regulation of protein synthesis and function. However, Chlamydia possess a limited number of canonical mechanisms of transcriptional regulation. Ser/Thr/Tyr phosphorylation of proteins in bacteria has been increasingly recognized as an important mechanism of post-translational control of protein function. We utilized 2D gel electrophoresis coupled with phosphoprotein staining and MALDI-TOF/TOF analysis to map the phosphoproteome of the EB and RB forms of Chlamydia caviae. Forty-two non-redundant phosphorylated proteins were identified (some proteins were present in multiple locations within the gels). Thirty-four phosphorylated proteins were identified in EBs, including proteins found in central metabolism and protein synthesis, Chlamydia-specific hypothetical proteins and virulence-related proteins. Eleven phosphorylated proteins were identified in RBs, mostly involved in protein synthesis and folding and a single virulence-related protein. Only three phosphoproteins were found in both EB and RB phosphoproteomes. Collectively, 41 of 42 C. caviae phosphoproteins were present across Chlamydia species, consistent with the existence of a conserved chlamydial phosphoproteome. The abundance of stage-specific phosphoproteins suggests that protein phosphorylation may play a role in regulating the function of developmental-stage-specific proteins and/or may function in concert with other factors in directing EB–RB transitions.
Introduction
Chlamydia are Gram-negative, obligate intracellular bacterial pathogens responsible for significant diseases in humans, economically important domestic animals including sheep and poultry, and a variety of wildlife (Horn, 2008). These pathogens undergo a biphasic developmental cycle that starts with the extracellular elementary body (EB) attaching to the surface of a susceptible cell (AbdelRahman & Belland, 2005). The EB then enters the cell via a pathogen-specified endocytic process, possibly mediated by a type 3 secretion system (T3SS) (Dautry-Varsat et al., 2005), and resides within a host-derived membrane (termed an inclusion) (Bastidas et al., 2013). Within the inclusion, the EB differentiates into the morphologically distinct, replicative form known as the reticulate body (RB) with replication proceeding by 9 h post-infection. The RBs divide via binary fission, differentiate back into EBs, and exit the host cell 40–72 h post-infection (the time is species dependent). Numerous studies analysing RNA types and abundance indicate that development proceeds through an early, middle and late phase of gene expression (Belland et al., 2003; Mäurer et al., 2007; Nicholson et al., 2003) and proteomic studies have revealed defined EB- and RB-specific protein profiles (Mukhopadhyay et al., 2006b; Saka et al., 2011; Skipp et al., 2005).
While differentiation is clearly a key process in infection, the exact mechanism(s) governing EB and RB transitions is unclear. More recently, a greater appreciation has grown regarding the role of Ser/Thr/Tyr phosphorylation as a global regulatory mechanism employed by bacteria (Mijakovic & Macek, 2012). While phosphorylation in bacteria is much less extensive than in eukaryotes, which often maintain >50 % of their proteome in a phosphorylated state (Olsen et al., 2010), phosphoproteome mapping of several bacterial species has demonstrated that bacteria can phosphorylate up to 13 % of their proteome (Gnad et al., 2010). Phosphorylation can modify protein function by acting as on/off or dimmer switches, altering localization and/or modulating protein interactions (Stülke, 2010). The ability of phosphorylation to modulate the activity of pre-formed proteins makes it an attractive mechanism for regulation of the EB to RB transition as EBs are not thought to actively produce new proteins prior to inclusion formation and differentiation.
Chlamydia encode two validated eukaryotic-like Ser/Thr protein kinases (PknD and Pkn1), a third predicted eukaryotic-like Ser/Thr protein kinase (Pkn5) and at least three putative protein phosphatases based on genome annotations (Johnson & Mahony, 2007; Verma & Maurelli, 2003). To assess the extent and possible roles of phosphorylation in chlamydial biology, we set out to map the phosphoproteome of the EB and RB forms of Chlamydia caviae GPIC (the causative agent of guinea pig inclusion conjunctivitis). C. caviae has homologues to 90 and 81 % of the predicted ORFs in Chlamydia pneumoniae (which causes pneumonia in humans) and Chlamydia trachomatis (which causes sexually transmitted infections and trachoma), respectively (Kalman et al., 1999; Read et al., 2003). In addition, the existence of a well-characterized C. caviae guinea pig infection model (Rank et al., 1985, 1995, 2003) and fully sequenced C. caviae genome (Read et al., 2003) combine to make C. caviae a model organism for studying the biology of Chlamydia.
Forty-two non-redundant phosphoproteins including 34 in EB samples and 11 in RB samples were identified. Only three phosphoproteins were present in both EBs and RBs. Of the 42 phosphoproteins identified, 41 had homologues in all sequenced Chlamydia. Consistent with the hypothesis that phosphorylation plays a role in mediating EB and RB transitions, EBs contained threefold more phosphoproteins than RBs, including key proteins involved in metabolism and pathogenesis. Together these data suggest that Chlamydia are capable of Ser/Thr/Tyr phosphorylation on a global level and that phosphorylation may play a role in regulating differentiation between the EB and RB developmental forms.
Methods
Cell culture conditions
L2 mouse fibroblasts were grown until confluent in T-175 culture flasks in Dulbecco's modified Eagle medium+GlutaMAX (DMEM) supplemented with 10 % heat-inactivated FBS at 37 °C with 5 % CO2. Cells were routinely monitored for Mycoplasma infection by creating a DNA lysate (Qiagen DNeasy kit) and screening by PCR using degenerate oligos that prime to the 16S rRNA of Mycoplasma species.
Cell infection and bacterial growth conditions
C. caviae GPIC strain SP6 (Binet et al., 2010) was used for all experiments. Bacterial stocks were tested for Mycoplasma contamination as described above. Confluent L2 cell monolayers were washed twice with DMEM to remove non-adherent cells and then infected with C. caviae GPIC SP6 diluted in sucrose phosphate glutamic acid buffer (SPG) [7.5 % (w/v) sucrose, 17 mM Na2HPO4, 3 mM NaH2PO4, 5 mM l-glutamic acid, pH 7.4] at an m.o.i. of 1 for EB harvests and an m.o.i. of 17 for RB harvests. Cells were rocked for 2 h at 37 °C with 5 % CO2 to allow for infection. FBS-DMEM supplemented with 1 × MEM non-essential amino acids solution and 1 μg cycloheximide ml − 1 (complete DMEM) was then added to the cells and incubated at 37 °C with 5 % CO2 until the desired harvest time. C. caviae titres were determined using the plaque assay (defined as p.f.u. ml − 1) (Matsumoto et al., 1998).
EB harvest
Infected L2 cells were incubated for 43 h at 37 °C with 5 % CO2. Medium containing non-adherent cells and free EBs was pooled and kept on ice. To isolate EBs from adherent cells, ice-cold DMEM was added to the flasks and cells were detached using sterile glass beads. The wash was added to the initial pooled medium and the infected cells were lysed on ice using sonication. Samples were centrifuged at 4 °C for 30 min at 15 000 g to pellet EBs. The EBs (pellet fraction) were then suspended in SPG and sonicated briefly to homogenize the suspension. The EB/SPG suspension was then centrifuged through 30 % renografin diluted in H2O at 4 °C for 40 min at 72 700 g. The pellet fraction was retained and suspended in SPG using sonication. The EB/SPG suspension was then centrifuged through a discontinuous gradient of 54/44/40 % renografin in water at 4 °C for 1 h at 50 500 g. EBs were isolated from the 54/44 % interface and diluted in SPG. This solution was then centrifuged at 4 °C for 1 h at 50 500 g and the pellet was suspended in SPG via sonication. The final EB/SPG stocks were stored at − 80 °C until use.
RB harvest
L2 cells were infected with C. caviae and incubated at 37 °C with 5 % CO2. After 15 h, the medium was decanted and the infected cells were washed with PBS to remove unattached cells and extracellular EBs. The adherent RB-containing cells were detached using glass beads in the presence of cold PBS containing 1 × phosphatase inhibitor cocktails 2 and 3 (Sigma), 1 × ProteoBlock protease inhibitor cocktail (Fermentas) and 250 units of Benzonase (Novagen). Cells were then gently lysed on ice using a dounce homogenizer. The RB-containing lysates were centrifuged at 4 °C for 10 min at 3000 g to remove unlysed cells. The supernatant was then centrifuged through 30 % renografin in PBS at 4 °C for 1 h at 45 500 g. The RB pellet was gently suspended in ice-cold PBS containing phosphatase and protease inhibitor cocktails and centrifuged through a discontinuous gradient consisting of 59/44/35 % renografin in PBS at 4 °C for 1 h at 45 500 g. The RB-containing material at the 44/35 % interface was collected and diluted into ice-cold SPG containing phosphatase and protease inhibitor cocktails and centrifuged at 4 °C for 25 min at 21 300 g. The final pellet was suspended in 2D gel wash buffer (10 mM Tris/HCl, 5 mM magnesium acetate, pH 8.0) and a small aliquot was taken for p.f.u. titre analysis to measure EB contamination. The remaining sample was centrifuged at 4 °C for 5 min at 16 100 g and the pellet was immediately processed for 2D gel analysis.
Sample preparation for 2D and 1D gel analysis
2D samples were prepared by washing the EB or RB sample once with 2D wash buffer. Pellets were then suspended in 2D lysis buffer (30 mM Tris/HCl, pH 8.8, 7 M urea, 2 M thiourea and 4 % CHAPS, 1 × phosphatase inhibitor cocktails 2 and 3) and sonicated on ice using three 2 s pulses. Samples were then shaken at room temperature for 30 min. Following lysis, samples were centrifuged at 4 °C for 30 min at 16 100 g. Supernatants were retained for 2D gel analysis (stored at − 80 °C until use), 1D sample preparation and protein quantification. Protein quantification was carried out using the Bio-Rad Protein Assay as directed by the manufacturer and assays were carried out using equivalent amounts of total protein for sample normalization. For 1D SDS-PAGE analysis, 2D samples were precipitated by mixing with nine volumes of methanol and then centrifuging for 5 min at 16 100 g. Pellets were retained, mixed with 1 × Laemmli buffer containing β-mercaptoethanol and boiled.
2D and 1D gel electrophoresis analysis
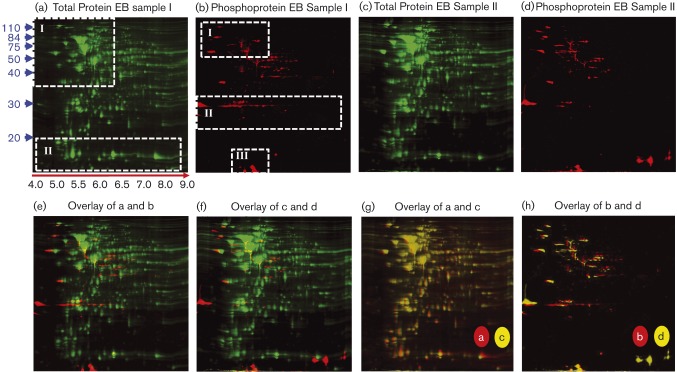
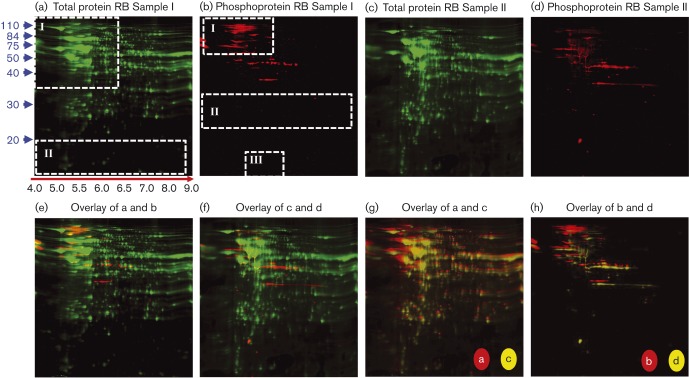
2D gel electrophoresis was carried out by Applied Biomics. Briefly, 200–300 μg of sample was initially separated using IEF on pH 4–9 gradient strips. Samples were then separated in the second dimension using SDS-PAGE, fixed and stained with Cy2 (GE Healthcare Life Sciences) for total protein and Pro-Q Diamond (Invitrogen) to detect phosphoproteins, and imaged using a Typhoon scanner (GE Healthcare Life Sciences). Two 2D gels were run using independently harvested EBs and RBs to provide biological replicates. Gel images were analysed using DeCyder software (GE Healthcare Life Sciences) and those phosphoprotein-containing spots chosen for MS analysis (based on a specific spot being present in both EB or RB biological replicates) were then excised from the gels using an Ettan Spot Picker (GE Healthcare Life Sciences) and stored at − 80 °C. To construct the gel images shown below in Figs 2(g,h) and 3(g,h), images were overlaid using the Olympus DP Manager software package (version 3.1.1.208).
Fig. 2. 2D gel electrophoresis analysis of C. caviae EBs. Biological replicates (Sample I and Sample II) were used to run two independent 2D gels (a and c). Samples were lysed using 2D lysis conditions and separated in the first dimension using IEF and in the second dimension using SDS-PAGE. Gels were fixed and stained with Cy2 to detect total protein (green in a and c) and Pro-Q Diamond to detect phosphoproteins (red in b and d). Total protein-stained and phosphoprotein-stained gel images from each independent sample were overlaid (e and f). An overlay of the total protein-stained gels from Sample I (a) and Sample II (c) is displayed in (g). In (h), the phosphoprotein-stained images from Sample I (b) and Sample II (d) were overlaid. Relative protein molecular mass markers are shown to the left of (a) in blue and relative IEF pH positions are shown below (a) (red arrow, black text). Quadrants of the gels highlighted for comparison in the Results section are shown in boxes with white, dashed borders (a and b).
Fig. 3. 2D gel electrophoresis analysis of C. caviae RBs. Other details are as for Fig. 2.
1D, 12 % SDS-PAGE gels were loaded with 2 μg of sample in each lane and gels were run at constant voltage. 1D gels were then either stained immediately with Coomassie brilliant blue stain or processed for Western blot analysis. Prestained protein molecular mass markers were used for protein band size determination.
Western blot analysis
After 1D SDS-PAGE, proteins were transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked in 5 % milk Tris-buffered saline (MTBS) and then incubated with either crude antisera from C. caviae_infected guinea pigs (provided by R. Rank, University of Arkansas) or mouse monoclonal anti-Hsp60 antibody (A57-B9, provided by Dr D. Rockey, Oregon State University) diluted 1 : 100 in MTBS. After incubation with the primary antibody, blots were washed with 0.5 % Tween TBS (TTBS) and incubated with horseradish peroxidase-conjugated anti-guinea pig IgG or anti-mouse IgG antibodies (GE Healthcare). Blots were then washed with TTBS, developed using Pierce SuperSignal West Pico chemiluminescent substrate (Thermo Scientific), and visualized using a FUJI LAS-3000 imaging system.
MALDI-TOF/TOF analysis
Peptide mass fingerprinting and MS/MS (collision-induced dissociation, CID) were performed by Applied Biomics. Briefly, 2D gel protein spots chosen for analysis were excised, washed to remove dyes and digested in-gel with trypsin, and the resulting peptides were extracted with trifluoroacetic acid extraction buffer. Samples were desalted using Zip-tip C18 cartridges (Millipore), mixed with alpha cyano-4-hydroxycinnamic acid and spotted on MALDI plates for MS analysis using MALDI-TOF and TOF/TOF modes on an Applied Biosystems Proteomics Analyser. Protein identification was determined by combining peptide mass fingerprinting mapping and peptide fragmentation mapping (CID) using GPS Explorer software (AB SCIEX) utilizing mascot 2.0 (Matrix Science) to search against the Chlamydia NCBI protein dataset (32 836 proteins) and the Mus musculus NCBI protein dataset (252 561 proteins) to identify host cell contaminant proteins. The program mascot 2.0 was used with default settings for raw data/peak conversion and peak-picking (signal to noise threshold of 50 and resolution of 50). Calibration was performed using Calibration mixture (Applied Biosystems) and no contaminant ions were excluded. Database search parameters were as follows: trypsin (enzyme specificity), one missed cleavage site permitted, searchable modifications (carbamidomethyl, oxidation, phosphorylation), 0.3 Da mass tolerance for precursor ions, 100 p.p.m. mass tolerance for fragment ions and confidence interval of greater than 95 %. The best protein match was based on protein score % confidence and ion score % confidence (with >95 % used as a significant identification) from MS and MS/MS analysis, respectively.
Bioinformatic analysis
Protein homology searches were performed using sequences derived from the C. caviae GPIC genome (accession number NC_003361) and NCBI blastp (default settings) (organisms are listed in Table S1, available in the online Supplementary Material). E-values greater than 1e − 7 were considered significant for homology analysis. Genome-wide homology comparisons were performed using the Comprehensive Microbial Resource Gene Homology Graph Tool found on the JCVI server (http://cmr.jcvi.org/cgi-bin/CMR/shared/MakeFrontPages.cgi?page = genomevs) using C. caviae GPIC as the reference genome. Protein homologues were aligned using ClustalW2 through the EMBL-EBI server (http://www.ebi.ac.uk/clustalw/) with default settings and the results were used to report per cent amino acid sequence identity. KEGG PATHWAY maps (http://www.genome.jp/kegg/pathway.html) and NCBI Protein Clusters [to identify Clusters of Orthologous Groups (COG) assignment] were used to sort proteins into functional classes.
Results
Purification and 1D gel analysis of C. caviae EBs and RBs
RBs and EBs were isolated at either 15 or 43 h, respectively, using density differential centrifugation. The time point for RB harvest was chosen to coincide with the eclipse phase of C. caviae GPIC strain SP6 growth when most of the bacteria exist as RBs (Binet et al., 2010). EBs were harvested at 43 h post-infection, which corresponds to the presence of maximum levels of intracellular EBs along with the beginning of cell lysis and EB release.
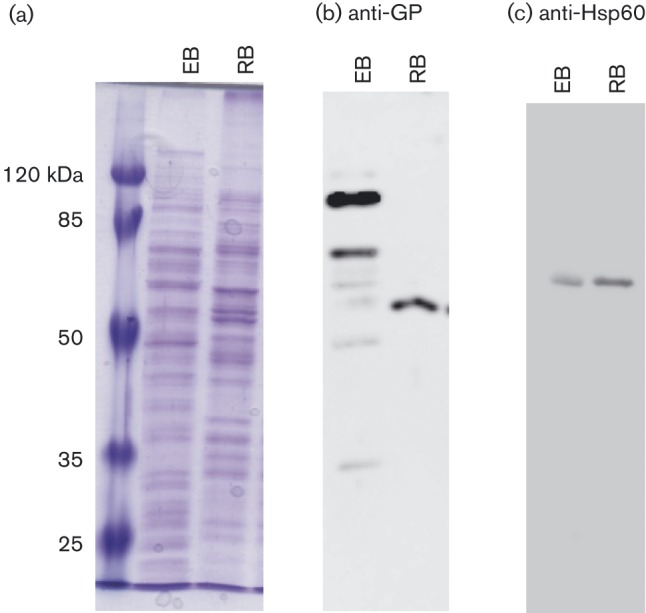
1D SDS-PAGE analysis was performed to assess sample quality and reproducibility prior to 2D gel analysis (Fig. 1a). Isolation of infectious EB particles was confirmed using the plaque assay. Based on p.f.u. titre results, RB samples did contain some EBs at an order of three logs less than the EB infectious input (data not shown). The differing global protein patterns in 1D (Fig. 1a) and 2D samples (Figs 2 and 3) and in Western blots of EB and RB samples probed with crude antisera from guinea pigs infected with C. caviae (Fig. 1b) are consistent with the samples being enriched for the EB or RB forms. Finally, Western blot analysis with anti-Hsp60 antibodies was performed to assess samples for a C. caviae-specific protein (Fig. 1c).
Fig. 1. 1D SDS-PAGE and Western blot analysis of C. caviae EBs and RBs. EB and RB samples were lysed using 2D lysis conditions and proteins were methanol-precipitated for 1D SDS-PAGE analysis. Samples were run on 12 % SDS-PAGE and then either stained with Coomassie brilliant blue (a) or transferred to nitrocellulose membranes for Western blot analysis (b and c). Membranes were probed either with crude antisera from C. caviae-infected guinea pigs (b) or with monoclonal anti-Hsp60 antibody A57-B9 (c) followed by horseradish peroxidase-conjugated anti-guinea pig or anti-mouse IgG antibodies, respectively. The protein sample source (EB or RB) is listed at the top of each panel. Molecular mass markers are shown to the left of (a).
2D gel electrophoresis and phosphoprotein profiling of the EB and RB developmental forms
EB and RB protein samples were separated on 2D gels and stained with Cy2 dye to detect total protein and with Pro-Q Diamond, which stains P-Ser, P-Thr, P-Tyr, P-His and P-Asp although P-His and P-Asp phospho-labels are rarely detected due to acid lability (Attwood et al., 2007), to specifically detect phosphorylated proteins. Two 2D gels were run using independent EB and RB harvests to provide biological replicates. Results for the EB and RB 2D gels are shown in Figs 2 and 3, respectively. Cy2-stained EB and RB 2D gels differed primarily in two quadrants (quadrants I and II, Figs 2a and 3a) consistent with the protein differences observed between samples using 1D gel analysis (Fig. 1a) and previous studies revealing the presence of different proteins/protein ratios present in the EB and RB developmental forms (Hatch et al., 1984; Mukhopadhyay et al., 2006b; Saka et al., 2011; Skipp et al., 2005). Phosphostaining of EB and RB gels also revealed differences between samples primarily in three regions (Figs 2 and 3b, quadrants I, II and III) with RB samples appearing to lack phosphoproteins below ∼35 kDa despite the presence of abundant Cy2-staining proteins below ∼35 kDa (Fig. 3a).
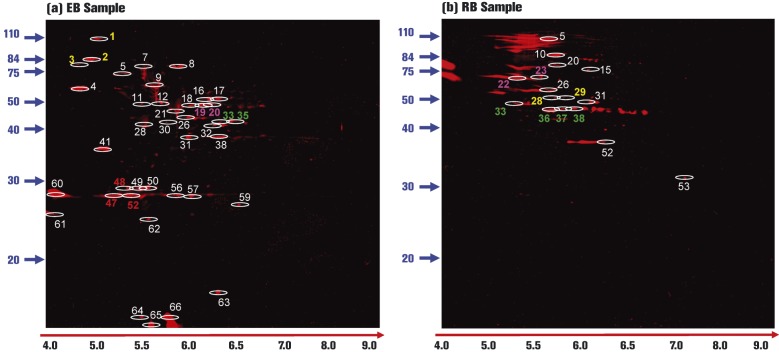
To select phosphoprotein-positive spots for MS analysis, EB and RB replicate sample gels were overlaid (Fig. 2h, EB Sample I versus EB Sample II; and Fig. 3h, RB Sample I versus RB Sample II) to identify phosphoprotein-containing spots present in both samples. Overall, 73 and 67 total phosphoproteins were detected in EB and RB samples, respectively (Table 1). Of these phosphoproteins, 44 and 52 spots were present in EB or RB biological gel replicates, respectively, based on gel overlay analysis and were subsequently chosen for MS-based protein identification. The locations of C. caviae phosphoproteins identified by MS/MS analysis are indicated in Fig. 4.
Table 1. Summary of C. caviae phosphoproteome 2D gel electrophoresis and MS/MS results.
| C. caviae developmental stage | ||
| EB | RB | |
| P-spots (total unique spots from EB or RB gels) | 73 | 67 |
| Overlapping P-spots* | 44 (60 %) | 52 (78 %) |
| C. caviae proteins† | 40 (91 %) | 16 (31 %) |
| Mouse proteins | 4 (9 %) | 36 (69 %) |
| Non-redundant‡ C. caviae proteins | 34 (85 %) | 11 (69 %) |
* P-spots present in either EB or RB biological replicate gels that were used for MS/MS analysis.
† Proteins identified using MS/MS. Percentages in parentheses refer to the number of C. caviae proteins/total proteins identified.
‡ Differentiating between the total C. caviae proteins identified and C. caviae proteins identified in more than one spot.
Fig. 4. Location of MS/MS-identified C. caviae EB and RB phosphoproteins. ProQ-Diamond-stained gel images were merged and analysed for phosphoprotein-containing spots present in EB biological replicate gels (a) or RB biological replicate gels (b). Phosphoprotein-containing spots were excised from the gel, digested with trypsin and identified using MS/MS. Only spots containing C. caviae proteins are highlighted. Non-marked spots were either not chosen for MS/MS identification or were found to be mouse contaminant proteins. In (a) and (b), spots identified with yellow, pink, green or red numbers were redundant proteins within EB or RB samples. White numbers designate non-redundant C. caviae proteins. Relative molecular mass markers are shown to the left of each panel and pH values are shown below each panel (red arrow, black type).
MS/MS identification of phosphoproteins
The list of identified C. caviae phosphoproteins is shown in Table 2. HemE, found in EBs, had not been previously identified in C. pneumoniae or C. trachomatis proteome studies utilizing global protein separation/MS methods (the C. caviae proteome has not been studied) (Mukhopadhyay et al., 2004, 2006a, b; Saka et al., 2011; Shaw et al., 2002; Skipp et al., 2005; Vandahl et al., 2001, 2002). Comprehensive MS/MS data results for each identified C. caviae phosphoprotein are listed in Table S2. Briefly, 56 C. caviae proteins were positively identified (40 EB phosphoproteins and 16 RB phosphoproteins). Of these proteins, 34 were non-redundant [proteins identified in multiple spots (redundant) were counted as one protein for this analysis) in EB samples and 11 were non-redundant in RB samples. Redundant EB proteins [CCA00030 (three spots), Pgk (two spots), CCA00005 (two spots) and PyrH (three spots)] and RB proteins [GroEL (two spots), AtpB (two spots) and Tuf (four spots)] are highlighted in Fig. 4. Three proteins, GroEL, DnaB and Tuf, were present in phosphorylated form in both EBs and RBs resulting in a total of 42 unique phosphorylated proteins in C. caviae of which 39 were identified in only the EB or RB form.
Table 2. C. caviae phosphoproteins identified using 2D gel electrophoresis coupled with phospho-specific protein staining and MALDI-MS/MS analysis.
| C. caviae GPIC | C. trachomatis | C. pneumoniae | ||||||
| Phosphoproteome spot identification | PD* | PD | ||||||
| Spot no. | Gene | Protein accession no. | Sample source† | Homologue‡ | EB | RB | Homologue | EB |
| 1, 2, 3 | CCA00030§ | 29839798 | EB | + | + | + | ||
| (cdsD) | ||||||||
| 4 | eno‖‖ | 29840718 | EB | + | + | + | ||
| 5 | CCA00015¶ | 29839784 | EB | + | + | + | ||
| 7 | dnaK | 29840008 | EB | + | + | + | ||
| 8 | proS | 29840011 | EB | + | ||||
| 9 | groEL† | 29840401 | EB | + | + | + | ||
| 11 | sucC‖‖ | 29840541 | EB | + | + | |||
| 12 | prfB# | 29839933 | EB | + | ||||
| 16 | dnaB † | 29839892 | EB | + | + | |||
| 17 | glgC‖‖ | 29839901 | EB | + | ||||
| 18 | CCA00035 (cdsN) | 29839803 | EB | + | + | |||
| 19, 20 | pgk‖‖ | 29839829 | EB | + | + | |||
| 21 | tuf† | 29840456 | EB | + | + | + | ||
| 26 | CCA00041 | 29839809 | EB | + | ||||
| 28 | sucB‖‖ | 29840183 | EB | + | + | |||
| 30 | CCA00732 | 29840489 | EB | + | ||||
| 31 | hemE | 29840635 | EB | |||||
| 32 | ompA | 29839815 | EB | + | + | + | ||
| 33, 35 | CCA00005 | 29839774 | EB | + | + | |||
| (aspC) | ||||||||
| 38 | gap‖‖ | 29839884 | EB | + | + | + | ||
| 41 | tal‖‖ | 29840447 | EB | + | + | + | ||
| 47, 48, 52 | pyrH | 29839812 | EB | + | + | |||
| 49 | CCA00083¶ | 29839851 | EB | + | + | + | ||
| 50 | tpiA‖‖ | 29840073 | EB | + | + | |||
| 56 | def | 29840076 | EB | + | + | + | ||
| 57 | adk | 29840294 | EB | + | + | + | ||
| 59 | CCA00595¶ | 29840353 | EB | + | ||||
| 60 | aaxB | 29840487 | EB | P | + | |||
| 61 | thiE | 29839970 | EB | A | A | |||
| 62 | ssb | 29840172 | EB | + | ||||
| 63 | CCA00630 | 29840388 | EB | |||||
| 64 | rsbV_2§ | 29840616 | EB | + | ||||
| 65 | CCA00330 | 29840094 | EB | |||||
| 66 | rsbV_1§ | 29840000 | EB | + | ||||
| 5 | CCA00282 | 29840049 | RB | + | + | + | ||
| (pmpG) | ||||||||
| 10 | fusA | 29839959 | RB | + | + | + | ||
| 15 | thrS | 29840712 | RB | + | + | |||
| 20 | tkt | 29840632 | RB | + | + | |||
| 22, 23 | groEL † | 29840401 | RB | + | + | + | ||
| 26 | tig | 29840674 | RB | + | + | + | ||
| 28, 29 | atpB | 29840441 | RB | + | + | |||
| 31 | dnaB † | 29839892 | RB | + | + | |||
| 33, 36, 37, 38 | tuf † | 29840456 | RB | + | + | + | ||
| 52 | ispH | 29840501 | RB | + | + | |||
| 53 | CCA00220 | 29839987 | RB | + | + | + | ||
* Proteomic data reflect the positive identification of proteins from Chlamydial species in other proteomic studies. Protein source (EB versus RB) is listed where appropriate.
† Phosphoproteins identified in both EB and RB samples.
P indicates presence of a pseudogene in C. trachomatis serovars and A indicates absence of a gene homologue across Chlamydia.
§ Proteins known to be phosphorylated in Chlamydia based on previous studies.
‖‖ Proteins involved in central metabolism, phosphorylated only in EBs.
¶ Hypothetical proteins that are unique to Chlamydia.
# Protein score confidence interval of < 95 % (see Table S2).
Of the phosphoproteins identified in this study, the Chlamydia homologues to the C. caviae CdsD, RsbV_1 and RsbV_2 proteins had been formerly identified as being phosphorylated using in vitro approaches (Hua et al., 2006; Johnson & Mahony, 2007). The identified phosphorylated peptide regions overlap with the S56 and S55 residues (RsbV_1 and RsbV_2, respectively) shown to be phosphorylated in the RsbV_1 and RsbV_2 proteins from C. trachomatis serovar D (Hua et al., 2006) (Table S2). C. caviae protein homologues previously shown to be phosphorylated using in vitro approaches that were not identified in this study include two functional kinases (PknD and Pkn1) (Verma & Maurelli, 2003) and the single chlamydial two-component regulator (CtcB/CtcC) (Koo & Stephens, 2003). To the best of our knowledge, only IncA and TARP (not identified in this study) have been shown to be phosphorylated in vivo using a Chlamydia tissue culture infection model (Clifton et al., 2004; Rockey et al., 1997). However, these proteins are secreted by Chlamydia and are believed to be phosphorylated by host cell kinases. As our study only analysed purified bacterial lysates, secreted proteins would not have been identified.
Mouse contaminant proteins (C. caviae were grown in L2 mouse fibroblasts) were identified in both EB and RB preparations. Many of the mouse protein contaminants detected are known to be phosphorylated (Gnad et al., 2010; Zhao et al., 2011), consistent with their identification in this study. The presence of host cell proteins is a common problem in Chlamydia proteome studies (Shaw et al., 2002; Skipp et al., 2005; Vandahl et al., 2001).
Conservation of phosphoproteins across Chlamydia
Sequencing and annotation of chlamydial genomes revealed that C. caviae shares approximately 90 and 81 % of its protein encoding genome with the human pathogens C. pneumoniae and C. trachomatis, respectively (Read et al., 2003). Based on this percentage of proteome homology, we anticipated that C. pneumoniae and C. trachomatis would share approximately 38/42 and 34/42 of the C. caviae phosphoproteome if it was selected under the same niche-specific pressure as the total genome. However, our findings show that >97 % of the C. caviae phosphoproteins are represented in both C. pneumoniae and C. trachomatis (Table 2) with only the C. caviae unique protein, ThiE, being absent (Read et al., 2003). A more detailed protein homology analysis further supports the high level of conservation of phosphorylated proteins amongst Chlamydia (Table S1). Notably, the Chlamydia ancestor Candidatus protochlamydia UWE25 has homologues to 87 % of the C. caviae phosphoproteins despite sharing only 74 % of the total C. caviae proteome (Horn et al., 2004).
Functional clustering of C. caviae phosphoproteins
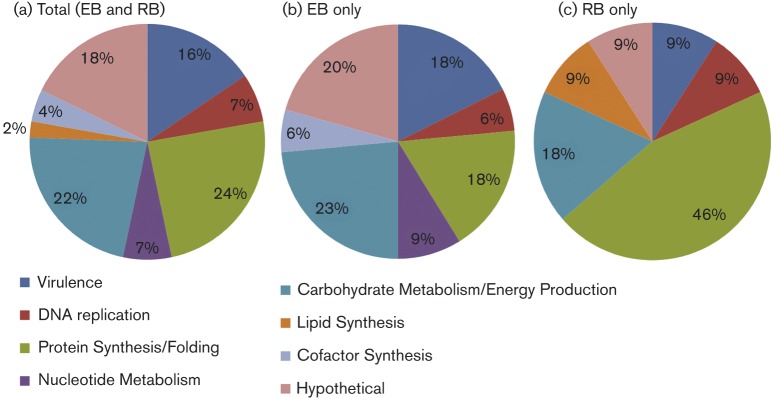
C. caviae phosphoprotein functional groups are shown in Fig. 5. The majority of phosphoproteins belong to protein classes involved in basic cellular processes including metabolism and protein synthesis. This was particularly true for RBs, in which 46 % of the phosphoproteome was devoted to these categories. While phosphoprotein classes in EBs were more widely distributed, 24 % of the phosphoproteome was still apportioned for these basic cellular processes. The C. caviae EB phosphoproteome contained several phosphorylated proteins located within the central carbon metabolic pathways including glycolysis, the TCA cycle, and the pentose phosphate pathway as well as glycogen metabolism (highlighted in Table 2). RBs contained only a single phosphorylated enzyme, transketolase, located within central carbon metabolism. Eight pan-Chlamydia hypothetical proteins (seven in EBs and one in RBs) including three hypothetical proteins unique to Chlamydia (CCA00015, CCA00083 and CCA00595 in EBs) were identified in this study.
Fig. 5. Functional classification of phosphoproteins. Phosphoproteins identified through MS/MS analysis were assessed for putative or known function using KEGG PATHWAY maps and NCBI Protein Clusters [for Clusters of Orthologous Groups (COG) assignments]. Functional groupings are listed below the pie charts and the percentage of phosphoproteins grouped into each functional class is shown in the pie charts. Phosphoproteins in EBs and RBs (a), EBs only (b) or RBs only (c).
Discussion
It has become widely accepted that bacteria, like eukaryotic cells, make use of global Ser/Thr/Tyr phosphorylation as a key mechanism to control protein activity, localization and interactions with other proteins (Mijakovic & Macek, 2012; Stülke, 2010). This post-translational method of regulation allows for more rapid adjustments to environmental stimuli and intra-bacterial metabolic fluxes than modulation of gene expression, which requires (at a minimum) altered levels of transcription and translation prior to phenotypic changes. Because of the rapidity by which phosphorylation acts, we hypothesized that Ser/Thr/Tyr phosphorylation could be a mechanism utilized by EBs to quickly emerge from a state of dormancy upon infection of a host cell.
Chlamydia species encode two eukaryotic-like Ser/Thr kinases [PknD (validated in C. pneumoniae and C. trachomatis) and Pkn1 (validated in C. trachomatis)] and one putative eukaryotic-like Ser/Thr kinase (Pkn5) (Verma & Maurelli, 2003). CdcD and IncG (the latter lacking a homologue in C. caviae) are in vitro targets for PknD and Pkn1, respectively, and at least six other phosphorylated proteins (CtcB/CtcC, anti-anti-sigma factors RsbV_1/RsbV_2, and the secreted proteins TARP and IncA) have been characterized (Clifton et al., 2004; Hua et al., 2006; Johnson & Mahony, 2007; Koo & Stephens, 2003; Rockey et al., 1997; Verma & Maurelli, 2003). However, TARP and IncA (the only two proteins shown to be phosphorylated in vivo) are probably phosphorylated by host kinases after secretion. Chlamydia species also encode three conserved protein phosphatases (CCA00399, CCA00964 and CCA00965), which would support a network of protein phosphorylation and dephosphorylation. However, global protein phosphorylation under in vivo conditions had not been explored.
We focused on determining the phosphoproteome of the C. caviae EB and RB forms to reveal developmental-stage-specific phosphorylation patterns. With this in mind, it is important to note the limitations of our approach and their impact on our conclusions. First, as with any global proteomic method, it is possible that phosphoproteins of low abundance were missed (Pro-Q Diamond stain detects 1–2 ng of multiphosphorylated proteins and 8 ng of a singly phosphorylated protein). Secondly, the use of a single RB harvest time point may have led to under-representation of the RB phosphoproteome due to differential protein expression (and possibly protein phosphorylation) during development. Thirdly, we were unable to conclusively identify phospho-Ser/Thr/Tyr residues in MS/MS mode for many of the dye-identified phosphoproteins. We did successfully detect phosphoproteins previously identified in other Chlamydia species (using in vitro approaches) along with mouse phosphoproteins identified using alternative phosphoproteomic methods, supporting the validity of our approach. Additionally, 2D gel approaches allow for acquisition of a ‘whole protein’ perspective, permitting the identification of proteins that exist in differentially phosphorylated forms.
The discovery that EBs contained threefold more phosphorylated proteins than RBs (34 versus 11) is consistent with prior proteome and global RNA studies demonstrating physiological differences between the EB and RB (Belland et al., 2003; Mäurer et al., 2007; Mukhopadhyay et al., 2006b; Saka et al., 2011; Skipp et al., 2005). The Hsp60 Western blot data (Fig. 1c) indicate the presence of increased levels of Hsp60 in RB versus EB samples. Using quantitative proteomics, Saka et al. (2011) reported that C. trachomatis RBs contain approximately fourfold more Hsp60 than EBs. As samples were normalized for total protein, our data support Saka et al.'s (2011) findings and are consistent with the hypothesis that the phosphoproteome skew observed was due to differing phosphorylation and not fewer RB proteins relative to total protein content compared with EB preparations. Proteins phosphorylated in EBs, but not in RBs, included proteins involved in central metabolism (highlighted in Table 2), nucleotide and co-factor metabolism, amino acid metabolism, and virulence (including two tbl3SS components). We hypothesize that phosphorylation inhibits the activity of the targeted EB proteins, allowing for rapid activation by dephosphorylation upon infection (Dworkin, 2015). Such a mechanism would also alleviate the need for protein synthesis in EBs, as they could be pre-packaged with all the requisite proteins needed for infection. Alternatively, it is possible that phosphorylation modulates protein function to meet the needs of the EB in the extracellular environment or during early infection (Fiuza et al., 2008; Jers et al., 2010; Parikh et al., 2009).
Only three phosphoproteins, DnaB, GroEL and Tuf, were identified in both EBs and RBs (Fig. 3 and Table 2). In EBs, GroEL and Tuf were present in single spots with relative pIs of 5.7 and 6.2, respectively. In contrast, in RBs both GroEL and Tuf were present in multiple spots located at the same molecular mass, but with lower pIs (5.4 and 5.8 for GroEL, and 5.3, 5.8, 6 and 6.2 for Tuf), typically associated with increasing levels of phosphorylation. These results could explain the altered pIs of GroEL and Tuf found in previous 2D gel proteome studies (Lundemose et al., 1990; Shaw et al., 1999, 2002; Vandahl et al., 2001). It is unknown whether the Chlamydia GroEL and Tuf respond to phosphorylation analogously to their protein homologues, which are altered for oligomerization or translation activity, respectively (Archambaud et al., 2005; Kumar et al., 2009). At least five other proteins (CdsD, Pgk, CCA00005 and PyrH in EBs, and AtpB in RBs) are present in more than one gel location (differing on the pI/molecular weight axis). Follow-up studies are needed to confirm that different locations in the 2D gels result from altered phosphorylation and not alternative protein processing or sample preparation.
The presence of few ( < 8 %) shared phosphoproteins between EBs and RBs suggests that protein function may be altered between these two forms. As our study was not designed to detect non-phosphorylated proteins, an alternative explanation is that the proteins are not present in both developmental forms. Where possible, we used existing Chlamydia species proteome data to determine the presence of proteins in EB and/or RB fractions (Table 1). It will be necessary to determine on a case-by-case basis whether proteins of interest are present in both EBs and RBs.
Two components of the tbl3SS apparatus, CdsD and CdsN (Peters et al., 2007), were phosphorylated only in EBs. CdsD, which is homologous to the outer ring protein YscD in Yersinia species, contains two fork-head-associated domains not present in the Yersinia YscD. These domains can be phosphorylated by PknD; however, it was not determined when CdsD is phosphorylated or whether phosphorylation actually occurs in vivo (Johnson & Mahony, 2007). In our study, CdsD was present in EB samples in three spots differing in both pI and molecular mass. Previous proteome data identified CdsD in C. trachomatis EBs and RBs indicating that these proteins are probably present in both developmental forms (Belland et al., 2003; Mäurer et al., 2007; Saka et al., 2011; Skipp et al., 2005). Thus, the absence of detectable phosphorylation of tbl3SS components in RBs suggests that the tbl3SS may be negatively regulated by phosphorylation. To the best of our knowledge, phosphorylation would be a unique mechanism for regulating the tbl3SS.
Our study supports the presence of global protein phosphorylation in C. caviae. The conservation of C. caviae phosphoproteins, kinases and phosphatases across Chlamydia species suggests that other chlamydiae also are capable of utilizing phosphorylation and may target the same proteins as C. caviae. Furthermore, the presence of different targets and the varied extent of phosphorylation between EBs versus RBs indicate that phosphorylation may be a mechanism of developmental regulation employed by Chlamydia. In-depth studies to examine the phosphorylation state of individual proteins throughout development (and possibly different phosphoproteome time point snapshots) are needed to better define phosphorylation time events in Chlamydia. In addition, mapping of the kinase/phosphatase/substrate network should identify targets for the development of Chlamydia-specific inhibitors. As proof of principle and supporting the importance of phosphorylation to chlamydial growth, a C. pneumoniae-specific PknD inhibitor has been shown to have antibacterial activity (Johnson et al., 2009). Finally, our study provides further evidence that even bacteria with reduced genomes take advantage of the Ser/Thr/Tyr protein phosphorylation tool kit.
Acknowledgements
This project was supported by NIAID grants 5R01AI044033 to A. T. M. and 1R15AI109566-01A1 to D. J. F. The opinions or assertions contained herein are those of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the Uniformed Services University of the Health Sciences.
Supplementary Data
Supplementary Data
Abbreviations:
- EB
elementary body
- RB
reticulate body
- T3SS
type 3 secretion system
References
- AbdelRahman Y.M., Belland R.J. (2005). The chlamydial developmental cycle FEMS Microbiol Rev 29 949–959 10.1016/j.femsre.2005.03.002 . [DOI] [PubMed] [Google Scholar]
- Archambaud C., Gouin E., Pizarro-Cerda J., Cossart P., Dussurget O. (2005). Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes Mol Microbiol 56 383–396 10.1111/j.1365-2958.2005.04551.x . [DOI] [PubMed] [Google Scholar]
- Attwood P.V., Piggott M.J., Zu X.L., Besant P.G. (2007). Focus on phosphohistidine Amino Acids 32 145–156 10.1007/s00726-006-0443-6 . [DOI] [PubMed] [Google Scholar]
- Bastidas R.J., Elwell C.A., Engel J.N., Valdivia R.H. (2013). Chlamydial intracellular survival strategies Cold Spring Harb Perspect Med 3 a010256 10.1101/cshperspect.a010256 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland R.J., Zhong G., Crane D.D., Hogan D., Sturdevant D., Sharma J., Beatty W.L., Caldwell H.D. (2003). Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis Proc Natl Acad Sci U S A 100 8478–8483 10.1073/pnas.1331135100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R., Bowlin A.K., Maurelli A.T., Rank R.G. (2010). Impact of azithromycin resistance mutations on the virulence and fitness of Chlamydia caviae in guinea pigs Antimicrob Agents Chemother 54 1094–1101 10.1128/AAC.01321-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D.R., Fields K.A., Grieshaber S.S., Dooley C.A., Fischer E.R., Mead D.J., Carabeo R.A., Hackstadt T. (2004). A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin Proc Natl Acad Sci U S A 101 10166–10171 10.1073/pnas.0402829101 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautry-Varsat A., Subtil A., Hackstadt T. (2005). Recent insights into the mechanisms of Chlamydia entry Cell Microbiol 7 1714–1722 . [DOI] [PubMed] [Google Scholar]
- Dworkin J. (2015). Ser/Thr phosphorylation as a regulatory mechanism in bacteria Curr Opin Microbiol 24 47–52 10.1016/j.mib.2015.01.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiuza M., Canova M.J., Patin D., Letek M., Zanella-Cléon I., Becchi M., Mateos L.M., Mengin-Lecreulx D., Molle V., Gil J.A. (2008). The MurC ligase essential for peptidoglycan biosynthesis is regulated by the serine/threonine protein kinase PknA in Corynebacterium glutamicum J Biol Chem 283 36553–36563 10.1074/jbc.M807175200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F., Forner F., Zielinska D.F., Birney E., Gunawardena J., Mann M. (2010). Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria Mol Cell Proteomics 9 2642–2653 10.1074/mcp.M110.001594 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch T.P., Allan I., Pearce J.H. (1984). Structural and polypeptide differences between envelopes of infective and reproductive life cycle forms of Chlamydia spp J Bacteriol 157 13–20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M. (2008). Chlamydiae as symbionts in eukaryotes Annu Rev Microbiol 62 113–131 10.1146/annurev.micro.62.081307.162818 . [DOI] [PubMed] [Google Scholar]
- Horn M., Collingro A., Schmitz-Esser S., Beier C.L., Purkhold U., Fartmann B., Brandt P., Nyakatura G.J., Droege M., other authors (2004). Illuminating the evolutionary history of chlamydiae Science 304 728–730. [DOI] [PubMed] [Google Scholar]
- Hua L., Hefty P.S., Lee Y.J., Lee Y.M., Stephens R.S., Price C.W. (2006). Core of the partner switching signalling mechanism is conserved in the obligate intracellular pathogen Chlamydia trachomatis Mol Microbiol 59 623–636 10.1111/j.1365-2958.2005.04962.x . [DOI] [PubMed] [Google Scholar]
- Jers C., Pedersen M.M., Paspaliari D.K., Schütz W., Johnsson C., Soufi B., Macek B., Jensen P.R., Mijakovic I. (2010). Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates Mol Microbiol 77 287–299 10.1111/j.1365-2958.2010.07227.x . [DOI] [PubMed] [Google Scholar]
- Johnson D.L., Mahony J.B. (2007). Chlamydophila pneumoniae PknD exhibits dual amino acid specificity and phosphorylates Cpn0712, a putative type III secretion YscD homolog J Bacteriol 189 7549–7555 10.1128/JB.00893-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.L., Stone C.B., Bulir D.C., Coombes B.K., Mahony J.B. (2009). A novel inhibitor of Chlamydophila pneumoniae protein kinase D (PknD) inhibits phosphorylation of CdsD and suppresses bacterial replication BMC Microbiol 9 218 10.1186/1471-2180-9-218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman S., Mitchell W., Marathe R., Lammel C., Fan J., Hyman R.W., Olinger L., Grimwood J., Davis R.W., Stephens R.S. (1999). Comparative genomes of Chlamydia pneumoniae C. trachomatis Nat Genet 21 385–389 10.1038/7716 . [DOI] [PubMed] [Google Scholar]
- Koo I.C., Stephens R.S. (2003). A developmentally regulated two-component signal transduction system in Chlamydia J Biol Chem 278 17314–17319 10.1074/jbc.M212170200 . [DOI] [PubMed] [Google Scholar]
- Kumar C.M., Khare G., Srikanth C.V., Tyagi A.K., Sardesai A.A., Mande S.C. (2009). Facilitated oligomerization of mycobacterial GroEL: evidence for phosphorylation-mediated oligomerization J Bacteriol 191 6525–6538 10.1128/JB.00652-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundemose A.G., Birkelund S., Larsen P.M., Fey S.J., Christiansen G. (1990). Characterization and identification of early proteins in Chlamydia trachomatis serovar L2 by two-dimensional gel electrophoresis Infect Immun 58 2478–2486 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Izutsu H., Miyashita N., Ohuchi M. (1998). Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma J Clin Microbiol 36 3013–3019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäurer A.P., Mehlitz A., Mollenkopf H.J., Meyer T.F. (2007). Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence PLoS Pathog 3 e83 10.1371/journal.ppat.0030083 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I., Macek B. (2012). Impact of phosphoproteomics on studies of bacterial physiology FEMS Microbiol Rev 36 877–892 10.1111/j.1574-6976.2011.00314.x . [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Miller R.D., Summersgill J.T. (2004). Analysis of altered protein expression patterns of Chlamydia pneumoniae by an integrated proteome-works system J Proteome Res 3 878–883 10.1021/pr0400031 . [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Miller R.D., Sullivan E.D., Theodoropoulos C., Mathews S.A., Timms P., Summersgill J.T. (2006a). Protein expression profiles of Chlamydia pneumoniae in models of persistence versus those of heat shock stress response Infect Immun 74 3853–3863 10.1128/IAI.02104-05 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Good D., Miller R.D., Graham J.E., Mathews S.A., Timms P., Summersgill J.T. (2006b). Identification of Chlamydia pneumoniae proteins in the transition from reticulate to elementary body formation Mol Cell Proteomics 5 2311–2318 10.1074/mcp.M600214-MCP200 . [DOI] [PubMed] [Google Scholar]
- Nicholson T.L., Olinger L., Chong K., Schoolnik G., Stephens R.S. (2003). Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis J Bacteriol 185 3179–3189 10.1128/JB.185.10.3179-3189.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., other authors (2010). Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis Sci Signal 3 ra3 . [DOI] [PubMed] [Google Scholar]
- Parikh A., Verma S.K., Khan S., Prakash B., Nandicoori V.K. (2009). PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity J Mol Biol 386 451–464 10.1016/j.jmb.2008.12.031 . [DOI] [PubMed] [Google Scholar]
- Peters J., Wilson D.P., Myers G., Timms P., Bavoil P.M. (2007). Type III secretion à la Chlamydia Trends Microbiol 15 241–251 10.1016/j.tim.2007.04.005 . [DOI] [PubMed] [Google Scholar]
- Rank R.G., Hough A.J.,, Jr, Jacobs R.F., Cohen C., Barron A.L. (1985). Chlamydial pneumonitis induced in newborn guinea pigs Infect Immun 48 153–158 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank R.G., Sanders M.M., Patton D.L. (1995). Increased incidence of oviduct pathology in the guinea pig after repeat vaginal inoculation with the chlamydial agent of guinea pig inclusion conjunctivitis Sex Transm Dis 22 48–54 10.1097/00007435-199501000-00008 . [DOI] [PubMed] [Google Scholar]
- Rank R.G., Bowlin A.K., Reed R.L., Darville T. (2003). Characterization of chlamydial genital infection resulting from sexual transmission from male to female guinea pigs and determination of infectious dose Infect Immun 71 6148–6154 10.1128/IAI.71.11.6148-6154.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read T.D., Myers G.S., Brunham R.C., Nelson W.C., Paulsen I.T., Heidelberg J., Holtzapple E., Khouri H., Federova N.B., other authors (2003). Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae Nucleic Acids Res 31 2134–2147 10.1093/nar/gkg321 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey D.D., Grosenbach D., Hruby D.E., Peacock M.G., Heinzen R.A., Hackstadt T. (1997). Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion Mol Microbiol 24 217–228 10.1046/j.1365-2958.1997.3371700.x . [DOI] [PubMed] [Google Scholar]
- Saka H.A., Thompson J.W., Chen Y.S., Kumar Y., Dubois L.G., Moseley M.A., Valdivia R.H. (2011). Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms Mol Microbiol 82 1185–1203 10.1111/j.1365-2958.2011.07877.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.C., Christiansen G., Birkelund S. (1999). Effects of interferon gamma on Chlamydia trachomatis serovar A and L2 protein expression investigated by two-dimensional gel electrophoresis Electrophoresis 20 775–780 . [DOI] [PubMed] [Google Scholar]
- Shaw A.C., Gevaert K., Demol H., Hoorelbeke B., Vandekerckhove J., Larsen M.R., Roepstorff P., Holm A., Christiansen G., Birkelund S. (2002). Comparative proteome analysis of Chlamydia trachomatis serovar A, D and L2 Proteomics 2 164–186 . [DOI] [PubMed] [Google Scholar]
- Skipp P., Robinson J., O'Connor C.D., Clarke I.N. (2005). Shotgun proteomic analysis of Chlamydia trachomatis Proteomics 5 1558–1573 10.1002/pmic.200401044 . [DOI] [PubMed] [Google Scholar]
- Stülke J. (2010). More than just activity control: phosphorylation may control all aspects of a protein's properties Mol Microbiol 77 273–275 10.1111/j.1365-2958.2010.07228.x . [DOI] [PubMed] [Google Scholar]
- Vandahl B.B., Birkelund S., Demol H., Hoorelbeke B., Christiansen G., Vandekerckhove J., Gevaert K. (2001). Proteome analysis of the Chlamydia pneumoniae elementary body Electrophoresis 22 1204–1223 . [DOI] [PubMed] [Google Scholar]
- Vandahl B.B., Birkelund S., Christiansen G. (2002). Proteome analysis of Chlamydia pneumoniae Methods Enzymol 358 277–288 . [DOI] [PubMed] [Google Scholar]
- Verma A., Maurelli A.T. (2003). Identification of two eukaryote-like serine/threonine kinases encoded by Chlamydia trachomatis serovar L2 and characterization of interacting partners of Pkn1 Infect Immun 71 5772–5784 10.1128/IAI.71.10.5772-5784.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., León I.R., Bak S., Mogensen M., Wrzesinski K., Højlund K., Jensen O.N. (2011). Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes Mol Cell Proteomics 10 M110000299 10.1074/mcp.M110.000299 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data