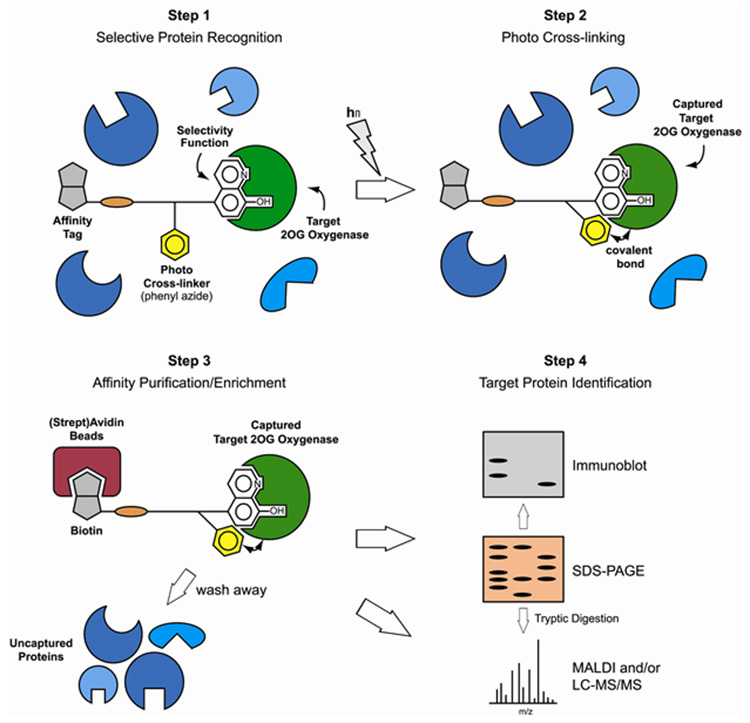
SUMMARY
2-Oxoglutarate (2-OG) dependent oxygenases have diverse roles in human biology. The inhibition of several 2-OG oxygenases is being targeted for therapeutic intervention, including for cancer, anemia and ischemic diseases. We report a small-molecule probe for 2-OG oxygenases that employs a hydroxyquinoline template coupled to a photoactivable cross-linking group and an affinity purification tag. Following studies with recombinant proteins, the probe was shown to cross-link to 2-OG oxygenases in human crude cell extracts, including to proteins at endogenous levels. This approach is useful for inhibitor profiling as demonstrated by cross-linking to the histone demethylase FBXL11 (KDM2A) in HEK293T nuclear extracts. The results also suggest that small-molecule probes may be suitable for substrate identification studies.
Keywords: 2-Oxoglutarate oxygenases, chemical probe, photo-affinity labelling, mass spectrometry, proteomics
Graphical abstract
INTRODUCTION
Oxygenases that employ 2-oxoglutarate (2-OG) as a co-substrate and ferrous iron as a cofactor have emerged as a large enzyme superfamily (Hausinger, 2004). In humans there are predicted to be > 60 oxygenases. To date, human oxygenases have been found to have roles in collagen biosynthesis, fatty acid metabolism, DNA/RNA repair and modifications, histone modification, and in the hypoxic response (Loenarz and Schofield, 2008).
In the hypoxic response in animals 2-OG oxygenases play important roles by catalysing post-translational hydroxylations of the hypoxia-inducible transcription factor (HIF) which orchestrates the expression of a large gene array. The oxygen dependence of these hydroxylases is proposed to enable them to act as oxygen sensors for the HIF system (Kaelin and Ratcliffe, 2008). trans-4-Prolyl-hydroxylation, catalyzed by the prolyl hydroxylase domain-containing enzymes, PHD1-3, of either of two prolyl-residues in the oxygen dependent degradation domain of HIF-α signals for degradation via the proteasome. HIF-α asparaginyl-hydroxylation is catalyzed by the Factor Inhibiting HIF (FIH) and reduces the interaction of HIF-α with transcriptional co-activator proteins (Kaelin and Ratcliffe, 2008). The up-regulation of HIF and/or increases in its activity (through inhibition of the HIF hydroxylases or by other means) may be beneficial for ischemic diseases, anemia, and gastrointestinal inflammatory diseases (Nagel et al., 2010). On the other hand, the therapeutic inhibition of HIF through increasing HIF hydroxylase activity (or by other means) is an approach to tumour treatment because HIF-α expression is increased in hypoxia.
2-OG oxygenases also play roles in fatty acid metabolism, i.e. in carnitine biosynthesis and chlorophyll metabolism, and in modification of nucleic acids via N-demethylation, t-RNA and 5-methylcytosine hydroxylation (for review see Loenarz and Schofield, 2008). 2-OG oxygenases (the JmjC family) have emerged as important in the modification of histones, by catalysing the N-demethylation of Nε-methyllysine residues. Different human subfamilies of 2-OG dependent Nε-lysine demethylases have been identified (Klose and Zhang, 2007), the members of which have a range of proposed roles, including in cellular differentiation and development. Mutations to two of the Jmj-C domain-containing demethylases (JARID1C and PHF8) are associated with X-linked human mental retardation, indicating that these 2-OG oxygenases are important for the normal neuronal function (Jensen et al., 2005; Laumonnier et al., 2005). The observation that JmjC proteins are often deleted, translocated, mutated, and aberrantly expressed in human cancers, and that some of these histone demethylases regulate the proliferation of cancer cell lines, suggests that their activation or the inactivation (some are proposed as onco-proteins and others as tumour suppressors) contributes to tumour development. For these reasons the therapeutic potential of JmjC histone demethylase inhibitors as anticancer agents is being considered (for review, see Spannhoff et al., 2009).
The diverse roles and substrates of 2-OG oxygenases make functional assignation work challenging. It is desirable to develop more efficient methods both for their identification in different cell types and for the identification of their substrates. One approach that has been applied to other enzyme families is the use of small-molecule ‘chemical’ probes, based on inhibitor templates specific for particular active sites (Kessler et al., 2001; Evans and Cravatt, 2006; Sadaghiani et al., 2007). However, to date the development of compounds that are selective for human 2-OG oxygenases over other enzyme families is at an early stage.
Despite recent advancements in mass spectrometric (MS) and bioinformatic methods, there remains a need to reduce the complexity of the proteome prior to detailed proteomic analyses. “Classical” approaches include 2D electrophoresis, affinity chromatography, and immuno-precipitation methods. Whilst these techniques are powerful, they suffer from limitations, e.g. 2D electrophoresis is not selective from a functional perspective and is not well-suited for the analysis of lipophilic proteins and affinity chromatography/immuno-precipitation methods normally require knowledge of the target proteins, and can be difficult to apply to complexes. Small-molecule based probes which are selective for specific protein families are complementary to the established techniques (Evans and Cravatt, 2006), and have long been used for profiling proteins, and particularly enzymes, in antibiotic research (e.g. penicillin binding proteins). More recently, they have been shown to be useful in profiling enzymes in eukaryotic cells. One strategy has employed the use of irreversible inhibitors derivatized with an affinity tag that enables efficient purification (Evans and Cravatt, 2006). A development of this strategy employs reversible inhibitors coupled to an affinity tag and a photoreactive group that “locks” the protein-inhibitor complex by irreversible cross-linking to enabling purification (Salisbury and Cravatt, 2008; Xu et al., 2009; Fisher et al., 2010).
Here we report on the development of a small-molecule probe for human 2-OG oxygenases (Table 1A) based on a reversibly binding inhibitor coupled to an affinity purification tag and a photoreactive group. We demonstrate the viability of the method for identifying 2-OG oxygenases in crude cell extracts and for profiling 2-OG oxygenase inhibitors.
Table 1. Structures (A) and Biological Validation (B) of Photoreactive Small-Molecule Probes used in the Study.
| ||||
|---|---|---|---|---|
| ||||
| Compound | R | JMJD2E | PHD2 | |
| FDH coupled assay | NMR-based binding assay | MS-based hydroxylation assay | ||
| IC50a | K D a | IC50a | ||
| DR014 |
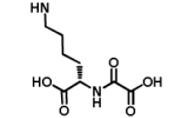
|
3870 μM | No binding @ 250 μM | No inhibition @ 300 μM |
| DR031 |
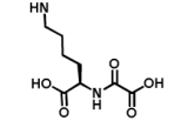
|
625 μM | No binding @ 250 μM | No inhibition @ 300 μM |
| DR025 |
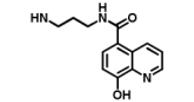
|
58.5 μM | 17.7 μM | 41.8 μM |
|
DR024 (Scaffold) |
OH | No inhibition @ 300 μM | No binding @ 250 μM | No inhibition @ 300 μM |
|
DR016 (Competitor) |
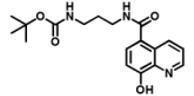
|
40.7 μM | 0.7 μM | 20.8 μM |
See also Figure S1A-C.
RESULTS
Probe Selection
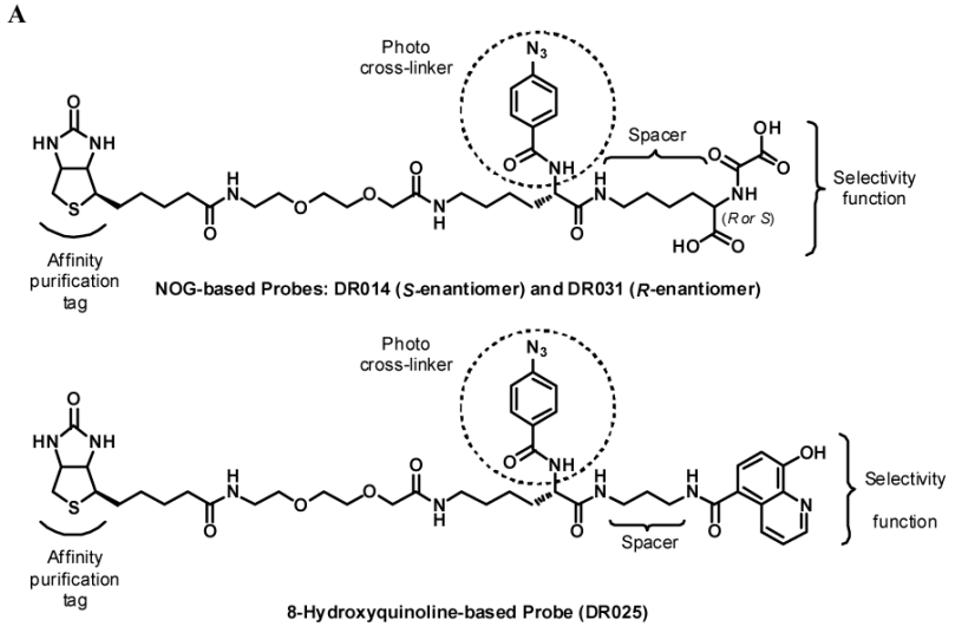
We began by preparing molecules with a common scaffold consisting of a substituted phenyl azide as photo cross-linking group capable of forming a covalent bond with the target protein after UV irradiation and a biotin derivative as an affinity purification tag (Table 1A). We chose this approach because it has been useful in analogous studies on kinases by Koester and co-workers (Fisher et al., 2010). We investigated two “inhibitor templates” in order to achieve selective and reversible binding to 2-OG oxygenases (selectivity function). In one series, we generated N-oxalylglycine (NOG) derivatives because of the close relationship of NOG with 2-OG, and because NOG/NOG derivatives inhibit a range of 2-OG oxygenases (Rose et al., 2010). Secondly, we synthesized 8-hydroxyquinoline (8-HQ) derivatives, because 8-HQ is a known iron chelator, and is present in inhibitors of 2-OG oxygenases including PHD2 and FIH (Warshakoon et al., 2006; Smirnova et al., 2010) and some histone demethylases (King et al., 2010) (Table 1A). As for the kinase work (Fisher et al., 2010), a spacer was introduced between the inhibitor / selectivity function and the phenylazide in order to reduce potential reactions of the photo cross-linker with the intention of enabling cross-linking close to but not within inhibitor / selectivity function binding site on the target oxygenases. Both epimers at the C-α chiral centre of the N-oxalyllysine group of the NOG-based probe were prepared because chirality at this centre is a means of achieving selective inhibition of some 2-OG oxygenases including the HIF hydroxylases (Table 1) (McDonough et al., 2005). Details of the synthesis of the probes are given in Supplemental Information.
We then investigated the capability of the probes to bind to recombinant PHD2 and JMJD2E (JMJD2E is likely not expressed, but its catalytic domain is representative of the JMJD2 histone demethylases). Initially, we tested for inhibition of / interaction with PHD2 using a MS-based hydroxylation assay (Flashman et al., 2010) and an NMR-based binding assay (Leung et al., 2010), and for inhibition of JMJD2E using a formaldehyde dehydrogenase (FDH) coupled assay (Rose et al., 2008) (Table 1B and Figure S1A-C).
The two NOG-based probes were at most only weakly active against both JMJD2E and, consistent with structural studies (McDonough et al., 2005), PHD2 (Table 1B and Figure S1C). In contrast, the 8-HQ derivative DR025 displayed promising binding/inhibition of both enzymes (IC50=58.4μM against JMJD2E and KD=17.7μM and IC50=41.8μM against PHD2), hence was selected for further work. In the absence of the selectivity function, the common scaffold DR024 (see Supplemental Information), was not observed to bind to PHD2 (NMR-based assay) and to inhibit JMJD2E (FDH coupled assay), and was therefore used as negative control in subsequent experiments (Table 1B). In contrast, the synthetic 8-HQ intermediate DR016 displayed a comparable binding/inhibition capacity to PHD2 and JMJD2E (Table 1B). This property made this compound useful for subsequent “competition” control experiments.
Photo Cross-Linking Experiments
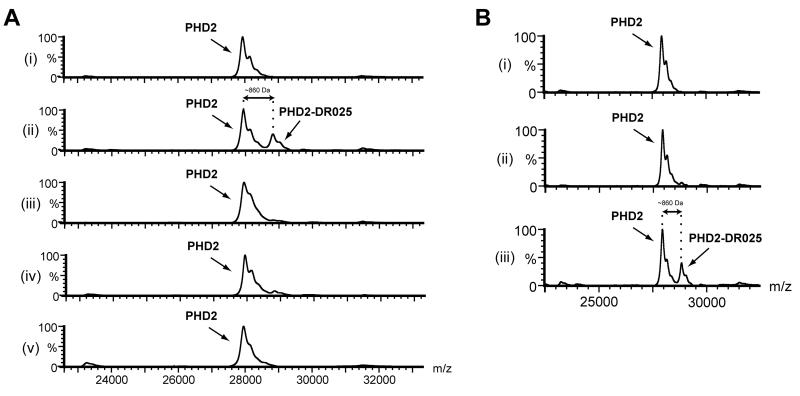
To test the suitability of DR025 as a cross-linker, we carried out studies with purified PHD2, using Mn(II) to substitute for Fe(II) because the PHD2-Mn(II) complex is not catalytically active. The extent of cross-linking was analyzed by protein MS through the observation of a mass shift approximately corresponding to the mass of the probe after the loss of N2. Prior to UV irradiation only a mass peak corresponding to PHD2 (~ 28 kDa) was observed (Figure 1A, i). After irradiation an additional peak with a mass shift of > 800 Da was observed, consistent with a covalent adduct formation with DR025 (m/z 861 for [M-N2]) (Figure 1A, ii). The extent of cross-linking by DR025 was substantially reduced in competition experiments (1:1 ratio with DR025) with a reported PHD2 inhibitor [(1-chloro-4-hydroxyisoquinoline-3-carbonyl)-amino]-acetic acid (Warshakoon et al., 2006) and with the 8-HQ containing DR016 (10:1 ratio with DR025) (Table 1B). These experiments provide evidence that this probe binds to the PHD2 active site (Figure 1A, iii and iv). No cross-linking was observed for the scaffold DR024 without the 8-HQ group (Table 1B) (Figure 1A, v). The presence of metal was important for efficient cross-linking (Figure 1B, ii and iii). The low level of cross-linking in the absence of Mn(II) (Figure 1B, ii) may be due to residual Fe(II) in the PHD2 sample (McNeill et al., 2005). Using the MS-method it was also possible to demonstrate selectivity for the formation of a covalent adduct between DR025 and PHD2 in protein mixtures containing additional enzymes (e.g. lysozyme, lactic dehydrogenase) unrelated to oxygenases (Figure S2A).
Figure 1. Cross-Linking between DR025 and PHD2 is Dependent on Irradiation and Mn(II).
(A) MALDI-MS spectra for photolabelling of PHD2181-426 by DR025. PHD2181-426 (5 μM) and DR025 (25 μM) were incubated (45 min, r.t.) in Tris buffer in the presence of Mn(II) (5 μM). After irradiation on ice (20 min, 365 nm) the resulting solutions were analyzed by MS. (i) and (ii) show results before and after irradiation; (iii) after irradiation in the presence of an inhibitor [(1-chloro-4-hydroxyisoquinoline-3-carbonyl)-amino]-acetic acid (Warshakoon et al., 2006) (25 μM) in a 1:1 concentration ratio with DR025; (iv) competition experiment with DR016 (250 μM) in a 10:1 molar ratio with DR025; (v) control with DR024 (25 μM); for selectivity and efficiency of PHD2 capture by DR025 see Figure S2. (B) Effect of Mn(II) on the efficiency of the PHD2181-426 capture by DR025. (i) and (iii) are relative to a mixture in Tris buffer of PHD2 (5 μM), DR025 (25 μM) and Mn(II) (5 μM) before and after irradiation, respectively. (ii) shows the effects of the irradiation on the same mixture without Mn(II).
The efficiency of cross-linking was investigated under conditions of varying wavelengths, concentrations of the target and the probe, incubation times and buffer. After some optimization, the following conditions were selected for further investigation: 20 minutes of irradiation at 365 nm with an energy of 5 mW/cm2, conditions which led to 35% of cross-linking relative to unmodified protein (Figure S2B). This level is in agreement with reported data for phenyl azide-mediated photo cross-linking; incomplete cross-linking is likely, at least in part, due to quenching of the reactive intermediates by solvent/buffer (Fleming, 1995).
Photo-affinity Labelling and Enrichment of Purified 2-OG Oxygenases
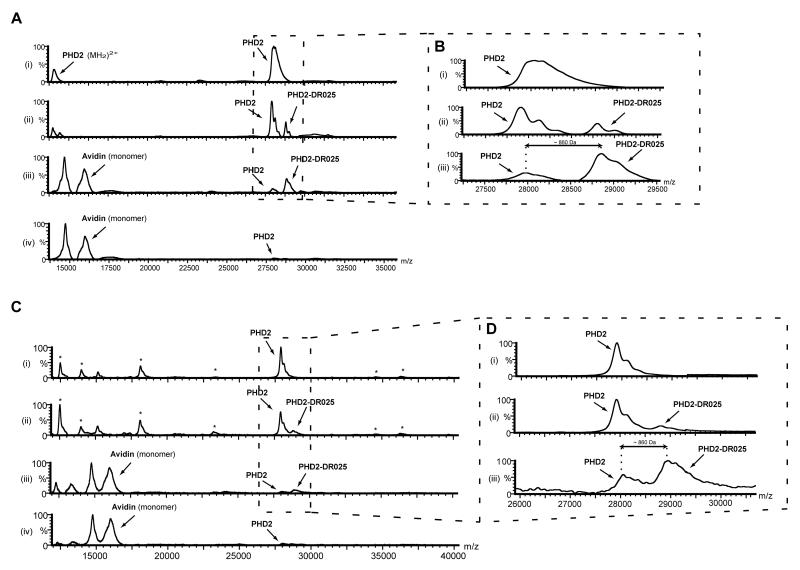
We then investigated the capability of the probe to enable enrichment of a target protein by the means of avidin-coated beads. Following irradiation the protein-probe mixture was incubated with avidin-coated agarose beads; after washing, the beads were then mixed with the MALDI matrix and analysed; after the affinity purification step there was enrichment of cross-linked PHD2 material (Figure 2A, ii and iii, and magnifications in Figure 2B). However, after cross-linking and affinity purification unmodified PHD2 was still observed (Figure 2A, iii and 2B, iii); Unmodified PHD2 was also observed, to a small extent, after the same treatment when using scaffold DR024 (Figure 2A, iv). This is likely due to relatively non-specific capture of PHD2 by the beads at the relatively high PHD2 concentrations used in these analyses (see below). Note that the limit of detection by MALDI-TOF MS requires protein concentrations (5μM in this case) that are likely higher than the endogenous levels of most proteins in biologically relevant samples.
Figure 2. Photo-Affinity Labelling and Enrichment of Purified PHD2 by DR025.
(A) MALDI spectra for the cross-linking of purified PHD2181-426 by DR025. PHD2181-426 (5 μM) and DR025 (15 μM) were incubated (45 min, r.t.) in Tris buffer in the presence of Mn(II) (5 μM). After irradiation on ice (20 min, 365 nm) the solutions were divided, with one part being analyzed by MALDI and the other incubated (30 min, r.t.) with avidin-coated agarose beads. After washing (see Experimental Procedures), the beads were mixed with the MALDI matrix and analysed. (i) before irradiation in the presence of DR025; (ii) after irradiation in the presence of DR025; (iii) after irradiation and avidin-bead mediated purification; (iv) after the irradiation in the presence of DR024 at the same concentration (15 μM) as DR025 and affinity purification. For the identification of the PHD2 region covalently cross-linked by DR025 see Figure 5 and the text. (B) Magnification of the PHD2181-426 region of spectra in (A). (C) MALDI spectra of the cross-linking of PHD2181-426 by DR025 in the presence of HEK293T cell lysates. PHD2181-426 (5 μM, 5.2 μg), Mn(II) (5 μM) and DR025 (15 μM) were incubated (45 min, r.t.) in Tris buffer in the presence of HEK293T cell lysates (~ 50 μg total protein). After UV treatment on ice (20 min, 365 nm) the resulting solutions were divided, with one part being analyzed by MALDI, and the other incubated (30 min, r.t.) with avidin-coated agarose beads. After washing, the beads were mixed with the MALDI matrix and spotted: (i) Before irradiation in the presence of DR025; (ii) after the irradiation in the presence of DR025; (iii) after irradiation and affinity purification in the presence of DR025; (iv), same as (iii) but in the presence of DR024 at the same concentration (15 μM) as DR025. (D) Magnification of the PHD2181-426 region of the spectra in (C).
We then carried out the analyses in the presence of HEK293T cell lysates as a background to test whether the probe can selectively target proteins in a complex mixture. Comparison of the spectra i-iii in Figure 2C shows that after affinity purification there is selective enrichment of the tagged PHD2 over background proteins (indicated with an asterisk). A limitation of this experiment was that the concentration of PHD2 was likely higher (5μM) as compared to other “background” proteins. As stated above, this may account for some of the apparent non-specific capture observed (Figure 2D, iii). Low levels of non-specific capture (but not cross-linking) were observed with the scaffold control DR024 (Figure 2C, iv).
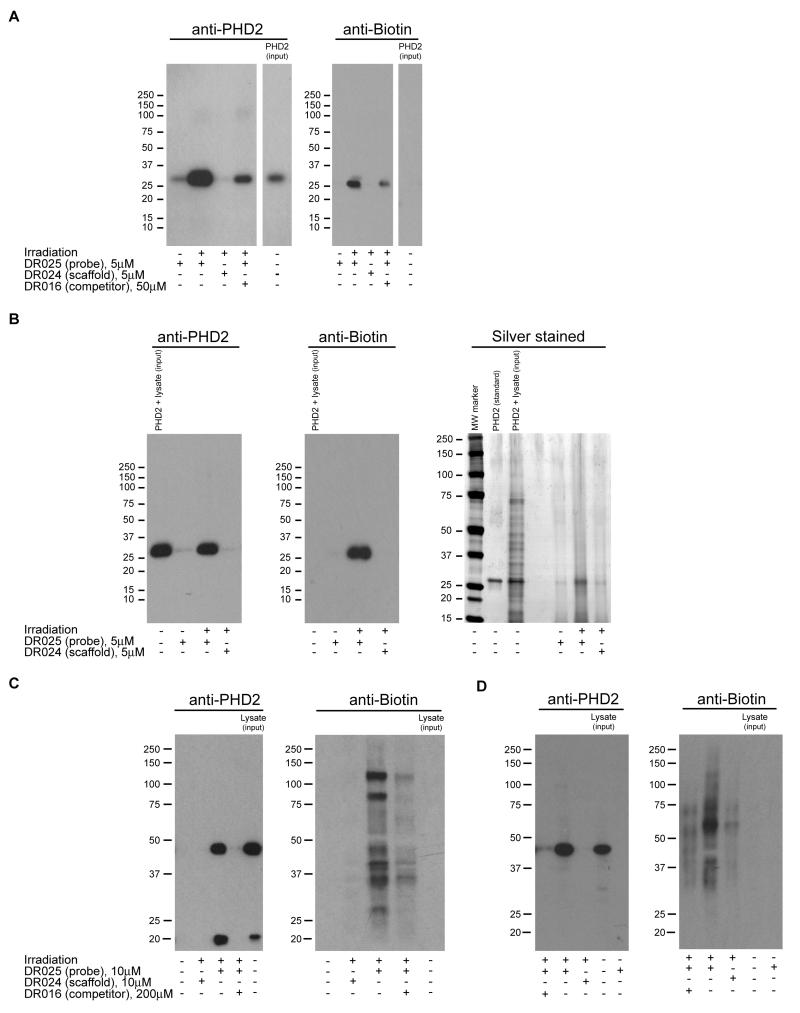
Labelling of purified PHD2 (alone and when added into HEK293T cell lysates) was also analysed by simultaneous Western blotting using anti-biotin and anti-PHD2 antibodies. As indicated in the anti-biotin blots (Figure 3A, right panel, and Figure 4B, central panel), biotinylation of PHD2 by DR025 was apparent in both experiments and was selective for PHD2 in the presence of the “background” proteins of HEK293T cell lysates (Figure 3B, central panel). Cross-linking was significantly reduced in the presence of a 10 fold excess of the 8-HQ competitor DR016 and absent in the control with the scaffold DR024 and in the experiment without irradiation. The anti-PHD2 blots confirmed that capture of PHD2 by DR025 was substantially reduced in the presence of DR016, in controls with the scaffold (DR024) alone, and without irradiation (Figure 3A and 3B, left panels). In the latter two experiments (i.e. DR024 alone and without irradiation), as previously found (Figure 2A-D), a low level of non-specific PHD2 capture was observed, as demonstrated by comparison of the corresponding lanes in the anti-biotin blots that show a complete absence of biotinylation (Figure 3A, right panel, and Figure 3B, central panel). It was notable that, compared to those with isolated proteins, non-specific PHD2 capture was reduced in the experiments with cell lysates, probably because of competition for ‘non-specific’ binding exerted by the other proteins (compare corresponding lanes in the left panels of Figures 3A and 3B). SDS-PAGE and silver staining (Figure 3B, right panel) supported the capability of DR025 to selectively capture PHD2 in the presence of irradiated HEK293T cell lysate proteins (Figure 3B, right panel). Identification of the labelled protein band at 28 kDa as the supplemented PHD2 was verified by LC-MS/MS analyses (see Experimental Procedures).
Figure 3. Photo-Affinity Labelling and Enrichment of Purified and Endogenous Human PHD2 by DR025.
(A) Western blot analysis for the capture of purified PHD2181-426 by DR025. PHD2181-426 (5 μM, 5.2 μg) and DR025 (5 μM) were incubated in the presence of Mn(II) (5 μM) at r.t.. After irradiation and purification by streptavidin-coated beads, proteins were separated by SDS-PAGE and analyzed by anti-PHD2 (polyclonal antibody from rabbit, left panel) and anti-biotin (right panel) immunoblotting. The input purified PHD2181-426 was ~ 4% of the protein corresponding to the other lanes. For an analogous experiment with JMJD2E see Figure S3A. (B) Immunoblotting and silver staining analysis showing capture of purified PHD2181-426 by DR025 in HEK293T cell lysates. The anti-PHD2 (polyclonal antibody from rabbit, left panel), the anti-biotin (central panel) and the silver stained SDS-PAGE gel (right panel) are relative to an experiment carried out under the same conditions as in (A) but in the presence of the HEK293T cell lysates (~ 43 μg total proteins). The input lysate supplemented with recombinant PHD2181-426 was ~ 4% of the total protein corresponding to the other lanes; the purified PHD2181-426 as standard was ~ 4% of the protein corresponding to the other lanes. For an analogous experiment with JMJD2E see Figure S3B. (C) Cross-linking with full length PHD2 in HEK293T cell lysates over-expressing full length PHD2. DR025 (10 μM) was incubated (45 min, r.t.) with an HEK293T cell lysate over-expressing human full length PHD2 (~ 40 μg total protein). After UV treatment (20 min, 365 nm) and purification by mean of avidin-coated agarose beads, proteins released from the beads were analyzed by SDS-PAGE and Western blots using anti-PHD2 (monoclonal antibody from mouse, left panel) and anti-biotin (right panel) antibodies. The input lysate over-expressing full length PHD2 was ~ 2% of the total protein corresponding to the other lanes. (D) Cross-linking of endogenous full length PHD2 in HEK293T cell lysates (i.e. not over-expressing PHD2). Conditions are as in (C) but employing HEK293T cell lysates (~ 200 μg total protein) not over-expressing PHD2. The input lysate was ~ 3% of the total protein corresponding to the other lanes.
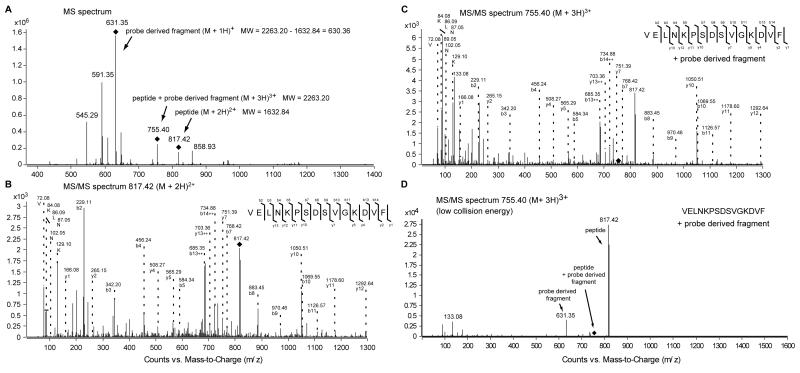
Figure 4. Identification by MS of the PHD2 Peptide Sequence Covalently Cross-linked by the Probe.
(A) MS spectrum of PHD2181-426 covalently cross-linked by DR025. PHD2181-426 (5 μM), Mn(II) (5 μM) and DR025 (25 μM) were incubated (45 min, r.t.) in Tris buffer. After irradiation (20 min, 365 nm) and purification by the means of streptavidin beads, photo cross-linked PHD2181-426 was digested on the beads by trypsin and analysed by LC-MS/MS. (B) MS/MS spectrum of the doubly charged precursor ion at m/z = 817.42 Da. The doubly charged precursor ion at m/z = 817.42 Da was identified as the peptide VELNKPSDSVGKDVF which represents the C-terminal region of PHD2181-426. The b- and y-fragment ion series, and the detected immoium ions are shown. (C) MS/MS spectrum of the triply charged precursor ion at m/z = 755.40 Da. The triply charged precursor ion at m/z = 755.40 Da exhibits the same fragmentation pattern as the co-eluting precursor at m/z = 817.42 Da with additional ions in the low molecular mass range. The b- and y-fragment ion series, and the detected immoium ions are shown as in (B). (D) MS/MS spectrum of the triply charged precursor ion at m/z = 755.40 Da with reduced collision energy. Under this conditions the triply charged precursor loses a singly charged fragment at m/z = 631.35 Da which generates the doubly charged precursor at m/z = 817.42 Da (MW = 1632.84 Da). For the MS/MS spectra in the low molecular range of the precursor ion masses associated to the cross-linked PHD2 see Figure S4.
We then tested the versatility of the probe for cross-linking to 2-OG oxygenases by carrying out experiments with the 2-OG dependent histone demethylase JMJD2E. Closely analogous results to those obtained with PHD2 were obtained with JMJD2E (Figure S3A and S3B), providing preliminary evidence for the suitability of DR025 as a general probe for the 2-OG oxygenase superfamily.
Validation of the Functional Probe by Detecting Endogenous Levels of Target Proteins
To investigate the utility of DR025 in a more biologically relevant context, we then carried out analyses of crude extracts prepared from human HEK293T cells (grown under normoxic conditions) that either over-expressed full length PHD2 or were untransfected (Figure 3C and 3D, respectively). The MALDI MS analysis method was not applicable, at least with our current capability, because of the complexity of the system and the low abundance of the target protein. In cell extracts from both untransfected and PHD2 over-expressing HEK293T cells, anti-PHD2 immunoblotting (left panel in the Figure 3C and 3D) revealed PHD2 capture by the DR025. A strong reduction in capture was observed in competition experiments using DR016, and a complete absence of any labelling was observed in controls with the inactive probe DR014 or without irradiation. These controls imply that the non-specific capture observed at the high PHD2 concentrations used in the preliminary analyses (Figures 2, 3A and 3B) is not, at least under the tested conditions, a problem when the target concentration is at relatively low “endogenous” levels. It was necessary to use a more sensitive chemiluminescence kit for immunoblotting to observe the same degree of capture in the lysates from the cells not over-expressing PHD2 than in those doing so (see Experimental Procedures for details). The two anti-biotin immunoblots (right panel in the Figure 3C and 3D) showed more than one band in the capture assay. Some of them were substantially reduced or absent in the competition (DR016) and scaffold (DR024) controls, indicating a specific enrichment of more than one protein as a result of the cross-linking process (see below). In the lanes with the input lysates and the experiments without irradiation, we did not detect any bands even from the naturally present biotinylated proteins, likely because the lysate was subjected to a “pre-clearing” with avidin-coated beads prior to probe treatment.
Mapping the PHD2-probe photo cross-linking site
We then investigated the photo cross-linking site of the probe DR025 with recombinant PHD2181-426 by performing photo cross-linking followed by purification and tryptic digestion/MS analysis to map the region(s) that are covalently modified. LC-MS/MS analysis of the digested photo cross-linked PHD2181-426 material revealed three co-eluting precursor ion masses that may contain cross-linked species (Figure 4A): (i) a doubly charged ion at m/z = 817.42 Da (MW = 1632.84 Da) was identified by MS/MS analysis as VELNKPSDSVGKDVF (Figure 4B), which corresponds to the C-terminal sequence of PHD2411-426; (ii) a triply charged precursor ion at m/z = 755.40 Da (MW = 2263.20 Da) exhibited the same fragmentation pattern under normal MS/MS conditions as the ion at 817.42 Da (MW = 1632.84 Da), but with additional ions in the low molecular range (Figure 4C), and (iii) a singly charged ion at m/z = 631.35 Da (MW = 630.35 Da) corresponding to the exact mass difference (MW = 2263.20 – 1632.84 = 630.36 Da) between the ions at 755.40 Da (MW = 2263.20 Da) and 817.42 (MW = 1632.84 Da). In the MS/MS spectrum of the precursor ion for the latter species (817.42 Da), we did not observe proteotypic immonium ions suggesting that it is not derived from a proteotypic peptide (Figure S4). We compared the MS/MS spectra in the low molecular range of all three precursor ions (Figure S4), and observed that fragment ions derived from the precursor at 631.35 Da (MW = 630.35 Da) were also present in the MS/MS spectrum of the triply charged precursor ion at 755.40 Da (MW = 2263.20 Da), but not in the spectrum of the doubly charged precursor ion at 817.42 Da (MW = 1632.84 Da). These observations imply that the C-terminal region of PHD2411-426 (VELNKPSDSVGKDVF) is reacted with DR025. We noted that during the ionisation, the majority of the cross-linked peptides undergo fragmentation releasing a non-proteotypic fragment (630.35 Da). To confirm this, we produced an MS/MS spectrum of the precursor at 755.40 Da [M+3H]3+ (MW = 2263.20 Da) with reduced collision energy (Figure 4D). Under these conditions, we observed only a doubly charged fragment ion at 817.42 Da (MW = 1632.84 Da) and a singly charged ion at 631.35 Da (MW = 630.35 Da) implying that the peptide does not (normally) fragment, and the DR025 derived fragment at 631.35 Da is separated in an intact form from the precursor ion at 755.40 Da (Figure 4D). Taken together, the spectra show that both precursors at 755.40 Da [M+3H]3+ (MW = 2263.20 Da) and 817.42 Da [M+2H]2+ (MW = 1632.84 Da) correspond to the same PHD2 derived peptide with and without a linked non-proteotypic adduct with a molecular weight of 630.35 Da. The precursor ion for the triply charged adduct at 755.40 Da was not observed in the non UV irradiated PHD2 protein control sample. The instability of the peptide-probe adduct under MS/MS conditions did not allow the exact assignment of the modified amino acid residue. Nonetheless, the results reveal that a major site of cross-linking of the probe as the C-terminal region of PHD2411-426.
Profiling of 2-OG Oxygenases in Human Cells
We investigated whether DR025 could be used to identify 2-OG oxygenases present in a human cell line (HEK293T cells grown under normoxic conditions) using MS (Wu and MacCoss, 2002). To differentiate between specifically enriched and ”non-specifically” captured proteins (see Table 2 and Experimental Procedures for details), we compared the results obtained for proteomic analyses in labelling experiments using DR025 with controls, including proteomic analyses of the digested input lysate. The identified 2-OG oxygenases were divided into three groups on the basis of the controls: Enrichment, Possible Enrichment, and No Enrichment. There was clear evidence for enrichment of PHD2 (Epstein et al., 2001), for two Nε-methyl histone lysyl demethylases JARID1C (Jensen et al., 2005) and FBXL11(Tsukada et al., 2006) and for the collagen lysyl hydroxylase LH3 (Risteli et al., 2009). These oxygenases were identified only in the labelling experiment with DR025, i.e. were not identified in the DR016 competition control, or in the other controls (scaffold DR024 and no irradiation). In the cases of PHD2 and LH3, we did not accrue evidence for them in the input lysate. For two other oxygenases, TET2 (Loenarz and Schofield, 2009) and the Nε-methyl histone lysyl demethylase PHF8 (Yu et al., 2010), we observed evidence for possible enrichment due to their identification when captured by the probe (DR025), but both were absent in the scaffold, no irradiation and input lysate controls. However, they were observed in the competition (DR016) control experiments but we cannot rule out the possibility that this is because of incomplete competition. Three 2-OG oxygenases were considered as likely not enriched because they were identified in at least one of the scaffold or no irradiation controls (Table 2).
Table 2. 2-OG Oxygenases Identified by DR025 in HEK293T Cell Lysatesa.
| ID |
2-OG Oxygenase |
DR025 (probe) |
DR016 (competition) |
DR024 (scaffold) |
No irradiation |
Lysate (input) |
Result b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| spect countsc | # pepd | % cove | spect counts | # pep | % cov | spect counts | # pep | % cov | spect counts | # pep | % cov | spect counts | # pep | % cov | |||
| Q9GZT9 | PHD2* | 2 | 1 | 4 | absent | absent | absent | absent | E | ||||||||
| P41229 |
JARID1C (KDM5C) |
2 | 2 | 1 | absent | absent | absent | 5 | 5 | 2 | E | ||||||
| Q9Y2K7 |
FBXL11* (KDM2A) |
1 | 1 | 0.9 | absent | absent | absent | 8 | 7 | 6 | E | ||||||
| O60568 | LH3 | 2 | 1 | 2 | absent | absent | absent | absent | E | ||||||||
| Q6N021 | TET2 | 2 | 2 | 1 | 9 | 9 | 4 | absent | absent | absent | PE | ||||||
| Q9UPP1 | PHF8 | 2 | 2 | 2 | 11 | 7 | 3 | absent | absent | absent | PE | ||||||
| Q15652 | JMJD1C | 5 | 1 | 0.6 | 27 | 14 | 5 | 21 | 14 | 5 | 29 | 20 | 8 | 22 | 14 | 7 | NE |
| Q9UGL1 |
JARID1B (KDM5B) |
1 | 1 | 1 | 8 | 8 | 6 | 11 | 11 | 7 | absent | 6 | 5 | 2 | NE | ||
| O94953 |
JMJD2B (KDM4B) |
4 | 2 | 2 | 18 | 8 | 7 | absent | 11 | 8 | 6 | absent | NE | ||||
DR025 (10 μM) was incubated with a whole lysate from HEK293T cells grown under normoxic conditions (~950 μg total protein, 45 min, room temperature). After irradiation (20 min, 365 nm) and purification by avidin-coated agarose beads, trypsin digestion, and LC-MS/MS analysis comparison with the human protein database were made. The columns reflect the following conditions: DR025 (probe), complete experiment in the presence of DR025; No irradiation, same as DR025 but without UV treatment; DR024 (scaffold), negative control experiment in the presence of scaffold DR024 (10 μM); DR016 (competitor), control experiment with the competitor DR016 (200 μM) in a 20:1 molar ratio with DR025; Lysate, input lysate, (5% of the total protein corresponding to the other columns).
E: Enrichment, enzyme not detected in the DR024 and No Irradiation negative controls; PE: Possible Enrichment, enzyme not detected in the DR024 and No irradiation negative controls, but detected with the DR016 competitor control; NE: No Enrichment, enzyme detected in at least one of the negative controls (DR024 or No Irradiation).
Spectral counts: number of MS/MS spectra identified.
Number of unique peptides identified by MS/MS.
Protein sequence coverage [%].
Note detection corresponds to the observation of at least 1 peptide with the correct predicted mass for the identified oxygenases.
See also Table S1.
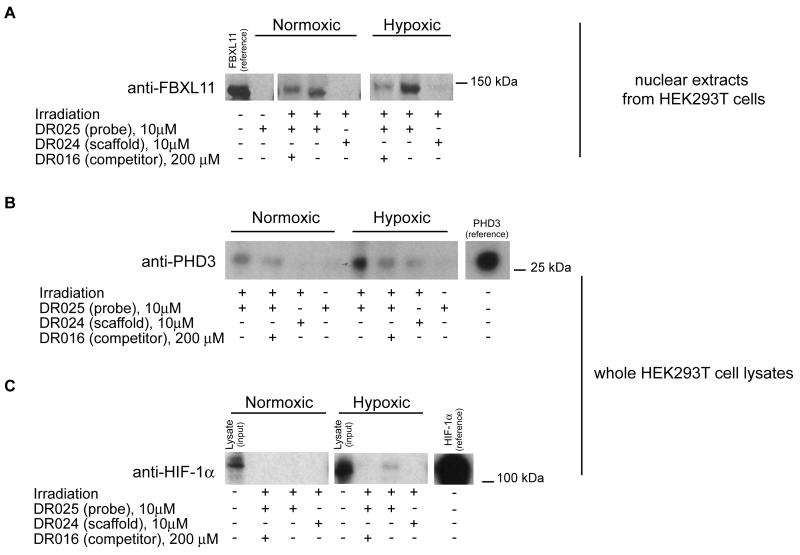
Overall, the MS results suggest that appropriately functionalised probes can capture and enrich endogenous levels of PHD2 and other 2-OG oxygenases. However, the identified 2-OG oxygenases had a low number of spectral counts relative to background proteins, which is in part due to the fact that these enzymes are not expressed at abundant levels in cells. Hence, to further validate the approach, we employed western blotting to confirm the enrichment of at least one 2-OG oxygenase other than PHD2. We focused our attention on the histone demethylase FBXL11 (KDM2A), an enzyme involved in epigenetic regulation (Tsukada et al., 2006). As shown Figure 5A (see also Figure S5A), Western blotting demonstrated that the probe was able to specifically capture FBXL11 in nuclear extracts of HEK293T cells grown under both normoxic conditions and, to a greater extent, under hypoxic conditions. The results with FBXL11 are important because they reveal the potential of the method to profile 2-OG oxygenase inhibitors.
Figure 5. Photo-Affinity Cross-Linking Experiments in Lysates from HEK293T Cells Grown under Normoxic and Hypoxic Conditions.
(A) Cross-linking of endogenous FBXL11 (KDM2A) in nuclear protein extracts of HEK293T cells. DR025 (10 μM) was incubated (45 min, r.t.) with nuclear extracts (~ 200 μg total protein) of HEK293T cells grown under normoxic (left) and hypoxic (right) conditions. After irradiation (20 min, 365 nm) and purification by streptavidin beads, the proteins released were analyzed by SDS-PAGE and subsequent anti-FBXL11 (polyclonal antibody from rabbit) immunoblotting. The reference protein was a nuclear protein extract of HEK293T cells over-expressing FBXL11 (KDM2A). For the entire immunoblot see Figure S5A. (B) Cross-linking of PHD3 at endogenous levels in HEK293T cell lysates. DR025 (10 μM) was incubated (45 min, r.t.) with cell lysate (~ 200 μg total protein) of HEK293T cells grown under normoxic (left side) and hypoxic (right side) conditions. After irradiation (20 min, 365 nm) and purification by streptavidin beads, released proteins were analyzed by SDS-PAGE and anti-PHD3 (monoclonal antibody from mouse) immunoblotting. The protein reference was a cell lysate (RCC4) over-expressing PHD3. For the entire immunoblot see Figure S5B. (C) Identification of endogenous HIF-1α in HEK293T cell lysates. DR025 (10 μM) was incubated (45 min, r.t.) with a whole lysate (~ 200 μg total protein) of HEK293T cells grown under normoxic (left) and hypoxic (1% O2) (right) conditions. After irradiation (20 min, 365 nm) and purification by streptavidin beads, released proteins were analyzed by SDS-PAGE and anti-HIF-1α (monoclonal antibody from mouse) immunoblotting. The input HEK293T lysate was ~ 3% of the total protein corresponding to the other lanes; the protein reference was a cell lysate (RCC4) over-expressing HIF-1α. For the entire immunoblot see Figure S5C.
To test whether 8-HQ is indeed an inhibitor of FBXL11 (KDM2A), we produced FBXL11 in a recombinant form and carried out tests using an assay that monitors formaldehyde production (see Experimental Procedures). The results revealed inhibition of FBXL11 with IC50 values in the micromolar range (IC50=9.1μM for DR016 and IC50=16.7μM for DR025, Figure S1D), but a lack of inhibition by the scaffold control DR024 (no inhibition at 100μM). These results reveal the potential of the method for profiling 2-OG oxygenases inhibitors.
Discrimination between Different Levels of PHD3 Expression Depending on Oxygenation Conditions
The finding that FBXL11 (KDM2A) was more efficiently captured by DR025 under hypoxic than normoxic conditions (Figures 5A and S5A) led us to then test the capability of the probe to discriminate between different target protein expression levels under different oxygenation conditions. We chose to analyze PHD3, which is known to be strongly induced by hypoxia in some cell lines (Pescador et al., 2005, Pollard et al., 2008; Appelhoff, et al., 2004). The anti-PHD3 immunoblot in HEK293T whole cell lysates prepared under normoxic or hypoxic conditions clearly showed that the probe DR025 (or a derivative thereof) is a valid tool to detect the difference of expression between normoxic and reduced oxygenation conditions (Figures 5B and S5B). On the basis of this evidence and studies on the inhibition of histone demethylation by 8-hydroxyquinoline derivatives, DR025 can be proposed as a potentially useful probe to profile different expression levels of 2-OG oxygenases in tissues.
Detection of the 2-OG Oxygenase (PHD2) Substrate HIF-1α at Endogenous Levels
Although we cannot rule out the possibility of other sites of cross-linking, the demonstration that the C-terminus of PHD2 is involved is interesting because this region interacts with its HIF-α substrate (Chowdhury et al., 2009). The available evidence suggests that extent to which PHD inhibitors block HIF-α substrate binding varies (Chowdhury et al., 2009). We therefore considered whether DR025 might be capable of “capturing” the HIF-α substrate of the PHDs along with the enzymes. As shown by anti-HIF-1α western blotting (Figures 5C and S5C) DR025 is able of capturing (by an unknown mechanism), albeit not strongly, endogenous levels of HIF-1α in HEK293T cell lysates at least when cultured under hypoxic conditions, where HIF-1α levels are increased relative to normoxic conditions (Figure 5C) (Kaelin and Ratcliffe, 2008).
DISCUSSION
Overall the results demonstrate the potential of a small molecule probe based approach for identifying 2-OG oxygenases in crude cell extracts via photo cross-linking and affinity purification coupled to MS-analyses. Importantly, we were able to demonstrate that the probe is able to capture human 2-OG oxygenases in cell extracts at endogenous levels (Figures 3D and 5). Thus, with appropriate development, such as by optimisation of the active site binding groups and cross-linking conditions, the photo cross-linking probe-method could be used for profiling 2-OG oxygenases from different cell types; This application is potentially useful for (patho)physiological analyses because levels of some 2-OG oxygenases (e.g. JMJD2C and PHD3) vary in diseased and hypoxic cells (Pollard et al., 2008). DR025 showed the capability to discriminate between different levels of expression of PHD3 and, to a lesser extent, of FBXL11 (KDM2A) in lysates from cells (HEK293T) grown under normoxic and hypoxic conditions (Figure 5A and 5B). These results support work showing that some 2-OG oxygenases, including PHD3 and, JMJD1A (Pollard et al., 2008) are induced by hypoxia.
Modifications of our lead probe designed to enable cell penetration by introducing post-lysis an affinity purification tag via “click” chemistry or other ligation techniques are possible, as reported in work on probes for proteases (Sadaghiani et al., 2007; Sieber et al., 2006). Although it is unlikely that DR025 itself will be a useful probe for all human 2-OG oxygenases, our results suggest that it may be useful for a significant subset. In principle, the probe approach could be also useful for identifying enzymes from the superfamily not already annotated as 2-OG oxygenases.
Various 2-OG oxygenases are being targeted for therapeutic intervention. γ-Butyrobetaine hydroxylase is a target for the clinically used compound Mildronate (Liepinsh et al., 2006) and the HIF-α hydroxylases and the histone demethylases are being explored as targets for anemia/ischemic disease and cancer, respectively. One application of probe methodology may be to profile oxygenases that a particular inhibitor/modulator targets inside a cell, including “off target” interactions of lead compounds. One problem with this approach is that a modification of a “small molecule” with relatively large photo cross-linking/affinity purification groups will inevitably modify binding characteristics, potentially leading to false negative results. However, given the high cost of pharmaceutical development, the effort of applying this, or related approaches, to potential leads in order to identify potentially deleterious interactions would seem to be small.
In support of the approach we found that DR025 cross-linked to the histone demethylase FBXL11 (KDM2A) in HEK293T cell lysates, as initially identified by MS analyses (Table 2), and confirmed by antibody analysis (Figure 5A). We subsequently demonstrated that DR025 and the 8-HQ derivative which acts as selectivity function (DR016), actually inhibit FBXL11 (KDM2A) to a significant extent (IC50=16.7μM for DR025 and IC50=9.1μM for DR016). This is the first reported inhibition study on FBXL11 (KDM2A) and supports the proposal that 8-HQs may be useful generic templates for the inhibition of 2-OG oxygenases (King et al., 2010).
Finally, we note that the photo cross-linking approach may have applications for functional assignment studies of 2-OG oxygenases and other enzymes. Analyses on purified PHD2 after cross-linking and affinity purification with DR025 demonstrate that cross-linking occurs between DR025 and the C-terminal region of PHD2411-426 (Figures 4 and S4). The C-terminus of PHD2 is involved in the HIF-α substrate binding (Chowdhury et al. 2009). This suggested that the probe may be useful in “capturing” a substrate from the cell-extract. Indeed, western blotting analyses revealed that HIF-1α was purified along with PHD2 at least from whole lysates of HEK293T cells cultured under hypoxic conditions (Figure 5C and S5C), consistent with the reported reduced degradation of HIF-1α under hypoxia. This preliminary evidence of an active site-directed probe capable of identifying endogenous levels of the natural substrate of a target enzyme, on the basis of our knowledge, is one of the first reported to date. As for the cross-linking to PHD2 itself, the mechanism of the apparent cross-linking to HIF-1α is uncertain; however, analysis of a PHD2-substrate structure (Chowdhury et al. 2009) suggests that it is possible that it occurs by cross-linking of probe with HIF-1α that is simultaneously bound to PHD2. An application of the chemical probe strategy reported here may therefore be to identify (new) substrates for 2-OG oxygenases (or indeed other protein interactors).
SIGNIFICANCE
2-Oxoglutarate (2-OG) dependent oxygenases catalyze a range of hydroxylation and N-methyl demethylation reactions that are important in oxygen sensing and the epigenetic control of gene expression. These enzymes are being targeted for modulation by small molecules as novel potential therapeutic targets for the treatment of anemia, ischemic diseases and cancer. We demonstrate that a small-molecule probe-based approach employing photo cross-linking and affinity purification is useful for the identification of 2-OG oxygenases present in human cell extracts and for identifying 2-OG oxygenases that interact with inhibitors. The approach was validated by studies on a transcription factor hydroxylase (PHD2, EGLN1) and by the finding that the probe binds to, and inhibits the Nε-methyl lysine histone demethylase FBXL11 (KDM2A). We also demonstrate the potential of this photo cross-linking small-molecule probe-based approach to “capture” the 2-OG oxygenase (PHD2) substrate HIF-1α, suggesting that it may be useful in substrate capture and discovery studies.
EXPERIMENTAL PROCEDURES
Synthesis of the Probes
Details of the synthesis of the probes DR014, DR031, DR025 are given in Supplemental Information, along with analytical data. The PHD2 inhibitor [(1-chloro-4-hydroxyisoquinoline-3-carbonyl)-amino]-acetic acid was synthesized as reported (Stubbs et al., 2009).
Photo Cross-Linking Experiments
The photo cross-linking procedure is described here for purified PHD2181-426 and DR025. For the other purified enzyme JMJD2E, experiments in the presence of non 2-OG enzymes (Figure S2A), experiment with HEK293T whole cell lysates as background, and tests in the presence of HEK293T whole cell lysates or nuclear protein extracts, capture was performed in an analogous manner. PHD2181-426 (5 μM) and MnCl2 (5 μM) were incubated with DR025 (25 μM) in Tris buffer (50 mM Tris, 100 mM NaCl, pH 7.4) with shaking at r.t. (45 min). The samples were then irradiated (20 min) on ice with UV light at 365 nm at an irradiance of 5 mW/cm2 (Spectrolinker XL-1500). Controls in the presence of the scaffold DR024 or of the competitive inhibitor DR016 were treated in the same way. After exposure to UV light, the samples were either directly analyzed by MALDI TOF MS or subjected to purification using (strept)avidin-coated beads.
Affinity Purification Experiments
The complete labelling and enrichment procedure is described here for purified PHD2181-426 and the probe (DR025) in the presence of the HEK293T cell lysates. In the experiments with the purified enzymes and in the presence of the only HEK293T whole cell lysates or nuclear protein extracts, the labelling and enrichment processes were performed in an analogous way. PHD2181-426 (7.5 μL of 25 μM in Tris buffer solution, final concentration 5 μM) and MnCl2 (1.5 μL of 125 μM in water, final concentration 5 μM) were incubated with DR025 (1.5 μL of 125 μM in water, final concentration 5 μM) in the presence of 27 μL of HEK293T whole cell lysates (1.6 μg/μL total protein) with shaking at r.t. for 45 min. The scaffold (DR024) control sample was prepared in the same way. The mixture was then UV irradiated as described above, and incubated with streptavidin-coated magnetic beads (50 μL, Dynabeads® MyOneTM Streptavidin C1, Invitrogen) for 30 min at r.t. on a shaker incubator. The beads were collected using a magnetic device (Dynal® DynaMagTM Spin, Invitrogen) and washed five times with washing buffer (10mM HEPES, pH 7.2, 1M NaCl, 1% Triton X-100, 2mM EDTA, 4mM dithiotreitol), then three times with water. In other experiments, after the photo cross-linking, the mixtures were incubated with avidin-coated agarose beads (Pierce® Monomeric Avidin Agarose, Thermo Scientific). In these cases the washing was performed with the same buffer but without any device and the supernatant was removed by pipetting. Beads were stored at −20 °C.
In the experiments aimed to label and enrich endogenous levels of target proteins, the final concentrations of probe (DR025) and scaffold control (DR024) were 10 μM and the total protein amounts in the whole HEK293T cell lysates and in the nuclear protein extracts were 200 μg. In the capture experiment carried out on the lysate obtained from HEK293T cells over-expressing PHD2 the total protein amount was 40 μg, while in the profiling of the 2-OG oxygenases present in the whole HEK293T cell lysate the total quantity of proteins was 950 μg per sample in the presence of 10 μM concentration of DR025 and DR024. The competition control tests were performed using the competitive inhibitor DR016 (final concentration 10 or 40 times higher than DR025) (see Figure legends for details). Negative control experiments were carried out by incubating with DR025 under the same conditions as the “capture” experiments but without irradiation.
Supplementary Material
ACKNOWLEDGEMENTS
C.J.S. was supported by the European Union, The Wellcome Trust, and the Biotechnology and Biological Research Council. B.M.K. and R.F. were supported by the Biomedical Research Centre (NIHR) Oxford, United Kingdom and a grant from Action Medical Research. M.A. was supported by the Swedish Research council, the Loo and Hans Ostermans Foundation for Geriatric Research and the Foundation for Geriatric Diseases at Karolinska Institutet. A.W. was recipient of an EMBO long term fellowship. A.M. was supported by Fondazione Roma. The nuclear protein extract of HEK293T cells over-expressing FBXL11 (KDM2A) as protein reference was a kind gift of Dr. Rob Klose. We thank Dr. Oliver King, Dr. Nathan Rose and Tristan Smart for assistance in recombinant protein production and peptide synthesis.
REFERENCES
- Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- Chowdhury R, McDonough MA, Mecinovic J, Loenarz C, Flashman E, Hewitson KS, Domene C, Schofield CJ. Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure. 2009;17:981–989. doi: 10.1016/j.str.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Epstein ACR, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and Mammalian Homologs Define a Family of Dioxygenases that Regulate HIF by Prolyl Hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem. Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- Fisher JJ, Graebner(nee’ Baessler) OY, Dalhoff C, Michaelis S, Schrey AK, Ungewiss J, Andrich K, Jeske D, Kroll F, Glinski M, et al. Comprehensive Identification of Staurosporine-Binding Kinases in the Hepatocyte Cell Line HepG2 using Capture Compound Mass Spectrometry (CCMS) J. Proteome Res. 2010;9:806–817. doi: 10.1021/pr9007333. [DOI] [PubMed] [Google Scholar]
- Flashman E, Davies SL, Yeoh KK, Schofield CJ. Investigating the dependence of the hypoxia-inducible factor hydroxylases (factor inhibiting HIF and prolyl hydroxylase domain 2) on ascorbate and other reducing agents. Biochem. J. 2010;427:135–142. doi: 10.1042/BJ20091609. [DOI] [PubMed] [Google Scholar]
- Fleming SA. Chemical reagents in photoaffinity labeling. Tetrahedron. 1995;51:12479–12520. [Google Scholar]
- Hausinger RP. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Kessler BM, Tortorella D, Altun M, Kisselev AF, Fiebiger E, Hekking BG, Ploegh HL, Overkleeft HS. Extended peptide-based inhibitors efficiently target the proteasome and reveal overlapping specificities of the catalytic beta-subunits. Chem. Biol. 2001;8:913–929. doi: 10.1016/s1074-5521(01)00069-2. [DOI] [PubMed] [Google Scholar]
- King ONF, Li S, Sakurai M, Kawamura A, Rose NR, Ng SS, Quinn AM, Rai G, Mott BT, Klose RJ, et al. Quantitative High-throughput Screening Identifies 8-Hydroxyquinolines as Cell-active Histone Demethylase Inhibitors. PLoSOne. 2010 doi: 10.1371/journal.pone.0015535. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007;8:307–317. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Holbert S, Ronce N, Faravelli F, Lenzner S, Schwartz CE, Lespinasse J, Van Esch H, Lacombe D, Goizet C, et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung IKH, Flashman E, Yeoh KK, Schofield CJ, Claridge TDW. Using NMR solvent water relaxation to investigate metalloenzyme-ligand binding interactions. J. Med. Chem. 2010;53:867–875. doi: 10.1021/jm901537q. [DOI] [PubMed] [Google Scholar]
- Liepinsh E, Vilskersts R, Loca D, Kirjanova O, Pugovichs O, Kalvinsh I, Dambrova M. Mildronate, an inhibitor of carnitine biosynthesis, induces an increase in gamma-butyrobetaine contents and cardioprotection in isolated rat heart infarction. J. Cardiovasc.. Pharmacol. 2006;48:314–319. doi: 10.1097/01.fjc.0000250077.07702.23. [DOI] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Oxygenases catalyzed 5-methylcytosine hydroxylation. Chem. Biol. 2009;16:580–583. doi: 10.1016/j.chembiol.2009.06.002. [DOI] [PubMed] [Google Scholar]
- McDonough MA, McNeill LA, Tilliet M, Papamicael CA, Chen Q-Y, Banerji, B., Hewitson KS, Schofield CJ. Selective inhibition of factor inhibiting hypoxia-inducible factor. J. Am. Chem. Soc. 2005;127:7680–7681. doi: 10.1021/ja050841b. [DOI] [PubMed] [Google Scholar]
- McNeill LA, Flashman E, Buck MRG, Hewitson KS, Clifton IJ, Jeschke G, Claridge TDW, Ehrismann D, Oldham NJ, Schofield CJ. Hypoxiainducible factor prolyl hydroxylase 2 has a high affinity for ferrous iron and 2-oxoglutarate. Mol. Bio. Syst. 2005;1:321–324. doi: 10.1039/b511249b. [DOI] [PubMed] [Google Scholar]
- Nagel S, Talbot NP, Mecinovic J, Smith TG, Buchan AM, Schofield CJ. Therapeutic manipulation of the HIF hydroxylases. Antioxid. Redox Signal. 2010;12:481–501. doi: 10.1089/ars.2009.2711. [DOI] [PubMed] [Google Scholar]
- Pescador N, Cuevas Y, Naranjo S, Alcaide M, Villar D, Landazuri MO, Del Peso L. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem. J. 2005;390:189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, Loenarz C, Mole DR, McDonough MA, Gleadle JM, Schofield CJ, Ratcliffe PJ. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1α. Biochem. J. 2008;416:387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- Risteli M, Ruotsalainen H, Salo AM, Sormunen R, Sipilä L, Baker NL, Lamandé SR, Vimpari-Kauppinen L, Myllylä R. Reduction of lysyl hydroxylase 3 causes deleterious changes in the deposition and organization of extracellular matrix. J. Biol. Chem. 2009;284:28204–28211. doi: 10.1074/jbc.M109.038190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Ng SS, Mecinovic J, Lienard BM, Bello SH, Sun Z, McDonough MA, Oppermann U, Schofield CJ. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. J. Med. Chem. 2008;51:7053–7056. doi: 10.1021/jm800936s. [DOI] [PubMed] [Google Scholar]
- Rose NR, Woon ECY, Kingham GL, King ONF, Mecinovic J, Clifton IJ, Ng SS, Talib-Hardy J, Oppermann U, McDonough MA, et al. Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometry screening and crystallographic approaches. J. Med. Chem. 2010;53:1810–1818. doi: 10.1021/jm901680b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani AM, Verhelst SHL, Bogyo M. Tagging and detection strategies for activity-based proteomics. Curr. Opin. Chem. Biol. 2007;11:20–28. doi: 10.1016/j.cbpa.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Salisbury CM, Cravatt BF. Optimization of activity-based probes for proteomic profiling of histone deacetylase complexes. J. Am. Chem. Soc. 2008;130:2184–2194. doi: 10.1021/ja074138u. [DOI] [PubMed] [Google Scholar]
- Sieber SA, Niessen S, Hoover HS, Cravatt BF. Proteomic profiling of metalloprotease activities with cocktails of active-site probes. Nat. Chem. Biol. 2006;2:274–281. doi: 10.1038/nchembio781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova NA, Rakhman I, Moroz N, Basso M, Payappilly J, Kazakov S, Hernandez-Guzman F, Gaisina IN, Kozikowski AP, Ratan RR, et al. Utilization of an in vivo reporter for high throughput identification of branched small molecule regulators of hypoxic adaptation. Chem. Biol. 2010;17:380–391. doi: 10.1016/j.chembiol.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem. 2009;4:1568–1582. doi: 10.1002/cmdc.200900301. [DOI] [PubMed] [Google Scholar]
- Stubbs CJ, Loenarz C, Mecinovic J, Yeoh KK, Hindley N, Lienard BM, Sobott F, Schofield CJ, Flashman E. Application of a Proteolysis/Mass Spectrometry Method for Investigating the Effects of Inhibitors on Hydroxylase Structure. J. Med. Chem. 2009;52:2799–2805. doi: 10.1021/jm900285r. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Warshakoon NC, Wu S, Boyer A, Kawamoto R, Sheville J, Renock S, Xu K, Pokross M, Zhou S, Winter C, et al. Structure-based design, synthesis, and SAR evaluation of a new series of 8-hydroxyquinolines as HIF-1α prolyl hydroxylase inhibitors. Bioorg. Med. Chem. Lett. 2006;16:5517–5522. doi: 10.1016/j.bmcl.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Wu CC, MacCoss MJ. Shotgun proteomics: tools for the analysis of complex biological systems. Curr. Opin. Mol. Ther. 2002;4:242–250. [PubMed] [Google Scholar]
- Xu C, Soragni E, Chou CJ, Herman D, Plasterer HL, Rusche JR, Gottesfeld JM. Chemical probes identify a role for histone deacetylase 3 in Friedreich’s ataxia gene silencing. Chem. Biol. 2009;16:980–989. doi: 10.1016/j.chembiol.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Wang Y, Huang S, Wang J, Deng Z, Zhang Q, Wu W, Zhang X, Liu Z, Gong W, et al. Structural insights into a novel histone demethylase PHF8. Cell Res. 2010;20:166–173. doi: 10.1038/cr.2010.8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.