Abstract
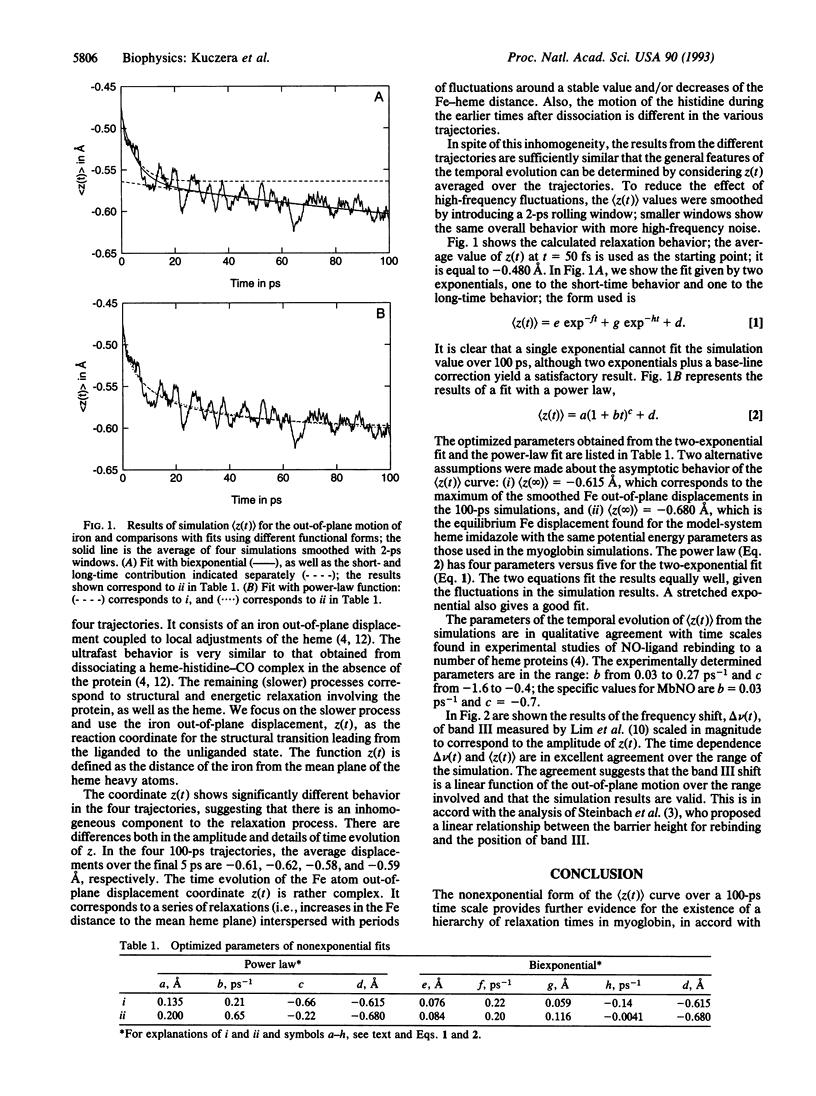
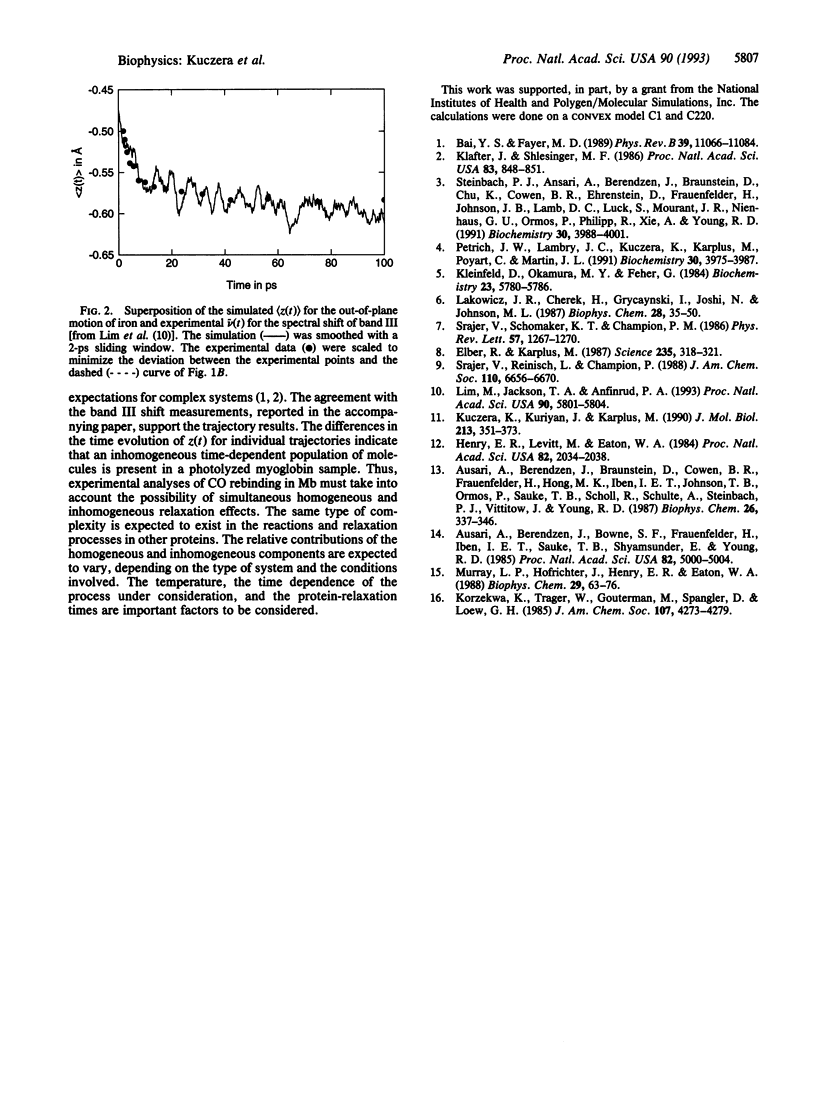
Molecular dynamics simulations of myoglobin after ligand photodissociation show that the out-of-plane motion of the heme iron has a rapid subpicosecond phase followed by a slower nonexponential process involving more global protein relaxation. Individual trajectories show rather different behavior, suggesting there is an inhomogeneous component to the relaxation. The calculated time dependence of the iron motion over 100 ps is in excellent agreement with the frequency shift of band III of the heme group [see Lim, M., Jackson, T. A. & Anfinrud, P. A. (1993) Proc. Natl. Acad. Sci. USA 90, 5801-5804]. If that the barrier to rebinding depends on the out-of-plane iron position, the time dependence obtained from the simulation can explain the nonexponential room-temperature geminate recombination of NO.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansari A., Berendzen J., Bowne S. F., Frauenfelder H., Iben I. E., Sauke T. B., Shyamsunder E., Young R. D. Protein states and proteinquakes. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A., Berendzen J., Braunstein D., Cowen B. R., Frauenfelder H., Hong M. K., Iben I. E., Johnson J. B., Ormos P., Sauke T. B. Rebinding and relaxation in the myoglobin pocket. Biophys Chem. 1987 May 9;26(2-3):337–355. doi: 10.1016/0301-4622(87)80034-0. [DOI] [PubMed] [Google Scholar]
- Bai YS, Fayer MD. Time scales and optical dephasing measurements: Investigation of dynamics in complex systems. Phys Rev B Condens Matter. 1989 May 15;39(15):11066–11084. doi: 10.1103/physrevb.39.11066. [DOI] [PubMed] [Google Scholar]
- Elber R., Karplus M. Multiple conformational states of proteins: a molecular dynamics analysis of myoglobin. Science. 1987 Jan 16;235(4786):318–321. doi: 10.1126/science.3798113. [DOI] [PubMed] [Google Scholar]
- Henry E. R., Levitt M., Eaton W. A. Molecular dynamics simulation of photodissociation of carbon monoxide from hemoglobin. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2034–2038. doi: 10.1073/pnas.82.7.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klafter J., Shlesinger M. F. On the relationship among three theories of relaxation in disordered systems. Proc Natl Acad Sci U S A. 1986 Feb;83(4):848–851. doi: 10.1073/pnas.83.4.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinfeld D., Okamura M. Y., Feher G. Electron-transfer kinetics in photosynthetic reaction centers cooled to cryogenic temperatures in the charge-separated state: evidence for light-induced structural changes. Biochemistry. 1984 Nov 20;23(24):5780–5786. doi: 10.1021/bi00319a017. [DOI] [PubMed] [Google Scholar]
- Kuczera K., Kuriyan J., Karplus M. Temperature dependence of the structure and dynamics of myoglobin. A simulation approach. J Mol Biol. 1990 May 20;213(2):351–373. doi: 10.1016/S0022-2836(05)80196-2. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., Cherek H., Gryczynski I., Joshi N., Johnson M. L. Analysis of fluorescence decay kinetics measured in the frequency domain using distributions of decay times. Biophys Chem. 1987 Oct;28(1):35–50. doi: 10.1016/0301-4622(87)80073-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M., Jackson T. A., Anfinrud P. A. Nonexponential protein relaxation: dynamics of conformational change in myoglobin. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5801–5804. doi: 10.1073/pnas.90.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray L. P., Hofrichter J., Henry E. R., Eaton W. A. Time-resolved optical spectroscopy and structural dynamics following photodissociation of carbonmonoxyhemoglobin. Biophys Chem. 1988 Feb;29(1-2):63–76. doi: 10.1016/0301-4622(88)87025-x. [DOI] [PubMed] [Google Scholar]
- Petrich J. W., Lambry J. C., Kuczera K., Karplus M., Poyart C., Martin J. L. Ligand binding and protein relaxation in heme proteins: a room temperature analysis of NO geminate recombination. Biochemistry. 1991 Apr 23;30(16):3975–3987. doi: 10.1021/bi00230a025. [DOI] [PubMed] [Google Scholar]
- Srajer V, V, Schomacker KT, Champion PM. Spectral broadening in biomolecules. Phys Rev Lett. 1986 Sep 8;57(10):1267–1270. doi: 10.1103/PhysRevLett.57.1267. [DOI] [PubMed] [Google Scholar]
- Steinbach P. J., Ansari A., Berendzen J., Braunstein D., Chu K., Cowen B. R., Ehrenstein D., Frauenfelder H., Johnson J. B., Lamb D. C. Ligand binding to heme proteins: connection between dynamics and function. Biochemistry. 1991 Apr 23;30(16):3988–4001. doi: 10.1021/bi00230a026. [DOI] [PubMed] [Google Scholar]



