Abstract
Influenza B viruses fall in two antigenically distinct lineages (B/Victoria/2/1987 and B/Yamagata/16/1988 lineage) that co-circulate with influenza A viruses of the H3N2 and H1N1 subtypes during seasonal epidemics. Infections with influenza B viruses contribute considerably to morbidity and mortality in the human population. Influenza B virus neutralizing antibodies, elicited by natural infections or vaccination, poorly cross-react with viruses of the opposing influenza B lineage. Therefore, there is an increased interest in identifying other correlates of protection which could aid the development of broadly protective vaccines. blast analysis revealed high sequence identity of all viral proteins. With two online epitope prediction algorithms, putative conserved epitopes relevant for study subjects used in the present study were predicted. The cross-reactivity of influenza B virus-specific polyclonal CD8+ cytotoxic T-lymphocyte (CTL) populations obtained from HLA-typed healthy study subjects, with intra-lineage drift variants and viruses of the opposing lineage, was determined by assessing their in vitro IFN-γ response and lytic activity. Here, we show for the first time, to the best of our knowledge, that CTLs directed to viruses of the B/Victoria/2/1987 lineage cross-react with viruses of the B/Yamagata/16/1988 lineage and vice versa.
Introduction
Influenza A viruses of the H1N1 and H3N2 subtypes and influenza B viruses cause annual outbreaks of respiratory tract disease in humans (WHO, 2014b). Seasonal recurrence of these viruses is a result of selection of variants that evade recognition by virus neutralizing antibodies induced by previous infections or vaccination (antigenic drift) (Chen & Holmes, 2008; Koel et al., 2013; Smith et al., 2004). In contrast to influenza A viruses, influenza B viruses are not further subdivided in antigenically distinct subtypes. In addition, the host range of influenza B viruses is more limited (Bodewes et al., 2013) than that of influenza A viruses, which infect a wide range of animal species and for which aquatic birds constitute a major reservoir of most subtypes (Boyce et al., 2009). As animal influenza A viruses of various subtypes have been shown to be able to cross the species barrier and can cause pandemic outbreaks, they continue to pose a threat for public health (Gao et al., 2013; van de Sandt et al., 2012).
Although no subtypes of influenza B viruses have been identified, two antigenically distinct lineages are distinguished based on their haemagglutinin (HA): the B/Victoria/2/1987 and B/Yamagata/16/1988 lineages (Rota et al., 1990). The B/Yamagata/16/1988 lineage was the dominant lineage until the mid-1980s when B/Victoria/2/1987 made a global appearance after its earlier detection in the 1970s in China (Chen et al., 2007). Viruses of both influenza B lineages co-circulated with influenza A/H3N2 and A/H1N1 viruses during various seasonal epidemics (Ambrose & Levin, 2012; CDC, 2009; McCullers et al., 2004). However, the relative contribution of these types and subtypes of influenza virus to the respective epidemics varied over the years. Influenza B viruses can be the major cause of seasonal epidemics or be almost completely absent (Ambrose & Levin, 2012; Heikkinen et al., 2014; Rota et al., 1990; Thompson et al., 2003).
Overall, influenza A/H3N2 virus infections are associated with the highest disease severity; however, infections with influenza B virus contribute considerably to morbidity and mortality in the human population (Ambrose & Levin, 2012; Feng et al., 2012; Simonsen et al., 1997; Thompson et al., 2003, 2004). Although influenza B viruses cause disease in all age groups, the burden of influenza B virus infections is highest amongst children and young adults (Ambrose & Levin, 2012; Heikkinen et al., 2014; Olson et al., 2007). To prevent severe disease and mortality, annual vaccination of individuals at high risk for influenza is recommended (WHO, 2014b). For many years trivalent seasonal influenza vaccines have been used. These vaccines contain components of three strains that match circulating influenza viruses antigenically: influenza A/H3N2 and A/H1N1, and one influenza B strain of either the B/Yamagata/16/1988 or B/Victoria/2/1987 lineage (WHO, 2014a). These vaccines aim at eliciting virus neutralizing strain-specific antibody responses. Unfortunately, antibodies directed against a virus of one lineage of influenza B poorly cross-react with viruses of the opposing influenza B lineage (Belshe et al., 2010; Shaw et al., 2002). The time-consuming process of vaccine production requires recommendation of vaccine strains months in advance of the upcoming influenza season (Russell et al., 2008). As only one strain of influenza B virus is included in most current trivalent seasonal influenza vaccines, vaccine effectiveness is reduced when the epidemic strain is of the opposing lineage (Belshe et al., 2010; Beran et al., 2009; Heinonen et al., 2011). The increased co-circulation of both influenza B lineages in the last decade has led to more frequent mismatches between the vaccine strain and the most dominant circulating influenza B lineage (Ambrose & Levin, 2012; Belshe, 2010; Belshe et al., 2010; CDC, 2014; Heikkinen et al., 2014). To address this problem, quadrivalent influenza vaccines, containing components of both influenza B lineages, have become available in some countries (CDC, 2013; FDA, 2012; WHO, 2014a). However, vaccine effectiveness may still be reduced in the case of unforeseen antigenic drift within either influenza B lineage (Belshe et al., 2010). This spurred an increased interest in identifying other correlates of protection, which could be relevant for future developments of broadly protective vaccines (van de Sandt et al., 2012). Of interest, antibodies cross-reactive with viruses of both the B/Yamagata/16/1988 and B/Victoria/2/1987 lineages have been demonstrated, but they contributed only to a limited extent to the overall antibody repertoire (Dreyfus et al., 2012). Here, we investigated the cross-reactivity of influenza B virus-specific CD8+ cytotoxic T-lymphocytes (CTLs). The main function of CTLs is to detect and eliminate virus-infected cells, thereby restricting viral replication and accelerating viral clearance (Sridhar et al., 2013; van de Sandt et al., 2012). Numerous studies have demonstrated that influenza A virus-specific CTLs contribute to heterosubtypic immunity against antigenically distinct influenza A virus strains. Influenza A virus-specific CTLs are predominantly directed to more conserved internal proteins (Assarsson et al., 2008; Hillaire et al., 2013; Jameson et al., 1998; Kreijtz et al., 2008; Lee et al., 2008; Quinones-Parra et al., 2014; van de Sandt et al., 2014a; Yewdell et al., 1985) and their contribution to cross-protective immunity has been demonstrated in various animal models (Flynn et al., 1998; Hillaire et al., 2011; Kreijtz et al., 2007, 2009; O'Neill et al., 2000; Weinfurter et al., 2011). Although in vivo evidence for the role of CTLs in protective heterosubtypic immunity in humans is limited (Epstein, 2006; McMichael et al., 1983; Slepushkin, 1959; Sridhar et al., 2013), several in vitro studies have demonstrated that human CTLs directed to seasonal influenza A viruses cross-react with possible pandemic influenza A viruses, including avian influenza viruses of the H5N1 and H7N9 subtype and swine origin vH3N2 viruses (Hillaire et al., 2013; Jameson et al., 1999; Kreijtz et al., 2008; Lee et al., 2008; Quinones-Parra et al., 2014; van de Sandt et al., 2014a). Virus-specific CTLs are also induced after influenza B virus infections (Robbins et al., 1989, 1995, 1997), but it is unknown to what extent human CTLs directed to an influenza B virus of one lineage can cross-react with viruses of the opposing lineage. Here, we show for the first time, to the best of our knowledge, that polyclonal CD8+T-cells directed to influenza B viruses of the B/Victoria/2/1987 lineage can cross-react with viruses of the B/Yamagata/16/1988 lineage and vice versa, although the antigen specificity of these cross-reactive CD8+T-cells was not defined. Furthermore, by using the prototypic viruses of both lineages (B/Victoria/2/1987 and B/Yamagata/16/1988) and more recent descendants (B/Netherlands/455/2011 and B/Netherlands/712/2011, respectively) we showed that these CD8+T-cells also recognize intra-lineage drift variants.
Results
Virus characterization
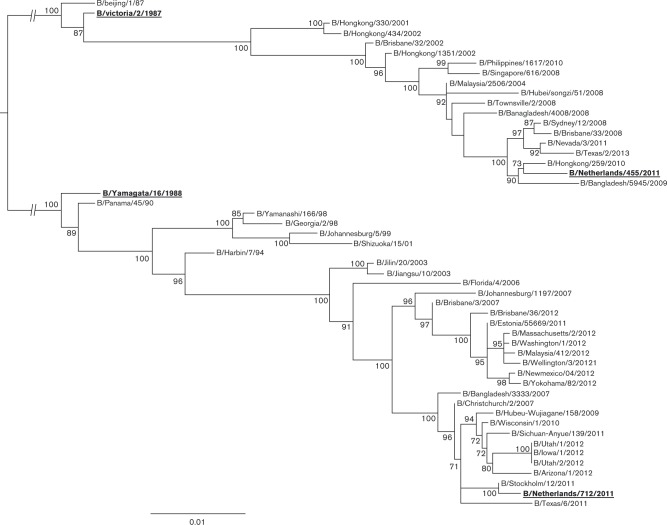
Phylogenetic analysis was performed to confirm the lineage of the influenza B viruses used in the present study. A dataset comprising the HA nucleotide sequences of 51 influenza B viruses isolated between 1987 and 2013 was used to determine the lineage of influenza viruses B/Netherlands/455/2011 and B/Netherlands/712/2011. A maximum-likelihood (ML) phylogenetic tree was reconstructed to study the nucleotide evolution of the HA gene segment of the influenza B/Yamagata/16/1988 and B/Victoria/2/1987 lineage. As expected, both prototypic strains B/Victoria/2/1987 and B/Yamagata/16/1988 were located near the base of the respective lineage. Based on this ML phylogenetic tree, it was concluded that the B/Netherlands/455/2011 virus belonged to the B/Victoria/2/1987 lineage, whilst B/Netherlands/712/2011 belonged to the B/Yamagata/16/1988 lineage (Fig. 1).
Fig. 1. Phylogenetic ML tree of the HA gene segment of human influenza B viruses. The phylogenetic ML tree was inferred from 51 HA nucleotide sequences of both influenza B virus lineages. Bootstrap values of 1000 replicates of ML trees are shown as percentages (values < 70 % are omitted). Bar roughly represents 1 % of nucleotide substitutions between close relatives. Bold type underlined sequences were used in the present study.
High amino acid sequence identity between both influenza B lineages
The percentage amino acid sequence identity of influenza A viral proteins has proven to be a good predictor of cross-reactivity of virus-specific T-cells with influenza A virus of various subtypes (van de Sandt et al., 2014a). Therefore, we wished to compare the overall amino acid sequence identity between the influenza B viruses used in the present study. blast analysis revealed that the sequence identity of all viral proteins was remarkably high (86–100 %) (Table 1).
Table 1. Percentage amino acid sequence identity between internal proteins of the influenza B viruses used in this study.
| Virus | Gene segment | Identity (%) | ||
|---|---|---|---|---|
| B/Victoria/02/1987 | B/Netherlands/712/2011 | B/Netherlands/455/2011 | ||
| B/Yamagata/16/1988 | PB2 | 99 | 99 | 99 |
| PB1 | 99 | 99 | 99 | |
| PA | 98 | 99 | 98 | |
| HA | 95 | 96 | 95 | |
| NP | 98 | 99 | 99 | |
| NA | 97 | 95 | 94 | |
| NB | 90 | 91 | 90 | |
| M1 | 100 | 99 | 100 | |
| BM2 | 93 | 98 | 96 | |
| NS1 | 99 | 94 | 93 | |
| NS2 | 100 | 98 | 98 | |
| B/Victoria/02/1987 | PB2 | 99 | 99 | |
| PB1 | 99 | 99 | ||
| PA | 98 | 98 | ||
| HA | 94 | 97 | ||
| NP | 98 | 98 | ||
| NA | 93 | 93 | ||
| NB | 87 | 86 | ||
| M1 | 99 | 100 | ||
| BM2 | 91 | 89 | ||
| NS1 | 94 | 93 | ||
| NS2 | 98 | 98 | ||
| B/Netherlands/712/2011 | PB2 | 99 | ||
| PB1 | 99 | |||
| PA | 99 | |||
| HA | 93 | |||
| NP | 99 | |||
| NA | 94 | |||
| NB | 93 | |||
| M1 | 99 | |||
| BM2 | 96 | |||
| NS1 | 97 | |||
| NS2 | 98 | |||
Prediction of conserved CD8+ T-cell epitopes
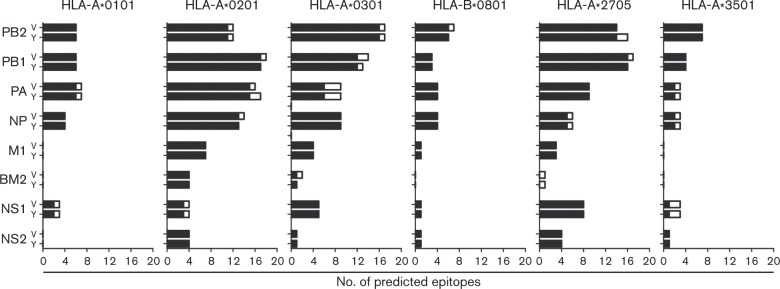
CD8+T-cell epitope prediction tools Syfpeithi and Immuneepitope were used to predict the presence of putative conserved CD8+T-cell epitopes in influenza B viral proteins. As the amino acid sequence identity between the respective strains was very high, this analysis was performed with the prototypic strains B/Victoria/2/1987 and B/Yamagata/16/1988 only. Epitope predictions were performed for the HLA alleles that corresponded with those of the study subjects (Table 2). Syfpeithi and Immuneepitope predicted a large number of putative epitopes in viral internal proteins of both B/Victoria/2/1987 and B/Yamagata/16/1988 (Fig. S1, available in the online Supplementary Material). Due to the large difference in the number of epitopes predicted by both programs, we considered an epitope a putative epitope when predicted by both programs. In addition, to be a putative conserved epitope, the epitope needed to be present in both viruses (Fig. 2). As shown in Fig. 2, most of the epitopes predicted by both programs were present in both virus strains. Only a small proportion of the predicted epitopes was unique for one of the two viruses. The data suggested that the majority of CTL epitopes were conserved between the two lineages of influenza B.
Table 2. HLA-A and HLA-B haplotypes of the study subjects.
| Group | Donor | HLA-A and HLA-B haplotypes |
|---|---|---|
| I | 8904 | HLA-A*0101, A*0201, B*0801, B*3501 |
| 6888 | ||
| 1578 | ||
| II | 7482 | HLA-A*0101, A*0201, B*0801, B*2705 |
| 2501 | ||
| 8801 | ||
| III | 6877 | HLA-A*0101, A*0301, B*0801, B*3501 |
| 9465 | ||
| 5891 |
Fig. 2. Epitopes predicted by Syfpeithi and Immuneepitope. Only epitopes predicted by both algorithms were considered to increase the fidelity of the prediction. Bars represent the total number of putative epitopes for B/Victoria/2/1987 (V) or B/Yamagata/16/1988 (Y); back bars represent the number of putative conserved epitopes present in both viruses, whilst white bars represent the number of putative epitopes present in one virus only.
To test the robustness of these prediction algorithms, we wished to establish the in vitro reactivity of influenza B virus-specific CD8+T-cells with the predicted epitopes. HLA-B*0801-restricted putative epitopes were selected as HLA-B*0801 was previously shown to be dominant in stimulating influenza B virus-specific CTLs (Boon et al., 2004) and the HLA-B*0801 allele was present in all the study subjects (Table 2). The HLA-B*0801 allele also gave us the opportunity to use three previously described peptides, i.e. NP263–271(ADRGLLRDI), NP413–421(ALKCKGFHV) and NP30–38(RPIIRPATL) (Robbins et al., 1997), of which only the NP30–38 epitope was predicted by our prediction algorithms. One study subject of each HLA group (Table 2; 6888, 8801 and 6877) was selected from which in vitro expanded polyclonal CD8+T-cells specific for B/Yamagata/16/1988 were tested for their reactivity with the predicted HLA-B*0801-restricted epitopes using peptide-loaded HLA class I-matched B-lymphoblastoid cell lines (BLCLs). To this end, we determined the IFN-γ production of the polyclonal CD8+T-cells in an ELISpot assay. Although donor 6877 responded to NP30–38 and M145–52, we did not observe a significant response to any of the other predicted or previously identified epitopes by donors 6877, 6888 and 8801, whilst all donors had a high response to BLCLs infected with the homologous virus (data not shown). These results clearly indicated that the prediction algorithms were not very reliable and therefore putative epitopes with other HLA restrictions were not tested.
Cross-reactivity of influenza B virus-specific CD8+ T-cells assessed by ELISpot
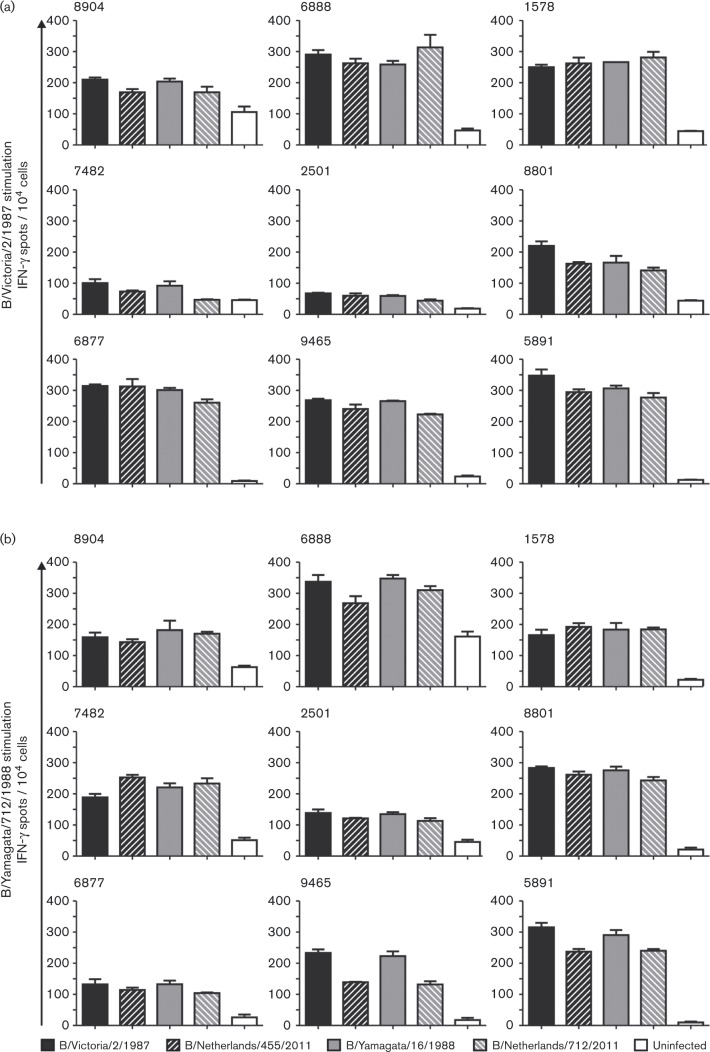
Next, we determined the extent of cross-reactivity of influenza B virus-specific CD8+T-cells with intra-lineage drifted variants and viruses of the opposing lineage. To this end, polyclonal CD8+T-cells derived from B/Victoria/2/1987 or B/Yamagata/16/1988 virus-stimulated peripheral blood mononuclear cell (PBMC) cultures were restimulated with HLA class I-matched BLCLs infected with the prototypic viruses (B/Victoria/2/1987 and B/Yamagata/16/1988) and the more recent viruses (B/Netherlands/455/2011 and B/Netherlands/712/2011, respectively). Activation of the polyclonal CD8+T-cells was assessed by measuring the number of IFN-γ-producing cells per 10 000 CD8+T-cells with the ELISpot assay (Fig. 3).
Fig. 3. Cross-reactivity of virus-specific polyclonal CD8+T-cells assessed by IFN-γ ELISpot. (a) B/Victoria/2/1987 virus-specific polyclonal CD8+T-cells or (b) B/Yamagata/16/1988 virus-specific polyclonal CD8+T-cells of nine study subjects were co-cultured with HLA-matched BLCLs infected with B/Victoria/2/1987, B/Netherlands/455/2011, B/Yamagata/16/1988 or B/Netherlands/712/2011.The number of IFN-γ-producing cells per 10 000 polyclonal CD8+T-cells was determined by ELISpot assay. Experiments were performed in triplicate; bar, sd. Uninfected BLCLs were used as negative controls. Identification number of the respective study subject is indicated in the left upper corner of each graph.
The reactivity of B/Victoria/2/1987 virus-specific CD8+T-cells is shown in Fig. 3(a). These cells of all study subjects responded to reactivation with the homologous B/Victoria/2/1987 virus, although two study subjects (7482 and 2501) were low responders. A similar response was observed after restimulation with the intra-lineage drift variant B/Netherlands/455/2011. In addition, after stimulation with a virus of the opposing lineage B/Yamagata/16/1988 or B/Netherlands/712/2011, a cross-reactive response was observed that did not substantially differ in magnitude from the response to viruses of the B/Victoria/2/1987 lineage, in most subjects. Fig. 3(b) shows the response of B/Yamagata/16/1988 virus-specific CD8+T-cells. Again, all study subjects responded to the restimulation with HLA-matched BLCLs infected with the homologous virus strain and also after stimulation with the intra-lineage drift variant B/Netherlands/712/1988. In addition, a cross-reactive response was observed after stimulation with HLA-matched BLCLs infected with both viruses of the opposing B/Victoria/2/1987 lineage (B/Victoria/2/1987 and B/Netherlands/712/2011), which was similar in magnitude as compared with the response to viruses of the B/Yamagata/16/1988 lineage.
Cross-reactivity of influenza B virus-specific CD8+ T-cells assessed by lytic activity
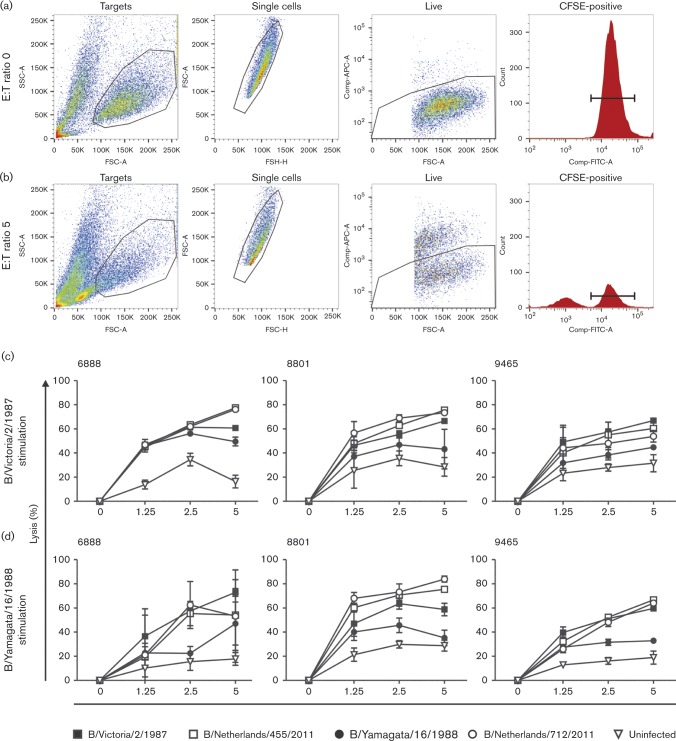
Next, we wished to assess the cross-reactive lytic capacity of these polyclonal CD8+T-cell populations. Based on the IFN-γ ELISpot results, we selected a strong responder from each HLA group of study subjects (6888, 8801 and 9465) to test the lytic capacity of the CD8+T-cells. To this end, in vitro expanded B/Victoria/2/1987 or B/Yamagata/16/1988 virus-specific polyclonal CD8+T-cells were incubated with carboxyfluorescein succinimidyl ester (CFSE)-labelled HLA-matched BLCLs infected with B/Victoria/2/1987, B/Yamagata/16/1988, B/Netherlands/455/2011 or B/Netherlands/712/2011. The gating strategy used for flow cytometry to detect lytic activity of the CD8+T-cells is shown in Fig. 4(a, b).
Fig. 4. Cross-reactivity of virus-specific polyclonal CD8+ T cells assessed by lytic activity.To assess the degree of lytic activity a gating strategy was used to determine the number of viable CFSE positive target cells in the (a) absence and (b) presence of virus-specific T cells using FlowJo software. (c) B/Victoria/2/1987 and (d) B/Yamagata/16/1988 virus-specific polyclonal CD8+ T cells from study subjects 6888, 8801 and 9465 were isolated after stimulation of PBMCs with the respective virus. Lytic activity against HLA-matched CFSE-labeled BLCLs infected with B/Victoria/2/1987, B/Netherlands/455/201, B/Yamagata/16/1988 and B/Netherlands/712/2011 was assessed. Uninfected BLCLs were used as negative controls. Experiments were performed in triplicate; bar, sd.
B/Victoria/2/1987 virus-specific CD8+T-cells displayed lytic capacity against cells infected with the homologous virus and the B/Netherlands/455/2011 virus of the same lineage in a effector-to-target cell (E : T) ratio-dependent fashion. In addition, cells infected with the heterologous viruses B/Yamagata/16/1988 and B/Netherlands/712/2011 of the opposing lineage were also lysed (Fig. 4c). A similar pattern of reciprocal lytic activity was observed for the B/Yamagata/16/1988 virus-specific CD8+T-cells (Fig. 4d). These results confirmed the cross-reactive nature of influenza B virus-specific CD8+T-cells observed in the ELISPOT assay.
Discussion
Influenza B viruses display less antigenic drift than influenza A viruses (Chen & Holmes, 2008; Lindstrom et al., 1999; Rota et al., 1992), yet they efficiently evade recognition by virus neutralizing antibodies present in the human population. This necessitates regular updates of the influenza B component of seasonal influenza vaccines. The circulation of influenza B viruses belonging to two antigenically distinct lineages further complicates the production of efficacious influenza vaccines. Inactivated influenza vaccines typically aim at the induction of virus neutralizing antibodies directed to the variable globular head region of the HA molecules of the respective influenza viruses. Consequently, there is interest in protective immune responses directed to more conserved proteins or regions thereof. Understanding humoral and cell-mediated immune responses to these conserved proteins may aid the development of more universal vaccines. Here, we assessed the cross-reactivity of influenza B virus-specific CD8+T-cells with viruses of the opposing lineage. It was concluded that influenza B virus-specific CD8+T-cells display a high degree of cross-reactivity with intra-lineage drift variants and viruses of the opposing lineage.
Although it was beyond the scope of this study to identify novel influenza B epitopes, we were interested whether predicted putative CTL epitopes were conserved between both influenza B lineages. The amino acid sequence identity of all viral proteins was very high between both lineages, which already suggested the existence of cross-reactive T-cell epitopes, as was also demonstrated for influenza A viruses (van de Sandt et al., 2014a). For the in silico prediction of epitopes, we excluded proteins encoded by the HA and NA gene segments, as these proteins undergo antigenic drift after positive selection by antibodies. Furthermore, it has been shown for influenza A viruses that these envelope proteins are minor targets for CTL responses (Lee et al., 2008). Two epitope prediction algorithms that are publicly available, i.e. Syfpeithi and Immuneepitope, were used in order to predict putative epitopes with the highest possible fidelity. A large number of putative epitopes was predicted by both programs, of which most were present in viruses of both lineages. However, the total number of epitopes predicted by both programs varied widely. The performance of these algorithms is not completely clear, and most likely false positives have also been predicted and false negatives omitted (Lundegaard et al., 2006; Roider et al., 2014). Indeed, out of six in vitro confirmed influenza B epitopes described previously (Robbins et al., 1989, 1995, 1997) that corresponded to the HLA alleles investigated in our study, only the NP30–38 RPIIRPATL epitope (HLA-B*0801 restricted) was predicted by both prediction programs. Other epitopes were solely predicted by Immuneepitope [NP85–94 KLGEFYNQMM and NP85–93 KLGEFYNQM (HLA-A*0201 restricted)] or Syfpeithi [NP413–421 ALKCKGFHV (HLA-B*0801 restricted)], and two epitopes [NP263–271 ADRGLLRDI (HLA-B*0801 restricted) and NP82–94 MVVKLGEFYNQMM (HLA-A*0201 restricted)] were not predicted at all. As these programs predict epitopes based on binding affinity of the epitope with the selected HLA allele, they do not take into account other possible factors that might play a role, such as the dissociation rate of the epitope (van der Burg et al., 1996), folding of the MHC class I molecules (Silver et al., 1991) or antigen processing (van de Sandt et al., 2012). To further test the robustness of the prediction algorithms, we determined the reactivity of polyclonal influenza B virus-specific polyclonal CD8+T-cells of three study subjects with the HLA-B*0801-restricted putative epitopes. We were unable to confirm any of the predicted epitopes, which included three previously described HLA-B*0801-restricted epitopes (NP30–38, NP263–271 and NP413–422). The lack of response to these previously described epitopes may be explained by mis-match of the HLA-C alleles of our study subjects. As the HLA-C alleles of our study subjects were not defined and Robbins et al. (1997) did not exclude the HLA-Cw7 allele as the presenting MHC class I molecule, it is possible that these epitopes are restricted by HLA-Cw7 instead of HLA-B*0801. This might also explain why only one study subject responded to the NP30–38 epitope (data not shown). Alternatively, differences in HLA makeup may have influenced immunodominance patterns (Boon et al., 2002). Thus, results obtained with epitope prediction algorithms should be interpreted with caution, which is in agreement with previous studies (Roider et al., 2014). Therefore, viral vectors expressing a single influenza B viral protein and/or overlapping peptide pools are considered more useful than in silico predictions for the identification of CD8+T-cell epitopes and establishing their immunodominance patterns.
The extent of cross-reactivity of influenza B virus-specific polyclonal CD8+T-cells was tested with two independent assays, i.e. IFN-γ ELISpot and CTL assays. In both assays, B/Victoria/2/1987 and B/Yamagata/16/1988 virus-specific CD8+T-cell populations displayed a high degree of cross-reactivity with drifted intra-lineage variants and across lineages, confirming that influenza B viruses indeed contain cross-reactive CTL epitopes. Differences in magnitude of the influenza B virus-specific CD8+T-cell responses between study subjects may reflect differences in HLA class I makeup (Boon et al., 2002) and/or differences in the history of influenza infections.
Most likely, subjects with alternative HLA alleles will also mount cross-reactive CD8+T-cell responses, considering the high level of sequence identity. Thus, in theory, infection with any influenza B virus will induce a cross-reactive influenza B virus-specific CD8+T-cell response. These cross-reactive CD8+T-cells may afford some degree of protection against a subsequent infection with an antigenically distinct influenza B virus, to which antibodies induced by previous infection will not be protective. Indeed, it was demonstrated during the pandemic of 2009 that, in the absence of virus neutralizing antibodies, the frequency of influenza A virus-specific CD8+T-cells correlated with a favourable disease outcome (Sridhar et al., 2013).
To address the problem of two co-circulating antigenically distinct influenza B lineages, quadrivalent seasonal influenza vaccines have become available that contain components of both influenza B lineages (CDC, 2013; FDA, 2012; WHO, 2014a). As quadrivalent vaccines elicit antibody responses against viruses of both lineages (Domachowske et al., 2013; Kieninger et al., 2013; Langley et al., 2013; Tinoco et al., 2014), they eliminate the risk that the incorrect B lineage is selected for inclusion in the vaccine. However, unforeseen antigenic drift within either influenza B lineage may affect vaccine effectiveness, although not as dramatically as a lineage mismatch (Belshe et al., 2010).
Vaccines that would induce cross-reactive T-cell-mediated immunity may offer another layer of protective immunity that is less sensitive to antigenic drift or circulation of an opposing lineage of influenza B virus. In particular, the use of live attenuated influenza vaccines has been shown to induce virus-specific CD8+T-cells, in contrast to inactivated vaccines (He et al., 2006; Hoft et al., 2011). Of interest, the viral proteins of live attenuated vaccines currently in use also display a high degree of sequence identity with recent circulating strains (93–100 % for B/Ann Arbor/1/66; data not shown), which supports the notion that these vaccines may also induce CD8+T-cell responses that cross-react with epidemic strains of the opposing lineage.
In conclusion, the present study shows for the first time, to the best of our knowledge, that human influenza B virus-specific CD8+T-cells are highly cross-reactive with influenza B viruses of the opposing lineage. Although quadrivalent influenza vaccines will be more commonly used in the near future, the induction of cross-reactive virus-specific T-cell responses may be a promising approach to broaden the protective efficacy of influenza vaccines, against both influenza A and B viruses. The induction of virus-specific CTL responses may be achieved with live attenuated influenza vaccines, especially in children (He et al., 2006). However, the administration of live attenuated influenza vaccines is restricted for certain high-risk groups (CDC, 2013; Fiore et al., 2010). The induction of virus-specific CD8+T-cell responses may also be achieved, e.g. by the use of specific adjuvants (Antrobus et al., 2014; Pérez-Girón et al., 2014; van de Sandt et al., 2014b) or novel antigen delivery systems (Altenburg et al., 2014; Berthoud et al., 2011; Daemen et al., 2005; Rimmelzwaan et al., 2000; Ulmer, 2002).
Methods
Cells
PBMCs were obtained from nine HLA-typed healthy blood donors (18–64 years of age) (Sanquin Bloodbank) by means of Lymphoprep (Axis-Shield) gradient centrifugation and were subsequently cryopreserved at − 135 °C. Study subjects were divided into three groups based on their HLA class I alleles (Table 2). The use of PBMCs for scientific research was approved by the Sanquin Bloodbank after informed consent was obtained from the blood donors.
Viruses
Prototypic influenza viruses B/Victoria/2/1987 (kindly provided by Vicki Gregory, WHO Collaborating Centre for Influenza, National Institute for Medical Research, London, UK) and B/Yamagata/16/1988 as well as the two more recent viruses, B/Netherlands/455/2011 and B/Netherlands/712/2011 belonging to each lineage, respectively, were propagated in Madin–Darby canine kidney cells at 37 °C. Culture supernatants were clarified by low-speed centrifugation and subsequently concentrated by ultracentrifugation, after which their infectious virus titres were determined as described previously (Rimmelzwaan et al., 1998).
Sequence analysis
Sequences of the eight gene segments of the above-described viruses were obtained as described previously (Westgeest et al., 2012) using segment-specific primers. Nucleotide sequences of the HA gene segments were used for phylogeny, and amino acid sequences of all gene segments were used to determine the amino acid sequence identity and for epitope prediction analysis, as described below.
Phylogeny
In addition to the above-mentioned viruses, an additional 47 human influenza B viruses, either used as vaccine strains or for which the lineage was previously confirmed by means of haemagglutination inhibition assay (Belshe, 2010; Belshe et al., 2010; Paiva et al., 2013; Rimmelzwaan et al., 2003; WHO, 2014a) and for which the complete HA nucleotide sequence was available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/genomes/FLU) or the Global Initiative on Sharing Avian Influenza Data (http://gisaid.org) influenza database, were selected for phylogenetic analysis. Previously published HA GenBank accession numbers are provided in Table S1. Nucleotide sequences of the HA gene segment of all 51 human influenza B viruses were aligned using the clustal w program running within BioEdit version 7.2.5 (Hall, 1999) and manually edited to maintain the correct reading frame. Nucleotides before the start codon and after the stop codon were removed. The nucleotide sequence alignment was used to determine the best-fit models of nucleotide substitution by jModelTest version 2.1.4 (Darriba et al., 2012; Guindon & Gascuel, 2003). The preferred ML-optimized model of nucleotide substitution, based on the Akaike information criterion, was TPM1uf+I+Γ4: Kimura three-parameter model (K81) (Kimura, 1981) with unequal base frequencies (uf), the proportion of invariant sites (I) and the Γ distribution of among-site rate variation with four categories estimated from the empirical data (Γ4). ML phylogenetic trees were reconstructed using the selected model of nucleotide substitution and PhyML version 3.1 (Guindon et al., 2010), performing a full heuristic search and subtree pruning and regrafting searches. The reliability of all phylogenetic groupings of each tree was determined through a non-parametric bootstrap resampling analysis with PhyML: 1000 replicates of ML trees were analysed by applying the TPM1uf+I+Γ4 model of nucleotide substitution. A detailed HA tree, including bootstrap values, is shown in Fig. 1. Trees were visualized through FigTree version 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/).
Amino acid sequence identity
blast analysis (http://blast.ncbi.nlm.nih.gov) was used to determine the amino acid sequence identity of all viral proteins of influenza B viruses used in this study, i.e. B/Victoria/2/1987, B/Yamagata/16/1988, B/Netherlands/455/2011 and B/Netherlands/712/2011 (Table 1).
Immunoinformatic analysis
Epitopes restricted for the HLA alleles of the study subjects were predicted based on the amino acid sequence of all internal proteins for B/Victoria/2/1987 and B/Yamagata/16/1988. For robustness, we used two online programs, i.e. Syfpeithi (http://www.syfpeithi.de) and Immuneepitope (http://tools.immuneepitope.org/mhci). In Syfpeithi, the MHC (HLA) type of the study subjects was selected in combination with prediction of epitopes of all possible lengths (all ‘mers’). The program indicated that putative epitopes were amongst the top 2 % of all top-scoring possibilities. An additional cut-off was set at a score of ≥ 19 as all known influenza A epitopes for these HLA alleles scored ≥ 19. In Immuneepitope, we also selected the HLA alleles of the study subjects in combination with all possible epitope lengths. The program indicated that most putative epitopes had an artificial neural network IC50 (nM) score of ≤ 500, so this was used as a cut-off value. Predicted amino acid sequences of putative epitopes are available on request.
Peptides
The HLA-B*0801-restricted putative epitopes predicted by Immuneepitope and Syfpeithi, and present in viruses of both lineages, were ordered as synthetic immunograde peptides (>85 % purity) (Eurogentec), in addition to previously in vitro confirmed HLA-B*0801-restricted influenza B epitopes NP263–271 ADRGLLRDI and NP413–421 ALKCKGFHV (Robbins et al., 1997).
In vitro expansion of influenza B virus-specific CD8+ T-cells
PBMCs obtained from HLA-typed healthy blood donors were stimulated with B/Victoria/2/1987 or B/Yamagata/16/1988 at m.o.i. 3, as described previously (Boon et al., 2002). Polyclonal CD8+T-cells were isolated from the expanded PBMC cultures 8–9 days after stimulation by means of CD8+ magnetically activated cell sorting according to the manufacturer's instructions (Miltenyi Biotec). These polyclonal CD8+T-cells were used as effector cells in IFN-γ ELISpot and lytic assays (see below).
Target cells
HLA-matched BLCLs were prepared as described previously (Rimmelzwaan et al., 2000). The cells (106) were incubated with or without 10 μM peptide for 1 h at 37 °C, and subsequently washed and resuspended in RPMI 1640 medium (Lonza) containing antibiotics and 10 % FBS (Sigma-Aldrich; R10F medium). Virus-infected target cells were prepared by inoculating BLCLs at m.o.i. 3 with B/Victoria/2/1987, B/Yamagata/16/1988, B/Netherlands/455/2011 or B/Netherlands/712/2011, or left untreated (negative control). After 1 h, cells were washed and resuspended in R10F medium, and then cultured for 16–18 h at 37 °C before being used for the stimulation of T-cells in IFN-γ ELISpot assays or as target cells in the CTL assays (see below).
IFN-γ ELISpot assay
The IFN-γ responses of in vitro expanded polyclonal CD8+T-cells were determined by ELISPOT assays, which were performed according to the manafacturer's recommendations (Mabtech). In brief, 10 000 in vitro expanded polyclonal CD8+T-cells were used as effector cells and incubated overnight with 30 000 peptide-loaded, virus-infected or untreated HLA class I-matched target cells, in triplicate. The mean number of spots was determined using an ELISpot reader and image analysis software (Aelvis).
CTL assay
To examine the lytic capacity of the in vitro expanded polyclonal CD8+T-cells, a CTL assay was used with CFSE-labelled target cells. In brief, 5 × 106 HLA class I-matched BLCLs were incubated with 50 μM CFSE (Sigma-Aldrich) for 5 min at 37 °C. These cells were subsequently inoculated with B/Victoria/2/1987, B/Yamagata/16/1988, B/Netherlands/455/2011 or B/Netherlands/712/2011 at m.o.i. 3 for 16–18 h. The infected and CFSE-labelled BLCLs were used as target cells and co-cultured with the in vitro expanded polyclonal CD8+ effector T-cells at E : T ratios of 5, 2.5 and 1.25. After a 3 h incubation period, dead cells were stained with ToPro3 (Invitrogen) for 10 min at 37 °C. Lysis in the target cell population was determined by flow cytometry using BD FACSDiva software (Becton Dickinson). Experiments were performed in triplicate. Percentage lysis was calculated by the formula: 100–[100 × (viable target cells in sample in presence of effector cells/viable target cells in absence of effector cells)].
Acknowledgements
R. A. M. F. received funding from the NIAID/NIH (HHSN272201400008C). G. F. R. received funding from EU grant FLUNIVAC (project ID 602604). A. D. M. E. O. was supported by the European Commission ERC project FLUPLAN (project ID 101920).
Supplementary Data
Supplementary Data
References
- Altenburg A.F., Kreijtz J.H., de Vries R.D., Song F., Fux R., Rimmelzwaan G.F., Sutter G., Volz A. (2014). Modified vaccinia virus ankara (MVA) as production platform for vaccines against influenza and other viral respiratory diseases Viruses 6 2735–2761 10.3390/v6072735 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C.S., Levin M.J. (2012). The rationale for quadrivalent influenza vaccines Hum Vaccin Immunother 8 81–88 10.4161/hv.8.1.17623 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antrobus R.D., Berthoud T.K., Mullarkey C.E., Hoschler K., Coughlan L., Zambon M., Hill A.V., Gilbert S.C. (2014). Coadministration of seasonal influenza vaccine and MVA-NP+M1 simultaneously achieves potent humoral and cell-mediated responses Mol Ther 22 233–238 10.1038/mt.2013.162 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E., Bui H.H., Sidney J., Zhang Q., Glenn J., Oseroff C., Mbawuike I.N., Alexander J., Newman M.J. (2008). Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans J Virol 82 12241–12251 10.1128/JVI.01563-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshe R.B. (2010). The need for quadrivalent vaccine against seasonal influenza Vaccine 28 (Suppl 4), D45–D53 10.1016/j.vaccine.2010.08.028 . [DOI] [PubMed] [Google Scholar]
- Belshe R.B., Coelingh K., Ambrose C.S., Woo J.C., Wu X. (2010). Efficacy of live attenuated influenza vaccine in children against influenza B viruses by lineage and antigenic similarity Vaccine 28 2149–2156 10.1016/j.vaccine.2009.11.068 . [DOI] [PubMed] [Google Scholar]
- Beran J., Wertzova V., Honegr K., Kaliskova E., Havlickova M., Havlik J., Jirincova H., Van Belle P., Jain V. (2009). Challenge of conducting a placebo-controlled randomized efficacy study for influenza vaccine in a season with low attack rate and a mismatched vaccine B strain: a concrete example BMC Infect Dis 9 2 10.1186/1471-2334-9-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoud T.K., Hamill M., Lillie P.J., Hwenda L., Collins K.A., Ewer K.J., Milicic A., Poyntz H.C., Lambe T. (2011). Potent CD8+T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1 Clin Infect Dis 52 1–7 10.1093/cid/ciq015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., Morick D., deMutsert G., Osinga N., Bestebroer T., vanderVliet S., Smits S.L., Kuiken T., Rimmelzwaan G.F. (2013). Recurring influenza B virus infections in seals Emerg Infect Dis 19 511–512 10.3201/eid1903.120965 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon A.C., de Mutsert G., Graus Y.M., Fouchier R.A., Sintnicolaas K., Osterhaus A.D., Rimmelzwaan G.F. (2002). The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype J Virol 76 582–590 10.1128/JVI.76.2.582-590.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon A.C., De Mutsert G., Fouchier R.A., Sintnicolaas K., Osterhaus A.D., Rimmelzwaan G.F. (2004). Preferential HLA usage in the influenza virus-specific CTL response J Immunol 172 4435–4443 10.4049/jimmunol.172.7.4435 . [DOI] [PubMed] [Google Scholar]
- Boyce W.M., Sandrock C., Kreuder-Johnson C., Kelly T., Cardona C. (2009). Avian influenza viruses in wild birds: a moving target Comp Immunol Microbiol Infect Dis 32 275–286 10.1016/j.cimid.2008.01.002 . [DOI] [PubMed] [Google Scholar]
- CDC (2009). Update: influenza activity – United States, September 28, 2008–April 4, 2009, and composition of the 2009–10 influenza vaccine MMWR Morb Mortal Wkly Rep 58 369–374 . [PubMed] [Google Scholar]
- CDC (2013). Prevention and control of seasonal influenza with vaccines. Recommendations of the Advisory Committee on Immunization Practices United States, 2013–2014 MMWR Recomm Rep 62 (RR-07),1–43 . [PubMed] [Google Scholar]
- CDC (2014). Past weekly surveillance reports http://www.cdc.gov/flu/weekly/pastreports.htm.
- Chen R., Holmes E.C. (2008). The evolutionary dynamics of human influenza B virus J Mol Evol 66 655–663 10.1007/s00239-008-9119-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.M., Guo Y.J., Wu K.Y., Guo J.F., Wang M., Dong J., Zhang Y., Li Z., Shu Y.L. (2007). Exploration of the emergence of the Victoria lineage of influenza B virus Arch Virol 152 415–422 10.1007/s00705-006-0852-6 . [DOI] [PubMed] [Google Scholar]
- Daemen T., de Mare A., Bungener L., de Jonge J., Huckriede A., Wilschut J. (2005). Virosomes for antigen and DNA delivery Adv Drug Deliv Rev 57 451–463 10.1016/j.addr.2004.09.005 . [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing Nat Methods 9 772 10.1038/nmeth.2109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domachowske J.B., Pankow-Culot H., Bautista M., Feng Y., Claeys C., Peeters M., Innis B.L., Jain V. (2013). A randomized trial of candidate inactivated quadrivalent influenza vaccine versus trivalent influenza vaccines in children aged 3–17 years J Infect Dis 207 1878–1887 10.1093/infdis/jit091 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C., Laursen N.S., Kwaks T., Zuijdgeest D., Khayat R., Ekiert D.C., Lee J.H., Metlagel Z., Bujny M.V. (2012). Highly conserved protective epitopes on influenza B viruses Science 337 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein S.L. (2006). Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature J Infect Dis 193 49–53 10.1086/498980 . [DOI] [PubMed] [Google Scholar]
- FDA (2012). FDA News Release: FDA approves first quadrivalent vaccine to prevent seasonal influenza http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm294057.htm.
- Feng L., Shay D.K., Jiang Y., Zhou H., Chen X., Zheng Y., Jiang L., Zhang Q., Lin H. (2012). Influenza-associated mortality in temperate and subtropical Chinese cities, 2003–2008 Bull World Health Organ 90 279–288B 10.2471/BLT.11.096958 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore A.E., Uyeki T.M., Broder K., Finelli L., Euler G.L., Singleton J.A., Iskander J.K., Wortley P.M., Shay D.K. (2010). Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010 MMWR Recomm Rep 59 (RR-8), 1–62 . [PubMed] [Google Scholar]
- Flynn K.J., Belz G.T., Altman J.D., Ahmed R., Woodland D.L., Doherty P.C. (1998). Virus-specific CD8+T cells in primary and secondary influenza pneumonia Immunity 8 683–691 10.1016/S1074-7613(00)80573-7 . [DOI] [PubMed] [Google Scholar]
- Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H. (2013). Human infection with a novel avian-origin influenza A (H7N9) virus N Engl J Med 368 1888–1897 10.1056/NEJMoa1304459 . [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood Syst Biol 52 696–704 10.1080/10635150390235520 . [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0 Syst Biol 59 307–321 10.1093/sysbio/syq010 . [DOI] [PubMed] [Google Scholar]
- Hall T.A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT Nucleic Acids Symp Ser 41 95–98. [Google Scholar]
- He X.S., Holmes T.H., Zhang C., Mahmood K., Kemble G.W., Lewis D.B., Dekker C.L., Greenberg H.B., Arvin A.M. (2006). Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines J Virol 80 11756–11766 10.1128/JVI.01460-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen T., Ikonen N., Ziegler T. (2014). Impact of influenza B lineage-level mismatch between trivalent seasonal influenza vaccines and circulating viruses, 1999–2012 Clin Infect Dis 59 1519–1524. [DOI] [PubMed] [Google Scholar]
- Heinonen S., Silvennoinen H., Lehtinen P., Vainionpää R., Ziegler T., Heikkinen T. (2011). Effectiveness of inactivated influenza vaccine in children aged 9 months to 3 years: an observational cohort study Lancet Infect Dis 11 23–29 10.1016/S1473-3099(10)70255-3 . [DOI] [PubMed] [Google Scholar]
- Hillaire M.L., van Trierum S.E., Kreijtz J.H., Bodewes R., Geelhoed-Mieras M.M., Nieuwkoop N.J., Fouchier R.A., Kuiken T., Osterhaus A.D., Rimmelzwaan G.F. (2011). Cross-protective immunity against influenza pH1N1 2009 viruses induced by seasonal influenza A (H3N2) virus is mediated by virus-specific T-cells J Gen Virol 92 2339–2349 10.1099/vir.0.033076-0 . [DOI] [PubMed] [Google Scholar]
- Hillaire M.L., Vogelzang-van Trierum S.E., Kreijtz J.H., de Mutsert G., Fouchier R.A., Osterhaus A.D., Rimmelzwaan G.F. (2013). Human T-cells directed to seasonal influenza A virus cross-react with 2009 pandemic influenza A (H1N1) and swine-origin triple-reassortant H3N2 influenza viruses J Gen Virol 94 583–592 10.1099/vir.0.048652-0 . [DOI] [PubMed] [Google Scholar]
- Hoft D.F., Babusis E., Worku S., Spencer C.T., Lottenbach K., Truscott S.M., Abate G., Sakala I.G., Edwards K.M. (2011). Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children J Infect Dis 204 845–853 10.1093/infdis/jir436 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson J., Cruz J., Ennis F.A. (1998). Human cytotoxic T-lymphocyte repertoire to influenza A viruses J Virol 72 8682–8689 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson J., Cruz J., Terajima M., Ennis F.A. (1999). Human CD8+ and CD4+T lymphocyte memory to influenza A viruses of swine and avian species J Immunol 162 7578–7583 . [PubMed] [Google Scholar]
- Kieninger D., Sheldon E., Lin W.Y., Yu C.J., Bayas J.M., Gabor J.J., Esen M., Fernandez Roure J.L., Narejos Perez S. (2013). Immunogenicity, reactogenicity and safety of an inactivated quadrivalent influenza vaccine candidate versus inactivated trivalent influenza vaccine: a phase III, randomized trial in adults aged ≥ 18 years BMC Infect Dis 13 343 10.1186/1471-2334-13-343 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1981). Estimation of evolutionary distances between homologous nucleotide sequences Proc Natl Acad Sci U S A 78 454–458 10.1073/pnas.78.1.454 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel B.F., Burke D.F., Bestebroer T.M., van der Vliet S., Zondag G.C., Vervaet G., Skepner E., Lewis N.S., Spronken M.I. (2013). Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution Science 342 976–979. [DOI] [PubMed] [Google Scholar]
- Kreijtz J.H., Bodewes R., van Amerongen G., Kuiken T., Fouchier R.A., Osterhaus A.D., Rimmelzwaan G.F. (2007). Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice Vaccine 25 612–620 10.1016/j.vaccine.2006.08.036 . [DOI] [PubMed] [Google Scholar]
- Kreijtz J.H., de Mutsert G., van Baalen C.A., Fouchier R.A., Osterhaus A.D., Rimmelzwaan G.F. (2008). Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus J Virol 82 5161–5166 10.1128/JVI.02694-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreijtz J.H., Bodewes R., van den Brand J.M., de Mutsert G., Baas C., van Amerongen G., Fouchier R.A., Osterhaus A.D., Rimmelzwaan G.F. (2009). Infection of mice with a human influenza A/H3N2 virus induces protective immunity against lethal infection with influenza A/H5N1 virus Vaccine 27 4983–4989 10.1016/j.vaccine.2009.05.079 . [DOI] [PubMed] [Google Scholar]
- Langley J.M., Carmona Martinez A., Chatterjee A., Halperin S.A., McNeil S., Reisinger K.S., Aggarwal N., Huang L.M., Peng C.T. (2013). Immunogenicity and safety of an inactivated quadrivalent influenza vaccine candidate: a phase III randomized controlled trial in children J Infect Dis 208 544–553 10.1093/infdis/jit263 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.Y., Ha L.A., Simmons C., de Jong M.D., Chau N.V., Schumacher R., Peng Y.C., McMichael A.J., Farrar J.J. (2008). Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals J Clin Invest 118 3478–3490 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom S.E., Hiromoto Y., Nishimura H., Saito T., Nerome R., Nerome K. (1999). Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes J Virol 73 4413–4426 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C., Nielsen M., Lund O. (2006). The validity of predicted T-cell epitopes Trends Biotechnol 24 537–538 10.1016/j.tibtech.2006.10.001 . [DOI] [PubMed] [Google Scholar]
- McCullers J.A., Saito T., Iverson A.R. (2004). Multiple genotypes of influenza B virus circulated between 1979 and 2003 J Virol 78 12817–12828 10.1128/JVI.78.23.12817-12828.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael A.J., Gotch F.M., Noble G.R., Beare P.A. (1983). Cytotoxic T-cell immunity to influenza N Engl J Med 309 13–17 10.1056/NEJM198307073090103 . [DOI] [PubMed] [Google Scholar]
- O'Neill E., Krauss S.L., Riberdy J.M., Webster R.G., Woodland D.L. (2000). Heterologous protection against lethal A/HongKong/156/97 (H5N1) influenza virus infection in C57BL/6 mice J Gen Virol 81 2689–2696 . [DOI] [PubMed] [Google Scholar]
- Olson D.R., Heffernan R.T., Paladini M., Konty K., Weiss D., Mostashari F. (2007). Monitoring the impact of influenza by age: emergency department fever and respiratory complaint surveillance in New York City PLoS Med 4 e247 10.1371/journal.pmed.0040247 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva T.M., Benega M.A., Silva D.B., Santos K.C., Cruz A.S., Hortenci M.F., Barbieri M.T., Monteiro M.M., Barbosa H.A., Carvalhanas T.R. (2013). Evolutionary pattern of reemerging influenza B/Victoria lineage viruses in São Paulo, Brazil, 1996–2012: implications for vaccine composition strategy J Med Virol 85 1983–1989 10.1002/jmv.23684 . [DOI] [PubMed] [Google Scholar]
- Pérez-Girón J.V., Belicha-Villanueva A., Hassan E., Gómez-Medina S., Cruz J.L., Lüdtke A., Ruibal P., Albrecht R.A., García-Sastre A., Muñoz-Fontela C. (2014). Mucosal polyinosinic-polycytidylic acid improves protection elicited by replicating influenza vaccines via enhanced dendritic cell function and T cell immunity J Immunol 193 1324–1332 10.4049/jimmunol.1400222 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Parra S., Grant E., Loh L., Nguyen T.H., Campbell K.A., Tong S.Y., Miller A., Doherty P.C., Vijaykrishna D. (2014). Preexisting CD8+T-cell immunity to the H7N9 influenza A virus varies across ethnicities Proc Natl Acad Sci U S A 111 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan G.F., Baars M., Claas E.C., Osterhaus A.D. (1998). Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro J Virol Methods 74 57–66 10.1016/S0166-0934(98)00071-8 . [DOI] [PubMed] [Google Scholar]
- Rimmelzwaan G.F., Nieuwkoop N., Brandenburg A., Sutter G., Beyer W.E., Maher D., Bates J., Osterhaus A.D. (2000). A randomized, double blind study in young healthy adults comparing cell mediated and humoral immune responses induced by influenza ISCOM vaccines and conventional vaccines Vaccine 19 1180–1187 10.1016/S0264-410X(00)00310-8 . [DOI] [PubMed] [Google Scholar]
- Rimmelzwaan G.F., deJong J.C., Bartelds A.I., Wilbrink B., Fouchier R.A., Osterhaus A.D. (2003). [The 2001/2002 influenza season and the vaccine composition for the 2002/ season] Ned Tijdschr Geneeskd 146 1846–1850 (in Dutch). [PubMed] [Google Scholar]
- Robbins P.A., Lettice L.A., Rota P., Santos-Aguado J., Rothbard J., McMichael A.J., Strominger J.L. (1989). Comparison between two peptide epitopes presented to cytotoxic T lymphocytes by HLA-A2.Evidence for discrete locations within HLA-A2 J Immunol 143 4098–4103 . [PubMed] [Google Scholar]
- Robbins P.A., Garboczi D.N., Strominger J.L. (1995). HLA-A*0201 complexes with two 10-Mer peptides differing at the P2 anchor residue have distinct refolding kinetics J Immunol 154 703–709 . [PubMed] [Google Scholar]
- Robbins P.A., Rota P.A., Shapiro S.Z. (1997). A broad cytotoxic T lymphocyte response to influenza type B virus presented by multiple HLA molecules Int Immunol 9 815–823 10.1093/intimm/9.6.815 . [DOI] [PubMed] [Google Scholar]
- Roider J., Meissner T., Kraut F., Vollbrecht T., Stirner R., Bogner J.R., Draenert R. (2014). Comparison of experimental fine-mapping to in silico prediction results of HIV-1 epitopes reveals ongoing need for mapping experiments Immunology 143 193–201 10.1111/imm.12301 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Wallis T.R., Harmon M.W., Rota J.S., Kendal A.P., Nerome K. (1990). Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983 Virology 175 59–68 10.1016/0042-6822(90)90186-U . [DOI] [PubMed] [Google Scholar]
- Rota P.A., Hemphill M.L., Whistler T., Regnery H.L., Kendal A.P. (1992). Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus J Gen Virol 73 2737–2742 10.1099/0022-1317-73-10-2737 . [DOI] [PubMed] [Google Scholar]
- Russell C.A., Jones T.C., Barr I.G., Cox N.J., Garten R.J., Gregory V., Gust I.D., Hampson A.W., Hay A.J. (2008). Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses Vaccine 26 (Suppl 4), D31–D34 10.1016/j.vaccine.2008.07.078 . [DOI] [PubMed] [Google Scholar]
- Shaw M.W., Xu X., Li Y., Normand S., Ueki R.T., Kunimoto G.Y., Hall H., Klimov A., Cox N.J., Subbarao K. (2002). Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000–2001 and 2001–2002 seasons Virology 303 1–8 10.1006/viro.2002.1719 . [DOI] [PubMed] [Google Scholar]
- Silver M.L., Parker K.C., Wiley D.C. (1991). Reconstitution by MHC-restricted peptides of HLA-A2 heavy chain with beta 2-microglobulin, in vitro Nature 350 619–622 10.1038/350619a0 . [DOI] [PubMed] [Google Scholar]
- Simonsen L., Clarke M.J., Williamson G.D., Stroup D.F., Arden N.H., Schonberger L.B. (1997). The impact of influenza epidemics on mortality: introducing a severity index Am J Public Health 87 1944–1950 10.2105/AJPH.87.12.1944 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepushkin A.N. (1959). The effect of a previous attack of A1 influenza on susceptibility to A2 virus during the 1957 outbreak Bull World Health Organ 20 297–301 . [PMC free article] [PubMed] [Google Scholar]
- Smith D.J., Lapedes A.S., de Jong J.C., Bestebroer T.M., Rimmelzwaan G.F., Osterhaus A.D., Fouchier R.A. (2004). Mapping the antigenic and genetic evolution of influenza virus Science 305 371–376. [DOI] [PubMed] [Google Scholar]
- Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., Bean T., Barclay W., Deeks J.J., Lalvani A. (2013). Cellular immune correlates of protection against symptomatic pandemic influenza Nat Med 19 1305–1312 10.1038/nm.3350 . [DOI] [PubMed] [Google Scholar]
- Thompson W.W., Shay D.K., Weintraub E., Brammer L., Cox N., Anderson L.J., Fukuda K. (2003). Mortality associated with influenza and respiratory syncytial virus in the United States JAMA 289 179–186 10.1001/jama.289.2.179 . [DOI] [PubMed] [Google Scholar]
- Thompson W.W., Shay D.K., Weintraub E., Brammer L., Bridges C.B., Cox N.J., Fukuda K. (2004). Influenza-associated hospitalizations in the United States JAMA 292 1333–1340 10.1001/jama.292.11.1333 . [DOI] [PubMed] [Google Scholar]
- Tinoco J.C., Pavia-Ruz N., Cruz-Valdez A., Aranza Doniz C., Chandrasekaran V., Dewé W., Liu A., Innis B.L., Jain V.K. (2014). Immunogenicity, reactogenicity, and safety of inactivated quadrivalent influenza vaccine candidate versus inactivated trivalent influenza vaccine in healthy adults aged ≥ 18 years: a phase III, randomized trial Vaccine 32 1480–1487 10.1016/j.vaccine.2014.01.022 . [DOI] [PubMed] [Google Scholar]
- Ulmer J.B. (2002). Influenza DNA vaccines Vaccine 20 (Suppl 2), S74–S76 10.1016/S0264-410X(02)00136-6 . [DOI] [PubMed] [Google Scholar]
- van de Sandt C.E., Kreijtz J.H., Rimmelzwaan G.F. (2012). Evasion of influenza A viruses from innate and adaptive immune responses Viruses 4 1438–1476 10.3390/v4091438 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sandt C.E., Kreijtz J.H., de Mutsert G., Geelhoed-Mieras M.M., Hillaire M.L., Vogelzang-van Trierum S.E., Osterhaus A.D., Fouchier R.A., Rimmelzwaan G.F. (2014a). Human cytotoxic T lymphocytes directed to seasonal influenza A viruses cross-react with the newly emerging H7N9 virus J Virol 88 1684–1693 10.1128/JVI.02843-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sandt C.E., Kreijtz J.H., Geelhoed-Mieras M.M., Vogelzang-van Trierum S.E., Nieuwkoop N.J., van de Vijver D.A., Fouchier R.A., Osterhaus A.D., Morein B., Rimmelzwaan G.F. (2014b). Novel G3/DT adjuvant promotes the induction of protective T cells responses after vaccination with a seasonal trivalent inactivated split-virion influenza vaccine Vaccine 32 5614–5623 10.1016/j.vaccine.2014.08.003 . [DOI] [PubMed] [Google Scholar]
- van der Burg S.H., Visseren M.J., Brandt R.M., Kast W.M., Melief C.J. (1996). Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability J Immunol 156 3308–3314 . [PubMed] [Google Scholar]
- Weinfurter J.T., Brunner K., Capuano S.V., III, Li C., Broman K.W., Kawaoka Y., Friedrich T.C. (2011). Cross-reactive T cells are involved in rapid clearance of 2009 pandemic H1N1 influenza virus in nonhuman primates PLoS Pathog 7 e1002381 10.1371/journal.ppat.1002381 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westgeest K.B., de Graaf M., Fourment M., Bestebroer T.M., van Beek R., Spronken M.I., de Jong J.C., Rimmelzwaan G.F., Russell C.A. (2012). Genetic evolution of the neuraminidase of influenza A (H3N2) viruses from 1968 to 2009 and its correspondence to haemagglutinin evolution J Gen Virol 93 1996–2007 10.1099/vir.0.043059-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2014a). WHO Recommendations on the Composition of Influenza Virus Vaccines http://www.who.int/influenza/vaccines/virus/recommendations/en.
- WHO (2014b). Influenza (Seasonal) Fact Sheet 211 http://www.who.int/mediacentre/factsheets/fs211/en/index.html.
- Yewdell J.W., Bennink J.R., Smith G.L., Moss B. (1985). Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes Proc Natl Acad Sci U S A 82 1785–1789 10.1073/pnas.82.6.1785 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data