Abstract
The Escherichia coli (E. coli) AlkB protein and its functional human homologues belong to a subfamily of 2-oxoglutarate (2OG)-dependent oxygenases (2OG oxygenases for simplicity) that enable the repair of cytotoxic methylation damage in nucleic acids and which catalyse t-RNA oxidations. DNA alkylation is a major mechanism of action for cytotoxic anti-cancer drugs. Thus, the inhibition of oxidative demethylation, catalyzed by these enzymes, has the potential to improve the efficacy of chemotherapies. Here we report that oligonucleotide aptamers constitute a new class of potent inhibitors of 2OG oxygenases. DNA aptamers can selectively bind to AlkB, with nanomolar affinity, and efficiently inhibit catalysis. The mechanism of inhibition was studied by capillary electrophoresis (CE) with laser-induced fluorescence (LIF) detection. Inhibition constants of the aptamers were determined and shown to correlate well with Kd values. The results of kinetic analyses imply that the aptamers bind AlkB away from the active site. Our findings should stimulate the development of oligonucleotide aptamers for human homologues of AlkB and further their study as potential enhancers of chemotherapy efficiency.
1. INTRODUCTION
Alkylating agents can cause DNA damage-induced apoptosis, and are widely used in anti-cancer chemotherapy. However, the efficiency of chemotherapeutic agents, which work through damaging DNA, is strongly reduced by DNA repair systems. Inhibition of nucleotide repair enzymes can be used in combination with anti-cancer alkylating agents, to allow for dose reduction and improvement of therapeutic efficiency.1 One mechanism of DNA repair involves enzymes from the subfamily of 2OG oxygenases. These enzymes catalyze the dealkylation of DNA or RNA, that have been modified by alkylating agents present in the environment or cellular-metabolism.2 2OG oxygenases acting on nucleic acids can also catalyze RNA hydroxylation and the oxidation of 5-methyl cytosine bases. 2OG oxygenases that modify nucleic acids are linked to diseases including obesity and cancer.3
The AlkB protein from E. coli is a well studied 2OG oxygenase. It is often used as a typical representative of the subfamily of 2OG oxygenases in functional studies on these enzymes and in the development of new methods for studying them.4-12 AlkB catalyzes demethylation of 1-methyladenine and 3-methylcytosine lesions in both single and double-stranded DNA. As with other 2OG oxygenases, AlkB catalysis employs 2OG and oxygen as co-substrates and produces CO2 and succinate as co-products (Fig. 1). Following oxidative decarboxylation of 2OG, a ferryl-intermediate is generated, which then reacts to give a hydroxylated intermediate. The further oxidation step of the enzymatic reaction releases formaldehyde, succinate and the dealkylated DNA product.
Figure 1.
A schematic of AlkB-catalyzed demethylation of DNA.
The AlkB-type cellular defense mechanism, which invloves the hydroxylation of alkyl groups from nucleic acid substrates, is conserved from bacteria to humans.4 In total, eight human AlkB homologues, and related nucleic acid oxygenases, are identified. Some of these enzymes are linked to diseases, including neurological disorders, obesity development, and various types of cancer.14-16 For example, ABH8 is reported to contribute to bladder cancer progression.17, 18 AlkBH2 and AlkBH3 expression are linked to human gastric cancer and non-small cell lung cancer.19-21 Further studies of AlkB and its human homologues require efficient and selective regulators of their enzymatic activity and reliable affinity probes that allow one to measure the expression levels of these proteins in complex biological matrixes.
As with many other proteins, antibodies are important reagents in studies on nucleic acid oxygenases.22 It has also been shown that ABH8 antibodies can inhibit catalysis, and can potentially be utilized for cancer diagnosis and treatment.23 Oligonucleotide aptamers (aptamers for simplicity) are single-stranded DNA or RNA structures that can recognize biomacromolecules with high affinity and selectivity. Aptamers are generally more stable than antibodies, as well as less expensive and easier to synthesize without the use of animals or cell culture. Nucleic acids offer multiple options for chemical or biochemical modification; accordingly, aptamers can usually be tagged with fluorophores without changing their native conformation or compromising their affinity.24
In addition to being efficient affinity probes, oligonucleotide aptamers (mostly RNA) have been shown to control the activity of enzymes with high affinity and specificity. Examples of typical target proteins include glycosylases, protein kinases, RNA polymerases, and others.25
Aptamers can be produced from highly diverse DNA or RNA libraries via systematic evolution of ligands by exponential enrichment (SELEX).26-27 Partitioning of binders from non-binders by non-equilibrium CE of equilibrium mixtures (NECEEM) enables rapid aptamer selection.28 In addition, NECEEM can be utilized as a general tool for measuring binding parameters of aptamer-target interaction (equilibrium dissociation constant, Kd, and the rate constant of complex dissociation, koff) and for the quantitative target detection in aptamer-based affinity analyses.28-32 Recently, we reported the use of NECEEM for: (i) the development of AlkB binding aptamers with affinity constants in the nM range and (ii) the quantitative aptamer-based detection of AlkB in vitro in the presence of E. coli cell lysates.33
Here, we report studies on the inhibition reaction of aptamers for AlkB. Our overall goal was to establish a bioanalytical approach for studying the mechanisms of inhibition of AlkB catalysis by aptamers and determining their inhibition constants. The developed methodology is general and should be applicable to the selection and characterization of aptamer-based inhibitors of other 2OG/Fe(II)-dependent oxygenases. Moreover, aptamer-based inhibitors can be developed for use in in vitro and potentially in vivo studies of other known nucleic acid modifying enzymes, and in the discovery of new members of this family.
2. MATERIALS AND METHODS
2.1. Materials
Uncoated fused-silica capillaries were purchased from Polymicro (Phoenix, AZ). USA). Synthetic fluorescently labelled DNA substrate (5’-TTCmTTTTTTTTTTTT-3’-fluorescein), and product (5’-TTCTTTTTTTTTTTT-3’-fluorescein) were synthesized by ATDbio (University of Southampton, U. K). AlkB from E. coli was purified according to the published procedure.4 A 1 mM stock solution of AlkB in 50 mM Tris HCl, 500 mM NaCl, 1 mM DTT at pH 7 was stored at −80 °C. Fluorescently-labelled DNA aptamers were synthesized and HPLC-purified HPLC by IDT, Coralville, IA. We analyzed the purity of aptamers by CE to confirm that it was less than 95%. All aptamers had a total length of 80 nucleotides, including 2 constant flanking regions of 20 nucleotides each; a fluorescein label was attached to the 5’-end. The aptamers were stored at −20 °C. All other chemicals were obtained from Sigma-Aldrich (Toronto, ON, Canada). All solutions were made using deionised water filtered through a 0.22 μm filter (Millipore, Nepean, ON, Canada).
2.2. Instrumentation
CE experiments were performed with a P/ACE MDQ instrument from Beckman Coulter (Fullerton, CA, USA) utilizing LIF detection (excitation at 488 nm and emission at 520 nm) and light absorption detection (280 nm). Uncoated fused-silica capillaries with a total length of 50 cm (40 cm to the detection window) and inner and outer diameters of 75 and 365 μm, respectively, were used. All samples were introduced into the capillary by pressure.
The capillary temperature was kept at 15 °C. Electrophoresis was carried out for a total of 30 min by an electric field of a specified strength with a positive electrode at the injection end of the capillary.
2.3. AlkB aptamers
We used previously selected DNA aptamers in their full-lengths including the “random” region and two constant flanking regions.33 In this study, the following aptamers were used: aptamer A2, TGCCTAGCGTTTCATTGTCCCTTCTTATTAGGTGATAATA, aptamer A3, CCCATATCGGTGAATGCACGAGCAACCGATTGACACGGG, aptamer A11, AGAAATTGGTACTGTATGAAACGGCAGCTGCACGTCGCG, and aptamer A24, GACTGCTGATGAGTCACTTTAACGTGGAGCAAAGATTAAA. The names, A2, A3, A11, and A24, are adapted from the original work.33 Aptamers used in the inhibition assays did not contain a fluorescent label, while the aptamers used in the affinity assays has a 6-carboxyfluorescein fluorescent label at their 5’-ends.
2.4. Assay of AlkB activity
The AlkB enzymatic activity assay was based on the quantification of the demethylated product from the following methylated substrate: 5′-TTCmTTTTTTTTTTTT-fluorescein-3′ (TTCm), where Cm is 3-methylated cytosine. The enzymatic reaction was initiated by the addition of AlkB, at a final concentration of 5 nM, to a mixture containing 50 mM Tris-HCl at pH 7.5, 4 mM L-ascorbic acid (AA), 160 μM 2OG, 80 μM (NH4)2SO4·FeSO4·6H2O as a source of iron (II), 20 to 400 nM TTCm, and 10,000 units/ml of catalase (all concentrations are final). To stop the reaction, EDTA was added at a final concentration of 5 mM to equilibrium mixtures containing the enzyme and substrate that had been incubated over different times. The initial reaction rate (nM/min) was measured as a slope of the initial (linear) part of the “product versus reaction time” curve. The initial reaction rate was determined for varying concentrations of the methylated DNA substrate; Km and kcat values were calculated by fitting the Michaelis-Menten equation to the experimental curve using the Origin Pro 8 SPO software (OriginLab Corporation, MA, USA).
2.5. Binding assay of AlkB aptamers
To determine Kd values of the aptamer-AlkB complexes, the reaction mixture containing 50-200 nM AlkB and 50-100 nM fluorescein-labelled aptamer (A2, A3, A11, or A24) was incubated at room temperature for 20 min. Then, this mixture was injected into a 50-cm-long capillary by a 0.5 psi × 10 s pressure pulse. An electric field of 400 V/cm was applied to the capillary with the positive electrode being at the inlet. The electrophoresis run buffer was 50 mM Tris-acetate, pH 8.2. Areas corresponding to the free and bound DNA were quantified by LIF detection, and the Kd values were determined using the NECEEM mathematics described elsewhere.31
The incubation buffer was 50 mM Tris-HCl, pH 7.5. The binding assay conditions were varied by adding the following components to the incubation buffer: a) 50 mM NaCl, b) 80 μM (NH4)2SO4·FeSO4·6H2O, 160 μM 2OG, and 4 mM AA, c) 150 nM TTCm, 80 μM FeSO4, 100 μM N-oxalylglycine (NOG), and 4 mM AA, and d) 150 nM TTCm, 3 mM CoCl2, 4 mM AA, and 15 mM 2OG. In the aptamer competition binding assays, the reaction mixture contained 200 nM AlkB, 100 nM fluorescein labelled A2 aptamer, and either 300 nM unlabelled A2 (A3) aptamer or 2 μM unlabelled A11 (A24) aptamer in the incubation buffer.
2.6. Inhibition assay
Inhibition assays were performed under the same conditions as the activity assay described above, but with addition of the inhibitor into the same buffer. The reaction product (P) and unreacted substrate (S) were separated by CE using a 20 mM Borax, 60 mM sodium dodecyl sulphate run buffer at pH 8.5 and quantitated with LIF detection. The initial rate of reaction was determined as a function of aptamer concentration at fixed TTCm substrate concentrations in the range of 20 to 300 nM. In the inhibition assays, we used the same four DNA aptamers described above, but without the 6-carboxyfluorescein fluorescent label at the 5’-end of the aptamer. Concentrations of aptamers were varied as following: a) A2 was present at concentrations ranging between 0.5 and 25 nM, b) A3, A11, and A24 were present at concentrations ranging between 30 and 250 nM. The concentration of NOG inhibitor was varied between 8 and 80 nM. For the determination of Ki values, the Lineweaver-Burk plot (Fig. 5) and Morrison fits were used (Fig. 4).
Figure 5.
Kinetic (Lineweaver-Burk) analysis of AlkB inhibition kinetics by DNA aptamers and, in-contrast, by NOG, a structural analogue of 2OG. Initial rates of AlkB-catalyzed demethylation of TTCm were determined at substrate concentrations ranging from 1 to 50 nM in the presence of 0, 30, 100, and 250 nM DNA aptamer (B-D graphs) or 0, 8, 16, and 80 nM NOG (graph A and E).
Figure 4.
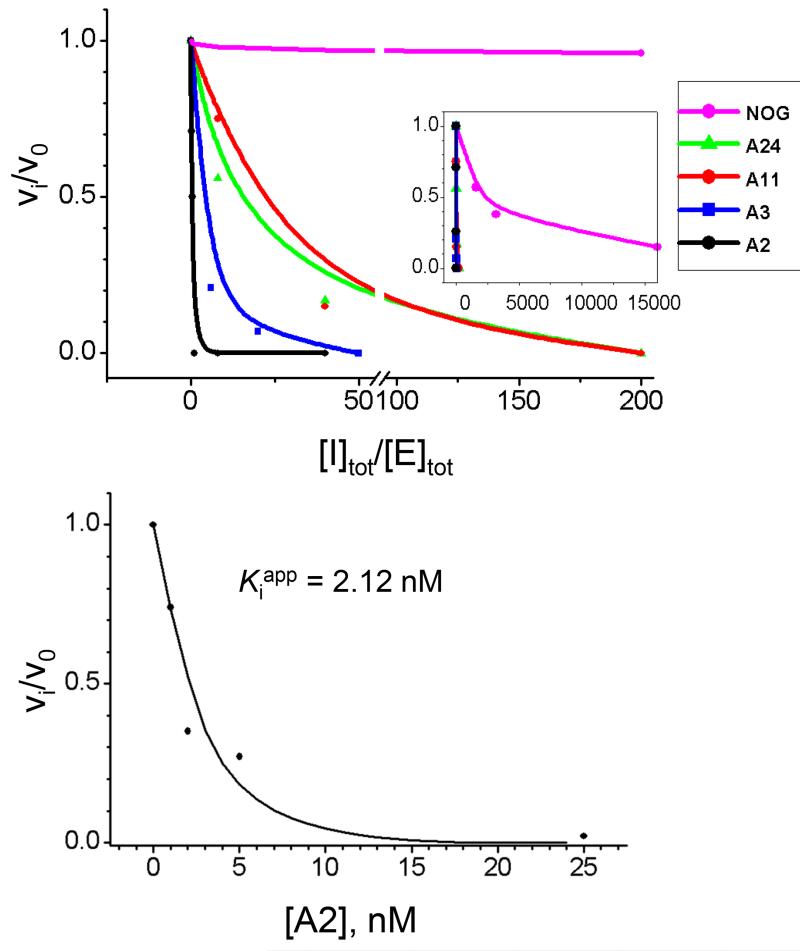
The top panel shows inhibitory activity of the A2 aptamer in comparison to those of A3, A11, and A24 aptamers and NOG. The inset shows the same graph with the expanded x axis. The bottom panel illustrates the determination of Ki for the A2 aptamer by using the Morrison fit.
3. RESULTS AND DISCUSSION
The AlkB-binding DNA aptamers studied here were isolated from a random-sequence DNA library by using the NECEEM method.33 The aptamers did not show any detectable non-specific binding to other proteins present in the E. coli cell lysate and were selective for the AlkB protein.33 Here, we utilized the NECEEM method to analyse the affinity of the AlkB aptamers towards AlkB and gauge their activity as inhibitors. Example of typical NECEEM electropherograms are shown in Fig. 2. The equilibrium mixture contained AlkB, a DNA aptamer, and the AlkB-aptamer complex. To determine Kd values, peak areas corresponding to unbound DNA and protein-DNA complex in the NECEEM electropherograms were analyzed. Since the DNA aptamers were fluorescently labeled, its complex with AlkB was also labeled and detectable by LIF. The equilibrium mixture was injected into an uncoated capillary with the positive electrode at the inlet. The mixture was then subjected to free-zone electrophoresis with a considerable electroosmotic flow that could carry all components of the equilibrium mixture to the capillary outlet. Unbound DNA, characterized by highly negative charge to size ratio, migrated slower than AlkB when the positive electrode is placed at the capillary inlet. The complex of the aptamer with the protein migrated between the unbound protein (not detected since it was not labeled) and the unbound DNA.
Figure 2.
The influence of Fe2+ and 2OG on the formation of an affinity complex between AlkB and the A2 aptamer. See text for details.
We used four out of the sixteen previously selected aptamers to investigate the aptamers ability to modulate AlkB catalysis.33 Four representative candidates were selected to cover a range of Kd values that included strong and moderate binders. The activity assay required different experimental conditions from those reported in the binding assay; thus, we tested the affinity of the strongest binder, aptamer A2, at varying complex formation conditions (Table 1). We found that the addition of 50 mM NaCl to the Tris-HCl incubation buffer increased the Kd value of the A2 aptamer by a factor of 4. Perhaps, increasing the ionic strength suppresses electrostatic interactions involving charged phosphate groups on DNA; such interactions are likely important for non-covalent binding between the protein and aptamer.
Table 1.
Kd (nM) and Ki (nM) values determined for AlkB aptamers.
| Conditions | A2 | A3 | A11 | A24 | |
|---|---|---|---|---|---|
| Kd | HEPES/NaCl# | 59.1±10.2 | 27.1±11.4 | 312±20 | 136±24 |
| HEPES | 46.1±14.4 | ||||
| Tris | 27.5 ± 2.5 | 29.3 ± 7.7 | 426 ± 88 | 117 ± 10 | |
| Tris/NaCl | 103 ± 6.4 | ||||
| Tris+Fe2+ | 16.5 ± 2 | ||||
| Tris+2OG | 15.0 ± 3.2 | ||||
| Tris+2OG+Fe2+ | 7.36 ± 1.26 | 32.8 ± 10.9 | 375 ± 94 | 131 ± 20 | |
| Tris+2OG/Co2+/TTCm | 37.8 ± 9.1 | ||||
| Tris/Fe2/NOG/TTCm | 24.8 ± 8.2 | ||||
| Ki | 4.42 ± 1.91 | 34.4 ± 6.4 | 224 ± 16 | 178 ± 10 |
Measured for PCR-amplified bacterial clones while the rest of the data are for synthetic aptamers.32
We then investigated the effect of adding Fe(II) and 2OG on aptamer binding to AlkB. The individual addition of either component resulted in a significant decrease in Kd, but a 2-fold decrease was observed when they were combined (Fig. 2). This observation may reflect the reported conformational changes that Fe(II) and 2OG have upon AlkB binding as observed by NMR.34 In contrast, the other three aptamers, A3, A11, and A24, did not change their affinity in the presence of the 2OG and iron (Table 1). The results may suggest that the binding sites on AlkB for these aptamers are different from the one that participates in A2 aptamer binding. However, because AlkB can catalyze the conversion of 2OG to succinate and CO2 without the substrate,5 these results are difficult to interpret unambiguously.
To investigate the proximity of the A2 binding site to the active site, we compared A2 aptamer binding to the enzyme-substrate complex with its binding to the AlkB protein without the substrate. Crystal structures of the AlkB protein and AlkB-ssDNA substrate complex reveal that the presence of the substrate widens the active site binding groove to allow hydrogen bonding with the DNA backbone.35 Accordingly, it was suggested that AlkB can exist in two distinct conformations that correspond to a “searching” DNA mode and a “repair DNA” mode. Therefore, if the aptamer binding site on the AlkB is close enough to the substrate binding region, the substrate-induced conformational change could also affect aptamer binding. We therefore employed NOG, a catalyatically inert 2OG analogue and Co(II), a similarly inert substitute for Fe(II) in AlkB catalysis.36 In the presence of TTCm substrate, the aptamers affinity for AlkB was the same as that without TTCm. (Table 1). Thus, the A2 aptamer displayed equal affinity for both the free enzyme and the ES complex, and the aptamers binding site is not affected by the presence of the substrate.
Table 1 summarizes the results of affinity assays performed for all four aptamers, showing a wide range of affinity constants. We confirmed the aptamer affinity ranking previously obtained by using PCR-amplified bacterial clones with the synthetic aptamers utilized in this study.33
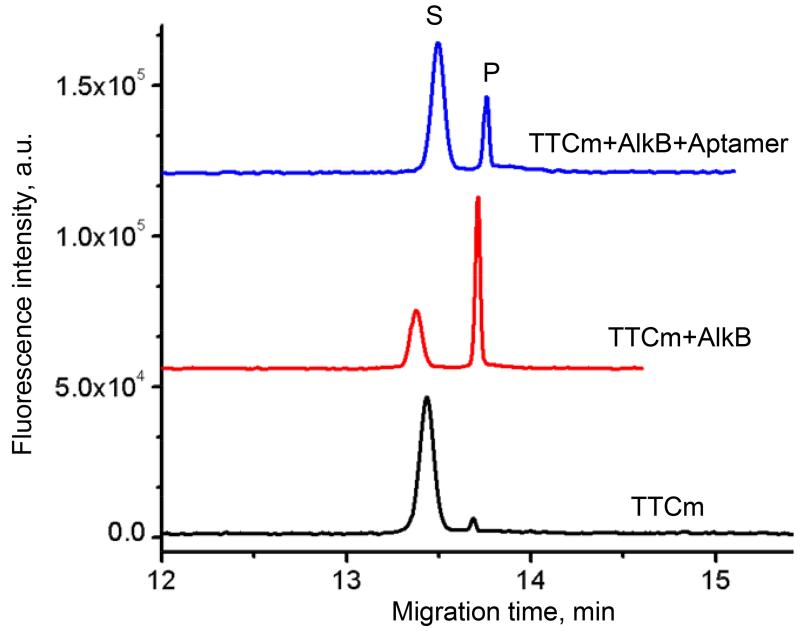
Strong binding of aptamers to AlkB protein suggest the possibility of controlling the AlkB proteins DNA repair activity.24 We thus studied possible inhibition using a detailed CE-based kinetic analysis. Crystallographic and other biophysical studies indicate that the formation of the AlkB enzyme-substrate complex occurs due to hydrophobic interactions, hydrogen bonding, and electrostatic interactions between the polyphosphate backbone of the DNA substrate and an electropositive DNA binding groove.35-38 Both DNA substrate and DNA aptamer are negative charge-carrying molecules that may compete for the same positive charges on the proteins surface. To further investigate the potential of aptamers for inhibition, we used a CE approach based on the separation and quantitation of the reaction product, TTC, and un-reacted substrate, TTCm. (a 15-nt DNA strand with a single methylated cytosine).10 To illustrate the method, we show in Fig. 3 electropherograms depicting the conversion of the methylated substrate into the demethylated product in the absence and in the presence of the A2 aptamer. The kinetic assay revealed a reduction in the amount of product formed in the presence of the aptamer (Fig. 3, top trace).
Figure 3.
Aptamer inhibition of demethylation of 100 nM methylated TTCm substrate by 5 nM AlkB (see Materials and Methods, sections 2.4 and 2.6). CE-based separation of the substrate (S) from the demethylated product (P) under the standard assay conditions is shown for the reaction mixture incubated for 1 min in the absence (middle line) and presence of 2 nM A2 aptamer (top line). The negative control (bottom line) was the reaction mixture before the addition of AlkB.
To study the mechanism of inhibition, kinetic analyses were performed in the presence of varying concentrations for each of the four aptamers. One of the strongest binders, aptamer A2 demonstrated exceptional inhibitory activity and completely stopped AlkB-catalyzed demethylation at concentrations comparable to the enzyme concentration. These results suggest that A2 is a tight binding inhibitor and, therefore, the data for this inhibitor was processed by using the Morrison fit.39 Our data analysis revealed that the Ki value for the A2 aptamer was as low as 2 nM (Fig. 4). We found that A2 inhibited the reaction in a DNA substrate-independent fashion. That suggests that A2 is a non-competitive inhibitor with respect to the substrate. The other three aptamers, A3, A11, and A24 appeared to be weaker inhibitors, and kinetic assay data were analyzed with the classical Lineweaver-Burk approach. The results revealed that each of these aptamers had caused significant reduction in the maximum reaction rate of AlkB catalysis, Vmax, with no change in Km (Fig. 5). These data indicate that A3, A11, and A24 act as non-competitive inhibitors of the AlkB proteins DNA repair activity with respect to both 2OG and the DNA substrate, and inhibit by binding the protein at a site distinct from the enzyme’s active site (at least with the tested substrate).
Inhibition of AlkB activity by aptamers was further compared with the established 2OG analogue, NOG, which competes with 2OG for AlkB binding. Graph E in Fig. 5 shows that NOG is a competitive inhibitor causing an increase in Km, with no change in Vmax. Competitive inhibition by NOG with respect to 2OG was expected and served as another confirmation that the mechanisms of AlkB inhibition were determined correctly. We also found a previously unknown non-competitive mechanism of inhibition by NOG with respect to the DNA substrate (Graph A in Fig. 5).
In contrast, aptamers had exhibited non-competitive inhibition in relation to the co-factor, 2OG in the same manner as to the substrate (graphs F-H). The non-competitive nature of inhibition by DNA aptamers with respect to DNA substrate was somewhat surprising as both molecules are very similar in their physical-chemical properties and could be expected to bind AlkB in a similar fashion. As judged by their Ki values the aptamers are about three orders of magnitude more potent inhibitors than NOG. For example, the Ki value for NOG was 11.6 μM, while that for A3 inhibitor was 34.3 nM. The inhibition mechanisms revealed by the Lineweaver-Burk analysis were also confirmed by curve fitting.
Although all four aptamers could bind and inhibit AlkB-catalyzed demethylation, A2 demonstrated a dramatically stronger inhibitory activity (Fig. 4, top panel). The observed difference between the inhibitory activity of A2 and other three aptamers correlates with their difference in binding affinity and sensitivity to the presence of 2OG and iron (II). Table 1 shows that the ranking of Ki values obtained for the four aptamers is in agreement with the ranking of Kd values for the AlkB-aptamer complexes, and that the A2 aptamer is the strongest binder and inhibitor.
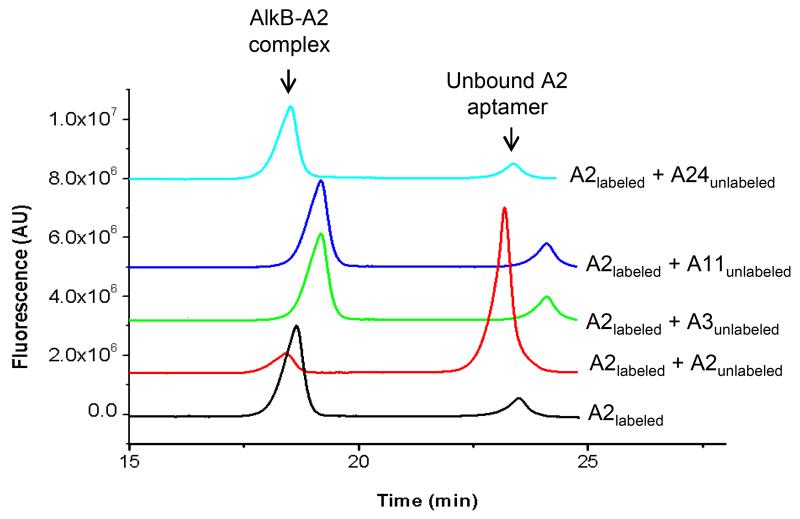
The potent inhibitory activity of the A2 aptamer and the dependence of this affinity on 2OG binding, suggests that A2 binds in a region different from the regions binding A3, A11, and A24 aptamers. To test this hypothesis, we tested the ability of the fluorescently labeled A2 aptamer to form a complex with AlkB in the presence of an excess of the other unlabeled aptamers. Figure 6 shows that the amount of the A2-AlkB complex did not change as the other aptamers (A3, A11, or A24) were added in a 20-fold excess (compare the bottom and three upper lines), while the addition of the unlabeled A2 aptamer, used as a positive control, almost completely suppressed the complex formation between AlkB and the fluorescently-labeled A2 (red line). The absence of competition between A2 and the rest of the aptamers suggests that A2 and the other three aptamers bind to different, non-overlapping sites. At present we do not know how the identified aptamers bind to AlkB, and it cannot be ruled out that they bind at a biologically relevant nucleic acid binding site that is not involved in binding our substrate.
Figure 6.
Competition of A3, A11, and A24 aptamers with A2 aptamer for the A2-binding site on AlkB. The A2 aptamer was fluorescently labeled and present at 100 nM while the competing aptamers were unlabeled and present at 2 μM. Displacement of the labeled A2 aptamer (100 nM) by the unlabeled A2 aptamer (300 nM) was used as a positive control (red trace).
The approach developed here for the evaluation of DNA aptamers, which can bind and specifically inhibit the activity of a target protein, can be potentially used for any 2OG/Fe(II)-dependent oxygenase. We note that not only do aptamers have the potential to decrease the catalytic rate of enzymes but they can also increase it. This methodology can be applied towards studies on aptamer-enzyme complexes, an aptamers’ ability to regulate protein activity in vivo, and for the discovery of new aptamer-based drug candidates.
4. CONCLUSIONS
We have studied DNA aptamers as potential inhibitors of AlkB-catalyzed DNA demethylation. Our investigations reveal the ability of DNA aptamers to be highly potent inhibitors of AlkB-catalyzed demethylation of DNA. The aptamers are non-competitive inhibitors with respect to 2OG and, at least with the tested sequences, non-competitive with respect to substrates. Not only can aptamers exhibit exquisitely selective binding properties but they are also able to modulate the function of AlkB when present in the nanomolar range of concentrations. Non-competitive inhibition allows low concentrations of aptamers to efficiently moderate the enzymatic activity. This might be an advantage for potential pharmaceutical applications of aptamers as inhibitors of human 2OG/Fe(II)-dependent oxygenases.
ACKNOWLEDGMENTS
The work was funded by NSERC Canada, the Biotechnology and Biological Sciences Research Council, and the Wellcome Trust.
ABBREVIATIONS USED
- AA
L-ascorbic acid
- A2
TGCCTAGCGTTTCATTGTCCCTTC-TTATTAGGTGATAATA
- A3
CCCATATCGGTGAATGCAC-GAGCAACCGATTGACACGGG
- A11
AGAAATTGGTACTGTATGAAACGGCAGCTGCACGTCGCG
- A24
GACTGCTGATGAGTCACTTTAACGTGGAGCAAAGATTAAA
- CE
capillary electrophoresis
- E. coli
Escherichia coli
- LIF
laser-induced fluorescence
- NECEEM
non-equilibrium capillary electrophoresis of equilibrium mixtures
- NOG
N-oxalylglycine
- 2OG
2-oxoglutarate
- P
product
- S
substrate
- SELEX
systematic evolution of ligands by exponential enrichment
- TTCM
5’-TTCmTTTTTTTTTTTT-3’-fluorescein
REFERENCES
- (1).Welford RWD, Hewitson KS, Mcneill LA, Schofield CJ, Schlemminger I. Anticancer agent combination using an AlkB inhibiting oxoglutoate analog or benzopyran derivative and an alkylating agent. WO 2004058252 A2 20040715 PCT Int. Appl. 2004
- (2).Koivisto P, Duncan T, Lindahl T, Sedgwick B. Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-methyladenine-DNA dioxygenase. J. Biol. Chem. 2003;278:44348–44354. doi: 10.1074/jbc.M307361200. [DOI] [PubMed] [Google Scholar]
- (3).Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. TBS. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- (4).Falnes PO, Klungland A, Alseth I. Repair of methyl lesions in DNA and RNA by oxydative demethylation. Neuroscience. 2007;145:1222–1232. doi: 10.1016/j.neuroscience.2006.11.018. [DOI] [PubMed] [Google Scholar]
- (5).Welford RWD, Schlemminger I, McNeil LA, Hewitson KS, Schofield CJ. The selectivity and Inhibition of AlkB. J. Biol. Chem. 2003;278:10157–10161. doi: 10.1074/jbc.M211058200. [DOI] [PubMed] [Google Scholar]
- (6).Roy TW, Bhagwat AS. Kinetic studies of Escherichia coli AlkB using a new fluorescence-based assay for DNA demethylation. Nucleic Acids Res. 2007;35:e147. doi: 10.1093/nar/gkm1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O’Connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J. Biol. Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- (8).Rose NR, McDonough MA, King ONF, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem. Soc. Rev. 2011;40:4364–4397. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- (9).Mishina Y, Duguid EM, He C. Direct reversal of DNA alkylation damage. Chem. Rev. 2006;106:215–232. doi: 10.1021/cr0404702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Liu L, Gerson SL. Targeted modulation of MGMT: Clinical implications. Clin. Cancer Res. 2006;12:328–331. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- (11).Karkhanina AA, Mecinovic J, Musheev MU, Krylova SM, Petrov AP, Hewitson KS, Flashman E, Schofield CJ, Krylov SN. Direct analysis of enzyme-catalyzed DNA demethylation. Anal. Chem. 2009;81:5871–5875. doi: 10.1021/ac9010556. [DOI] [PubMed] [Google Scholar]
- (12).Yi CQ, Yang CG, He CA. A non-heme iron-mediated chemical demethylation in DNA and RNA. Accounts Chem. Res. 2009;42:519–529. doi: 10.1021/ar800178j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Tsujikawa K, Koike K, Kitae K, Shinkawa A, Arima H, Suzuki T, Tsuchiya M, Makino Y, Furukawa T, Konishi N, Yamamoto H. Expression and sub-cellular localization of human ABH family molecules. J. Cell Mol. Med. 2007;11:1105–1116. doi: 10.1111/j.1582-4934.2007.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Shan X, Tashiro H, Lin CL. The identification and characterization of oxidized RNAs in Alzaheimer’s desease. Oncogene. 2003;21:8886–8894. doi: 10.1523/JNEUROSCI.23-12-04913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gerken T, Girard CA, Tung YCL, Webby CL, Saudek V, Hewitson KS, Yeo GSH, McDonough MA, Cunliffe S, McNeill LA, Galvanovskis J, Rorsman P, Robins P, Prieur X, Coll AP, Ma M, Jovanovic Z, Farooqi IS, Sedgwick B, Barroso L, Lindahl T, Ponting CP, Ashcroft FM, O’Rahilly S, Schofield CJ. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, Klungland A. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 2010;30:1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. Human AlkB homolog ABH8 is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Shimada K, Nakamura M, Anai S, De Velasco M, Tanaka M, Tsujikawa OY, Konishi N. A novel human analogue ABH8, contributes to bladder cancer progression. Cancer Res. 2009;69:3157–3164. doi: 10.1158/0008-5472.CAN-08-3530. [DOI] [PubMed] [Google Scholar]
- (19).Gao WL, Li L, Xu P, Fang J, Xiao S, Chen S. J. Gastroen. Hepatol. 2011;26:577–584. doi: 10.1111/j.1440-1746.2010.06531.x. [DOI] [PubMed] [Google Scholar]
- (20).Wu SS, Xu W, Liu S, Chen B, Wang XL, Wang Y, Liu SF, Wu JQ. Down-regulating of AlkBH2 increases cisplatin sensitivity in H1299 lung cancer cells. Acta Pharmacol. Sin. 2011;32:393–398. doi: 10.1038/aps.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Tasaki M, Shimada K, Kimura H, Tsujikawa K, Konishi N. AlkBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. Brit. J. Cancer. 2011;104:700–706. doi: 10.1038/sj.bjc.6606012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Westbye MP, Feyzi E, Aas PA, Vagbo CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk NB, Slupphaug G, Otterlei M, Krokan HE. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J. Biol. Chem. 2008;283:25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Shimada K, Nakamura M, Anai S, De Velasco M, Tanaka M, Tsujikawa K, Ouji Y, Konishi N. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 2009;69:3157–3164. doi: 10.1158/0008-5472.CAN-08-3530. [DOI] [PubMed] [Google Scholar]
- (24).Goulko AA, Li F, Le XC. Bioanalytical applications of aptamer and molecular-beacon probes in fluorescence-affinity assays. Trends Anal. Chem. 2009;28:878–892. [Google Scholar]
- (25).Vuyisich M, Beal PA. Controlling protein activity with ligand-regulate RNA aptamers. Chem. Biol. 2002;9:907–913. doi: 10.1016/s1074-5521(02)00185-0. [DOI] [PubMed] [Google Scholar]
- (26).Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- (27).Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- (28).Berezovski MV, Musheev MU, Drabovich AP, Jitkova JV, Krylov SN. Non-SELEX: selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat. Protoc. 2006;1:1359–1368. doi: 10.1038/nprot.2006.200. [DOI] [PubMed] [Google Scholar]
- (29).Berezovski M, Drabovich A, Krylova SM, Musheev M, Okhonin V, Petrov A, Krylov SN. Non-equilibrium capillary electrophoresis of equilibrium mixtures (NECEEM): a universal tool for the development of aptamers. J. Am. Chem. Soc. 2005;127:3165–3176. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- (30).Berezovski M, Krylov SN. Nonequilibrium capillary electrophoresis of equilibrium mixtures – a single experiment reveals equilibrium and kinetic parameters of protein-DNA interactions. J. Am. Chem. Soc. 2002;124:13674–13675. doi: 10.1021/ja028212e. [DOI] [PubMed] [Google Scholar]
- (31).Drabovich AP, Berezovski M, Musheev MU, Krylov SN. Selection of smart small-molecule ligands: the proof of principle. Anal. Chem. 2009;81:490–494. doi: 10.1021/ac8023813. [DOI] [PubMed] [Google Scholar]
- (32).Drabovich AP, Okhonin V, Berezovski M, Krylov SN. Smart aptamers facilitate multi-probe affinity analysis of proteins with ultra-wide dynamic range. J. Am. Chem. Soc. 2007;129:7260–7261. doi: 10.1021/ja072269p. [DOI] [PubMed] [Google Scholar]
- (33).Krylova SM, Karkhanina AA, Musheev MU, Bagg EAL, Schofield CJ, Krylov SN. DNA aptamers as analytical tools for the quantitative analysis of DNA-dealkylating enzymes. Anal. Biochem. 2011;414:261–265. doi: 10.1016/j.ab.2011.03.010. [DOI] [PubMed] [Google Scholar]
- (34).Bleijlevens B, Shivarattan T, Flashman E, Yang Y, Simpson P, Koivisto P, Sedgwick B, Schofield CJ, Matthews SJ. Dynamic states of the. DNA repair enzyme AlkB regulate product release. EMBO Rep. 2008;9:872–877. 26. doi: 10.1038/embor.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Schofield CJ, Zhang Z. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struc. Biol. 1999;9:722–731. doi: 10.1016/s0959-440x(99)00036-6. [DOI] [PubMed] [Google Scholar]
- (36).Holland PJ, Hollis T. Structural and mutational analysis of Escherichia coli AlkB provides insight into substrate specificity and DNA searching. PloS One. 2010;5:e8680. doi: 10.1371/journal.pone.0008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:71–78. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Koivisto P, Duncan T, Lindahl T, Sedgwick B. Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-Methyladenine-DNA dioxygenase. J. Biol. Chem. 2003;278:44348–44354. doi: 10.1074/jbc.M307361200. [DOI] [PubMed] [Google Scholar]
- (39).Morrison JF. Kinetics of the reversible inhibition of enzyme-catalyzed reactions by tight-binding inhibitors. BBA-Enzymology. 1969;185:269–286. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]