Abstract
Transcription of genes encoding small structured RNAs such as tRNAs, spliceosomal U6 snRNA and ribosomal 5S RNA is carried out by RNA polymerase III (Pol III), the largest yet structurally least characterized eukaryotic RNA polymerase. The cryo-EM structures of the S. cerevisiae Pol III elongating complex at 3.9 Å resolution and the apo Pol III enzyme in two different conformations at 4.6 and 4.7 Å resolution, respectively, allow for the first time to build a 17-subunit atomic model of Pol III. The reconstructions reveal the precise orientation of the C82/C34/C31 heterotrimer in close proximity to the stalk. The C53/C37 heterodimer positions residues involved in transcription termination close to the non-template DNA strand. In the apo Pol III structures, the stalk adopts different orientations coupled with closed and open conformations of the clamp. Our results provide novel insights into Pol III-specific transcription and the adaptation of Pol III towards its small transcriptional targets.
Keywords: RNA polymerase III, Pol III, electron cryo-microscopy, tRNA, transcription
In eukaryotes, transcription of structured, small RNAs such as tRNAs, spliceosomal U6 snRNA, ribosomal 5S RNA and 7 SL RNA is mediated by RNA polymerase III (Pol III)1. Transcriptional activity of Pol III is elevated in cancer cells, and the tumor suppressors p53 and Rb and the proto-oncogene Myc directly regulate Pol III mediated transcription2.
Pol III is the largest of the three RNA polymerases containing 17 subunits with a total molecular weight of 0.7 MDa, while RNA polymerase I (Pol I) and RNA polymerase II (Pol II), comprise 14 and 12 subunits, respectively. The architecture of the 10-subunit core is conserved among all eukaryotic RNA polymerases3. In Pol III it comprises the two largest subunits C160 and C128 that form the active site. Five additional core subunits ABC27, ABC23, ABC14.5, ABC10α and ABC10β are shared between Pol I, Pol II and Pol III. Subunits AC40 and AC19 are common between Pol I and Pol III and are closely related to their Pol II counterparts, Rpb3 and Rpb11. The Pol III core is completed by subunit C11 involved in transcription termination4,5 and RNA cleavage activity5. Subunit C11 corresponds to subunit A12.2 in Pol I6 and both subunits are homologous to a fusion protein that comprises the N-terminal domain of Pol II subunit Rbp9 and the C-terminal domain of TFIIS5.
The Pol III stalk comprises subunits C17 and C25 and is involved in transcription initiation3 and in binding of single-stranded RNA7. The C53/C37 heterodimer is involved in transcription initiation and termination4,8,9 and is distantly related to Pol I A49/A34.5 and Pol II TFIIF3,10,11. The C53/C37 heterodimer occupies a large density on the Pol III core opposite of the stalk12 and extends into the DNA-binding cleft as shown by chemical crosslinking8,13. Finally, the C82/C34/C31 heterotrimer forms a stable subcomplex14,15 that is positioned on the C160 clamp domain in close proximity to the stalk12,16. The C82/C34/C31 heterotrimer is involved in transcription initiation3,17 and recruitment of Pol III to TFIIIB18,19. Subunits C82 and C34 in the heterotrimer are distantly related to TFIIEα and TFIIEβ, respectively10, further supporting the hypothesis that during evolution general transcription factors such as TFIIF and TFIIE became stably associated Pol I and Pol III subunits. Maf1 is a global repressor of Pol III conserved in eukaryotes that under stress conditions becomes dephosphorylated and translocates to the nucleus where it directly binds Pol III20, 21.
In the last decade, detailed structural studies of Pol I and Pol II contributed to elucidate basic mechanisms of DNA-dependent RNA transcription22-25. However, structural insight into Pol III mediated transcription has been limited to date as the highest resolution Pol III structures only extend to 10 Å for apo Pol III26 and to 16.5 Å and 19 Å for the Pol III elongation complex21,26. Here, we present the first atomic structures of apo Pol III and elongating Pol III obtained by single particle electron cryo-microscopy (cryo-EM). Our reconstructions consolidate biochemical data, allow the detailed comparison with the Pol I and Pol II enzymes, and provide additional insight into Pol III specific transcription.
Overall architecture of Pol III
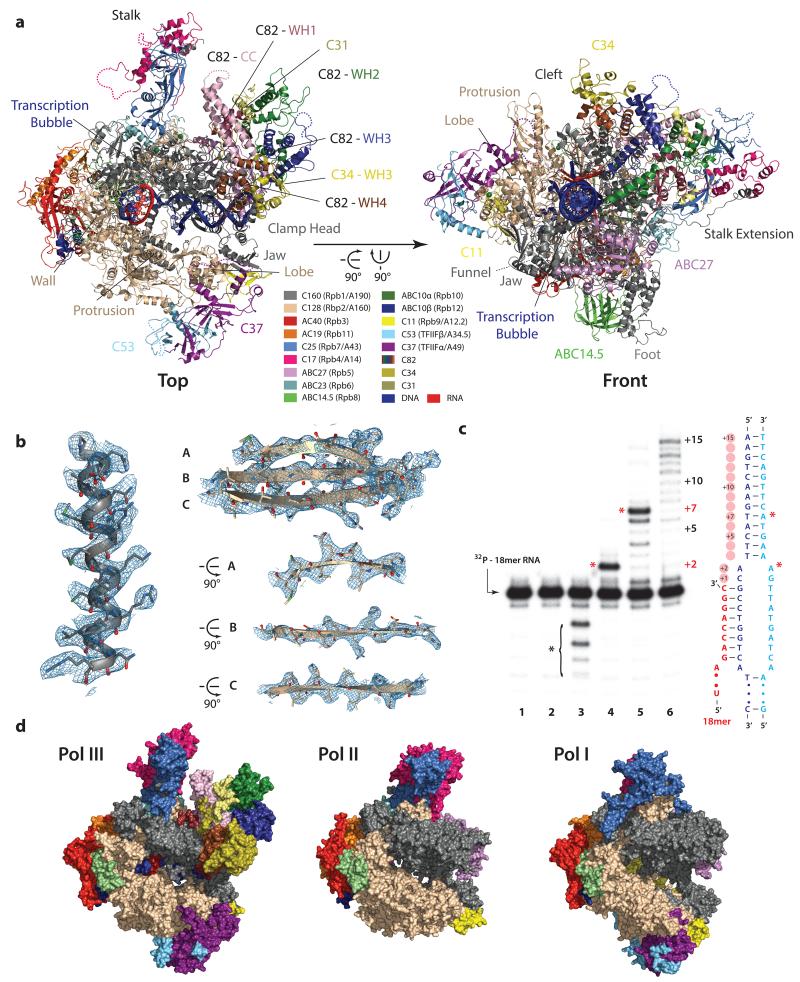
Saccharomyces cerevisiae Pol III was purified as previously described27 yielding pure, homogenous and transcriptionally active enzyme (Fig. 1). To shed light onto the Pol III architecture and to further investigate Pol III mediated transcription, we acquired cryo-EM images of Pol III bound to an assembled transcriptionally active bubble (elongating Pol III, Fig. 1c) and of native unbound Pol III (apo Pol III) on an FEI Titan Krios equipped with a Falcon II direct electron detector (Methods, Extended Data Fig. 1). The final map of elongating Pol III was determined using maximum-likelihood based 3D classification in RELION28 at an average resolution of 3.9 Å (Extended Data Fig. 1), but extends beyond 3.5 Å in the Pol III core (Extended Data Figs. 1c, 2, 3). This EM density was used to build and refine an atomic model of the complete 17-subunit structure of elongating Pol III (Fig. 1a, Extended Data Fig. 4, Table 1). Representative densities (Fig. 1b, Extended Data Fig. 2) and the relatively isotropic resolution (Extended Data Fig. 1c, Table 2) demonstrate that the quality of the cryo-EM density map is comparable to electron density maps of Pol I and Pol II obtained by X-ray crystallography at nominally higher resolutions (Extended Data Fig. 3). The overall architecture of the Pol III core is conserved with respect to Pol I and Pol II (Fig. 1d). However, the clamp head part of subunit C160 is enlarged compared to its Pol I and Pol II counterparts (Extended Data Fig. 5). In addition, C160 contains an extended foot that different as Pol I and Pol II forms a large interface with the shared subunit ABC14.5. Furthermore, a C-terminal extension of C160, unique to Pol III, protrudes from the core and together with the C160 N-terminus contacts the stalk. Similarly, the second largest subunit C128 shows an overall conserved fold, but contains an extended protrusion that increases the depth of the DNA-binding cleft in comparison to Pol I and Pol II (Extended Data Fig. 5). The Pol III cryo-EM structure also includes the Pol III-specific C82/C34/C31 heterotrimer and the C53/C37 heterodimer both showing several unexpected features as discussed below.
Figure 1. Cryo-EM structure of RNA polymerase III.
a, Top and front view of Pol III, individual elements and domains are labeled. Dotted lines indicate regions that are not included in the model. The color code is presented in the corresponding boxes. b, Representative densities of the Pol III core with the fitted model demonstrate the high detail visible in the final cryo-EM structure. c, RNA extension assay demonstrates RNA elongation and cleavage activity of Pol III. The transcription bubble used for the activity assays and for the cryo-EM structure determination is depicted at the right (see also Methods). Lane 1: annealed transcription bubble with 32P labeled RNA (18mer). Lane 2: with NTP mix. Lane 3: with Pol III but without NTPs showing the intrinsic RNA cleavage activity of Pol III, cleavage products are denoted by a black asterisk. Lanes 4 and 5: with Pol III and NTPs excluding ATP (Lane 4) or CTP (Lane 5) showing nucleotide-specificity and elongation arrest at +2 (no ATP) or +7 (no CTP) denoted by red asterisks. Lane 6: with Pol III and NTPs, the +15 run-off shows full-length extension. d, Surface view of the elongating Pol III structure (this study) compared to Pol I (PDB 4c3i) and Pol II (PDB 1wcm). Homologous subunits in Pol I and Pol II are colored based on Pol III and as indicated in a.
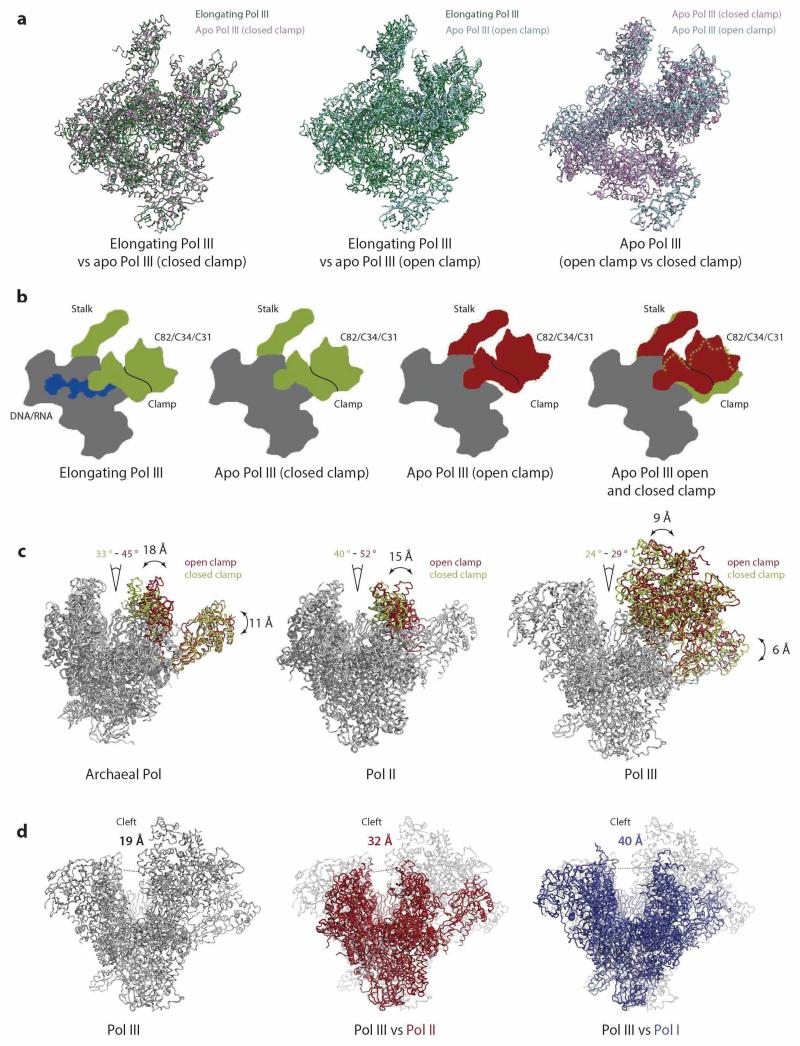
The apo Pol III dataset yielded two major 3D classes showing distinct conformations at 4.6 Å and 4.7 Å resolution (Extended Data Figs. 1b, c). One reconstruction is very similar to the elongating Pol III (rmsd = 0.43 Å4813CA), whereas the second reconstruction shows an altered orientation of the stalk, the heterotrimer and a more open cleft resulting in a larger difference with elongating Pol III (rmsd = 2.73 Å4795 CA). The two apo Pol III conformations presumably result from “closed” and “open” states of the clamp domain as discussed below (Extended Data Fig. 6a-c).
A narrow cleft encloses DNA
In both apo Pol III and the elongating Pol III structures we observe a characteristic narrower cleft in comparison to Pol I and Pol II (Extended Data Fig. 6d). The DNA duplex is embedded into the Pol III cleft and extends from downstream base pair (bp) +14 towards the active site until the upstream bp −9 (Fig. 2a). The DNA duplex is anchored between the jaw and lobe domains and the oppositely positioned extended clamp head. In addition, the WH2 and WH3 domains of C82 (see below) lie in close proximity and further stabilize downstream DNA (Fig. 2b). Subunit ABC27 completes this enclosure by inserting a proline-containing loop into the minor groove between bps +11 to +14, thus threading the DNA duplex towards the active center. In the crystal structure of elongating Pol II bound to a transcription bubble that comprises upstream and downstream DNA duplexes (53-mer DNA oligonucleotides)29, the proline-containing loop also protrudes into the minor groove of downstream DNA, while upstream DNA interacts with a hairpin-loop/wedge in Pol II subunit Rbp2 (residues 862-874) also present in Pol III subunit C128 (residues 794-806) as well as in Pol I. Global recognition of upstream and downstream DNA therefore appears to be conserved among the three eukaryotic RNA polymerases.
Figure 2. Transcription of Pol III and association with DNA/RNA duplex.
a, The left panel shows an elongating Pol III ribbon model with the segmented density of the transcription bubble displayed at 4.5 Å for better visibility. DNA and RNA densities are shown in blue and red, respectively. The transcription bubble is shown in stick representation. The downstream DNA duplex is embedded in the cleft, an eight basepair DNA/RNA hybrid was built based on the density. The two right panels show close-ups at two orthogonal views. b, The downstream DNA duplex is tightly bound between the jaw (grey), the clamp head (grey), subunit ABC27 (pink) and two C82 WH domains (WH2 - green and WH3 - blue). c, Cross-section of the elongating Pol III density at 3.9 Å with colored density corresponding to the transcription bubble. The density for the downstream DNA duplex exceeds even beyond bp +15 in the map, although it is much weaker compared to DNA density in the cleft, thus likely corresponding to an unstably stacked second DNA duplex. d, Close-up view of the active site of Pol III (left) and Pol II (right, PDB 1y1w). An extended rudder that points towards a stretch of the protrusion (residues 390-400) and a buried fork loop 1 suggest that the DNA/RNA hybrid in the Pol III core is less tightly bound compared to Pol II (right panel), where fork loop 1 protrudes into the core and together with the rudder and the wall forms a barrier. Similar as in Pol II, the trigger loop is unstructured in Pol III.
The template DNA strand unwinds at base pair +2 and a characteristic A-type DNA-RNA hybrid forms at positions −1 to −9 (Fig. 2a). Additional density in the RNA exit channel likely corresponds to the emerging and thus more flexible single stranded RNA (Fig. 2c). No clear density for upstream DNA is visible, indicating flexibility of the emerging duplex. Another noticeable feature is the strong density of the downstream DNA double strand in comparison to the much weaker density for the DNA/RNA duplex (Figs. 2a-c). This is in contrast to the Pol II elongation complex, where both densities are of equal quality29-31. Strikingly, the DNA/RNA duplex is tightly associated with the Pol II elongation complex between the wall and fork loop 1/rudder whereas in Pol III the rudder and fork loop 1 reach toward the protrusion, thereby reducing the association of the DNA/RNA hybrid with the Pol III active center (Fig. 2d). While the DNA/RNA hybrid appears to be only loosely associated with the active site, we observe a tight enclosure of the downstream DNA duplex at the entrance of the DNA-binding cleft.
Pol III heterotrimer protrudes into the cleft
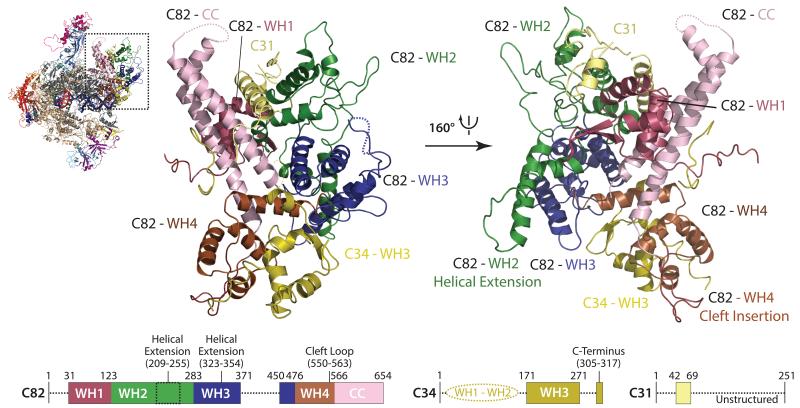
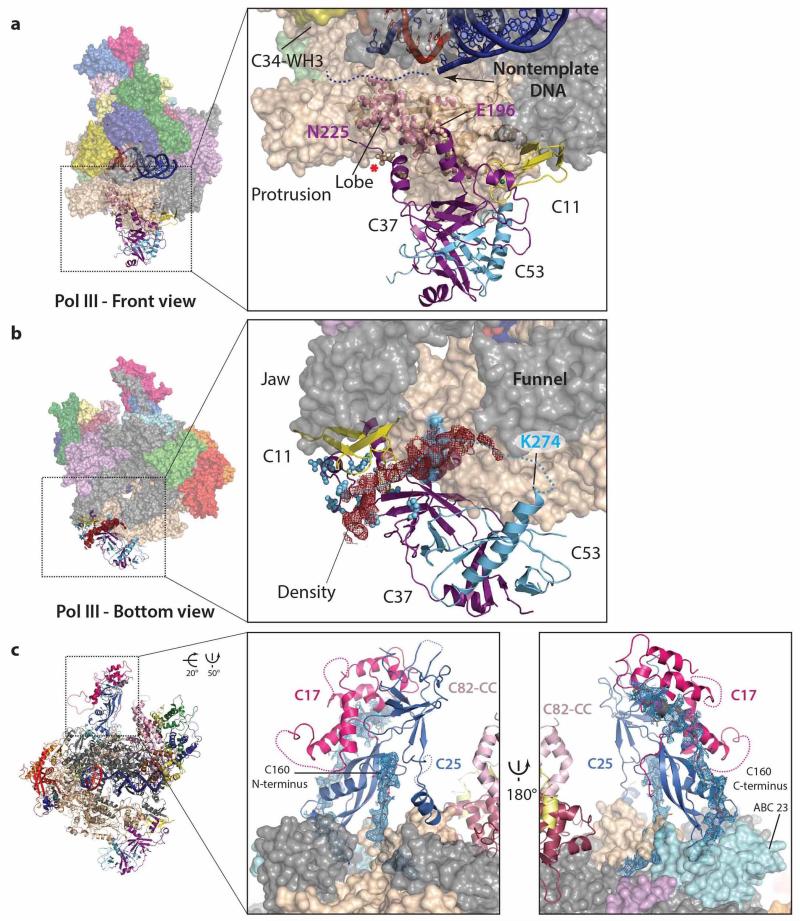
Although the crystal structure of the human C82 orthologue hRPC6232 is available, its integration into the C82/C34/C31 heterotrimer and the precise orientation of the heterotrimer within the complete Pol III enzyme could not be clarified by previous cryo-EM reconstructions due to their limited resolution12,21,26. In addition, the functional roles of the together seven winged-helix (WH) domains present in subunits C82 and C34 and often found in transcription factors as DNA-binding or protein-protein interaction modules33 are still poorly understood. Our cryo-EM structure shows how the heterotrimer packs onto the clamp head by forming a large, hydrophobic interface through various WH domains (C82-WH1/WH4 and C34-WH3; Fig. 3). In this case the WH domains serve as protein-protein interacting domains, although they might still contact DNA during open complex formation in the transcription initiation process. The coiled-coil domain of C82 protrudes towards the stalk and the remaining C82-WH2 and WH3 are facing away from the core. Strikingly, C82-WH2 and C82-WH3 align with the clamp head to reach around bp +15 towards downstream DNA. Furthermore, an additional long ‘cleft loop’ extending from C82-WH4 passes through a canyon in the clamp head into the DNA-binding cleft close to DNA bp +7 (Extended Data Fig. 7a). Subunit C34 comprises three WH domains that span from C82 towards the protrusion crossing the DNA-binding cleft16. TFIIEα/β also possesses three WH domains and in the cryo-EM structure of the human Pol II pre-initiation complex crosses the DNA-binding cleft34. The two N-terminal C34 WH domains are not visible in the Pol III cryo-EM density like the A49 tandem WH domain in the Pol I crystal structure24,25, while the third WH domain tightly associates with C82 and is located at the periphery of the heterotrimer. The C34 C-terminal region following the third WH domain passes the C82-WH4 and contacts the C82 coiled-coil domain (Fig. 3). The third subunit of the heterotrimer, namely C31, was predicted to be largely unstructured and associated between C82 and the stalk, as shown by crosslinking analysis16. However, we were able to build a mainly helical element of C31 (residues 42-69) demonstrating that C31 extends along the surface from the C34 C-terminus towards C82-WH4 over the C82 coiled-coil domain to reach the stalk, where it becomes disordered. Additional density close to the interface of C82-WH1 and WH2 and between the heterotrimer and the stalk most likely corresponds to C-terminal stretches of C31, but no sequence could be unambiguously assigned to it (Extended Data Fig. 7b). Nevertheless, the topology of C31 and the extended interaction interface with at least three WH domains, the C82 coiled-coil domain and the stalk confirms the previously reported role of C31 in connecting the heterotrimer to the Pol III core and stalk17,35,36.
Figure 3. Architecture of the Pol III-specific heterotrimer.
The panel shows the C82/C34/C31 heterotrimer as ribbon representation in top (left) and bottom (right) view. Schematic representations of C82 (left), C34 (middle) and C31 (right) depict domain boundaries. Structured and disordered regions are marked with solid and dotted lines, respectively.
Transcription termination by Pol III heterodimer
Previous EM and crosslinking studies of Pol III and the C53/C37 subcomplex positioned a conserved dimerization module at the lobe of Pol III12,13,21,26 similar to TFIIF and A49/A34.5 in Pol II37 and Pol I24,25, respectively. On the other hand, extensions of C53 and C37 crosslink close to the active site, the stalk and the heterotrimer8,13,16. Our structure better characterizes the interaction network of the C53/C37 heterodimer with other Pol III subunits. Strikingly, C37 shows an extended contact surface with C11 and the cleft when compared to TFIIFα and A49 (Fig. 4a) consistent with its proposed role in C11 association with the core9. Our cryo-EM structure also rationalizes the role of the C53/C37 heterodimer in Pol III transcription termination that only requires a stretch of 5-7 thymines in the non-template DNA strand for efficient transcription termination38. Subunit C37 extends towards the DNA-binding cleft where it positions a flexible loop (residues 197-224) that has been shown to contact C34 and the Pol III-specific TFIIIB subunit Bdp113,39 before folding back into a helix (residues 230-240; Extended Data Fig. 8a). Deleting the five residues (R226, L227, T228, G229, S230) leading into this helix produces a terminator read-through phenotype in S. cerevisiae4, and in S. pombe the corresponding region was identified as a hotspot for terminator read-through mutations40. The same residues have also been cross-linked to C12813 and in the cryo-EM structure are packed onto a section of the C128 lobe that when deleted also results in a termination read-through phenotype41. In addition, these five residues are in close proximity to the non-template DNA strand that is flexible in the Pol III structure (Extended Data Fig. 8a). Direct interactions between subunit C37 presumably involve these five residues and the first four thymines of the non-template DNA strand and result in a conformational switch of Pol III towards a metastable pre-termination complex, while the presence of a fifth thymine results in transcription termination4. In contrast, Pol I and Pol II require additional cis and/or trans acting factors for transcription termination42 and accordingly TFIIF or A49/A34.5 do not possess similar features (Fig. 4a, Extended Data Fig. 6c).
Figure 4. Architecture and Pol III-specific function of the C53/C37 heterodimer and C11.
a, Model of the Pol III C53/C37 heterodimer shown in ribbon representation bound to the Pol III core (left), Pol II homologue TFIIFα/β bound to the Pol II core (center, PDB 4v1n) and Pol I homologue A49/A34.5 bound to the Pol I core (right, PDB 4c3i). The red asterisk (left panel) marks the position of the five residues that upon deletion lead to a terminator read-through phenotype13. Schematic representations of C53 and C37 show the domain boundaries of the dimerization domain (DD) and additional elements. Dotted lines indicate unstructured regions. b, Conformation of subunit C11 in Pol III (left), subunit Rpb9 in Pol II (middle), and subunit A12.2 in Pol I. Subunits C11, Rpb9 and A12.2 are depicted with yellow surface rendering; C53/C37, TFIIFα/β and A49/A34.5 are depicted in ribbon representation, all other subunits are colored in grey. Arrows indicate the potential movement of the C11 C-terminal TFIIS domain, the red dotted circle indicates the linker that connects the C11 N- and C-terminal domains.
The C-terminus of C37 folds back and forms a hydrophobic interface with the C53 and C37 core consistent with the crucial role of the C37 C-terminus in assembling and stabilizing the C53/C37 heterodimer9. C53 is located below the lobe and extension 2 and its C-terminal dimerization domain is indeed tightly associated with the C37 dimerization domain (Fig. 4a). The remaining 270 residues of the N-terminal region of C53 are flexible and could not be attributed to any density, although crosslinking experiments suggest that it is involved into a complex interaction network8,13. Interestingly, weak (but continuous) density in the cleft between C11 and C37 could be identified as a part of C53 based on specific photo-crosslinks13, although the density is too weak for model building and sequence assignment (Extended Data Fig. 8b). This observation implies that the N-terminal extension of C53 travels from the C160 funnel back to the periphery of the heterodimer in close proximity to the previously described C37/C11 interacting region before moving towards the cleft, the stalk and the heterotrimer8,13,16.
The TFIIS-like domain of C11 is flexible
The core subunit C11 is responsible for Pol III’s intrinsic RNA cleavage activity43 that is likely mediated by its conserved TFIIS-like domain5. Notably, the orthologous Pol I subunit A12.2 also contains a C-terminal TFIIS-like domain that associates to the active site24,25. Both C11 and A12.2 also harbor a conserved N-terminal Zn-binding domain with high homology to Pol II Rbp9. The N-terminal Rpb9-like domain of C11 is anchored between C37, the C128 lobe domain and the jaw domain of subunit C160 at a similar position as the N-terminal domains of Rpb9 and A12.2 (Fig. 4b). In contrast, density corresponding to the C-terminal TFIIS-like domain was not observed in the elongating Pol III structure, but was only identified in both apo Pol III structures where it occupies a cleft between Rpb5 and the C160 funnel as observed at a low density threshold (Extended Data Fig. 7c). The position of the C11 C-terminal domain is far away from the position of the corresponding A12.2 C-terminal domain suggesting that the C11 C-terminal domain is mobile and only temporally recruited to the catalytic center (in analogy to TFIIS in Pol II). A long linker (residues 37-61) that connects both domains presumably accounts for the required mobility (Fig. 4b). C11 mutations at the extended interface between C37 and C11 N-terminal domain induce terminator read-through transcripts, while mutations of the C-terminal domain of C11 affect RNA 3’ cleavage reflecting the involvement of the flexibly linked N- and C-terminal domains of C11 in two termination-related, yet distinct activities44.
Pol III stalk relays conformational changes
The Pol III stalk subunits C25 and C17 are homologous to Pol II Rpb7/Rpb4 and Pol A43/A147,36. Consistent with previous low-resolution EM studies12,21,26, the Pol III cryo-EM structure confirms that the closed HDRC domain fold observed in the S. pombe C25/C17 heterodimer45 (PDB 3ayh) describes the conformation of the stalk on active Pol III most appropriately. Furthermore, we see a tight interaction network of the Pol III stalk and the C160 N- and C-terminal extensions, the latter contacting and positioning the C17 HDRC domain on C25 (Extended Data Fig. 8c). This leads to a tight anchoring of the stalk to the core, which is additionally strengthened by the Pol III-specific helix of C25 that extends from the stalk and contacts the clamp.
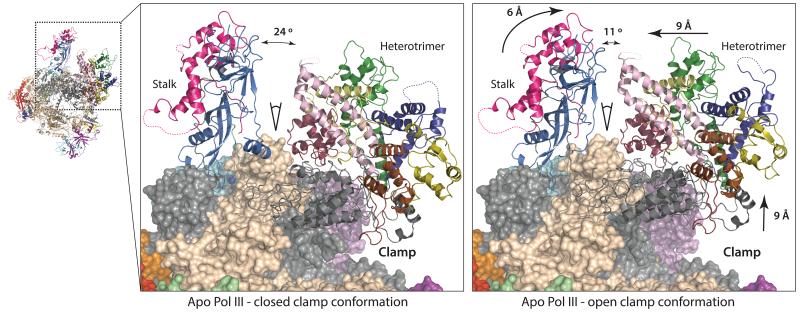
Strikingly, the open and closed clamp conformations of apo Pol III (Extended Data Fig. 6b) demonstrate two structurally distinct conformations of the stalk, the clamp head and the heterotrimer (Fig. 5). A flexible clamp has been also reported in the bacterial RNA polymerase, where it was shown that the clamp is predominantly open in the unbound conformation, then closes during initiation and elongation46. Furthermore, the archaeal RNA polymerase and Pol II contain a flexible clamp and in both systems the status of the clamp is associated with the stalk30,34,47. In contrast to archaeal RNA polymerase and Pol II, in Pol III the clamp movement is less pronounced resulting in a narrower Pol III cleft compared to other RNA polymerases even in the open clamp conformation (Extended Data Fig. 6c, d). Here, we show both clamp conformations in Pol III visualized from the same sample and speculate that a moving stalk in Pol III can mediate the observed conformational changes. The open clamp conformation opens the cleft and Pol III could thus better associate with target DNA, whereas a transition to the closed clamp positions the heterotrimer next to the unwound DNA and could enable C82 and C34 mediated promoter opening and subsequent elongation.
Figure 5. Conformational changes in apo Pol III.
‘Closed clamp’ (left) and ‘open clamp’ (right) conformations of apo Pol III. The C82/C34/C31 heterotrimer, the stalk and the clamp are shown in ribbon representation, the core in surface representation. Arrows in the right panel and corresponding values indicate movements of the stalk, the heterotrimer and the clamp head relative to the closed clamp state.
METHODS
Sample preparation
The 38 nucleotide template DNA strand (AAGTCAAGTACTTACGCCTGGTCATTACTAGTACTGCC), non-template DNA strand (GGCAGTACTAGTAAACTAGTATTGAAAGTACTTGACTT) and the 18 nucleotide RNA (UAUGCAUAAAGACCAGGC) were designed as previously described31, except that a two nucleotide overhang on both ends and one GC base pair was removed. HPLC grade single-stranded oligonucleotides (Eurofins MWG Synthesis GmbH) were diluted in 15 mM Tris, pH 7.5, 150 mM (NH4)2SO4. Template and non-template strands were heated to 95 °C and slowly cooled to room temperature allowing formation of the 11 nucleotide mismatch double-stranded DNA. In a second step, RNA was added, the mixture heated to 45 °C, and slowly cooled to 4 °C. Finally, 5 mM MgCl2 and 10 mM DTT were added to the annealed transcription bubble.
Pol III was purified endogenously from Saccharomyces cerevisiae as previously described27. For apo Pol III, the buffer was exchanged to EM-Buffer (15 mM Tris pH 7.5, 150 mM (NH4)2SO4, 10 mM DTT) and diluted to 0.2 mg/ml for grid freezing. To form the Pol III elongation complex, the buffer was exchanged to EM-Buffer and 5 mM MgCl2 were added. Pol III was then diluted to 1 mg/ml and incubated with 5x excess of transcription bubble for 1 h at 7 °C. The elongating Pol III complexes were then diluted 5 fold for grid freezing.
RNA extension assay
Labeling of the RNA and annealing was performed as previously described24 except that the full transcription bubble (see above) was used as template. An excess of transcription bubble was incubated with Pol III in EM-Buffer for 1 h at 4 °C. The RNA elongation was initiated by addition of NTPs in EM-Buffer with additional 10 mM MgCl2 at 28 °C for 20 min. The reaction was stopped by adding 1× 8 M urea loading buffer and subsequent heating to 95 °C for 5 min. The samples were analyzed on a denaturing polyacrylamide gel (17% PAGE, 7 M urea) using a FLA7000 phosphoimager (Fujifilm).
Electron microscopy
A total of 2.5 μl of 0.2 mg/ml Pol III and ~0.2 mg/ml elongating Pol III were applied to glow-discharged Quantifoil grids (400 mesh holey carbon 1.2/1.3 copper for elongating Pol III and molybdenum for apo Pol III) using an FEI Vitrobot at 95% humidity and 20 °C (Vitrobot Mark III, FEI Eindhoven, Netherlands). The sample was incubated for 15 s on the grid before blotting for 9 s (double-sided, blot force 3) and immediately flash-frozen in liquid ethane.
Micrographs were acquired at 300 kV using an FEI Titan Krios equipped with a Falcon II direct detector at a magnification of 75,000, corresponding to a pixel size of 1.084 Å. Image acquisition was performed with EPU Software (FEI Eindhoven, Netherlands) and micrographs were collected at an under-focus varying between 1.4 and 4.2 μm. We collected a total of seven frames accumulating to a dose of 42 e−/Å2 (apo Pol III) and 45.5 e−/Å2 (elongating Pol III) over 2.0 s exposure for apo Pol III and 1.6 s for elongating Pol III. In total, 2,398 and 2,639 micrographs were collected for elongating Pol III and for apo Pol III, respectively.
Image processing
The seven frame micrographs were aligned using MOTIONCORR50. The resulting frame stacks and total exposure images (7 frame sums) were used for further processing. The contrast-transfer function of the micrographs was determined using CTFFIND351 and Thon rings were manually inspected for astigmatism. Particle picking was performed on 20 Å low-pass filtered micrographs (Extended Data Fig. 1a). Initially, a subset of 100 micrographs was chosen from the elongating Pol III dataset and a total of 14,942 particles were selected manually using EMAN2 boxer swarm tool52. 2D classes from RELION28 obtained with the subset were used as templates for RELION’s autopicking procedure on the full datasets. The selected particles were sorted by the particle sorting routine implemented in RELION 1.3 and manually inspected. All further processing was performed with RELION 1.3 (Extended Data Fig. 1b). As starting models for 3D classification, we used the available 10 Å map of apo Pol III (EMDB-1804; apo Pol III) and the 16.5 Å map of elongating Pol III (EMDB-1803) low-pass filtered to 60 Å. For the elongating Pol III, the initial dataset of 800,000 particles was classified in 3D into 10 classes. Four classes were selected based on high particle abundance and occupancy of peripheral density at positions corresponding to the C82/C34/C31 and C53/C37 subcomplexes. The remaining 461,594 particles were subjected to a 3D sub-classification (for Pol III, 3D classification was initiated at this point with 515,397 particles). From 10 calculated classes, the most populated class with 121,241 particles for apo Pol III and 75,751 particles for elongating Pol III was further refined. We then employed RELION’s movie processing routine (including the ‘particle polishing’ step) using all seven frames. The procedure deblurs images by particle-based motion correction and minimizes effects of radiation damage by B-factor like amplitude weighting of the subframes53. With the particle-polished dataset, we performed another 3D classification using locally restrained alignment to resolve structural heterogeneity in the particle subset, which resulted in two classes with 68,818 and 52,423 particles for the apo Pol III data set and two classes at 49,543 and 26,208 particles for the elongating Pol III data set, respectively. Both apo Pol III classes were refined and finally, we obtained two reconstructions, class 1 (closed clamp apo Pol III) with a resolution of 4.6 Å and class 2 (open clamp apo Pol III) at 4.7 Å. For the elongating Pol III, class 1 showed improved alignment and yielded a map with increased resolvability compared to the 75,751 particle reconstruction, whereas class 2 showed no apparent conformational difference to class 1, but showed reduced resolvability, corresponding to less well aligned particles. Class 1 was subsequently refined to a resolution of 3.9 Å according to FSC 0.143 criterion54. In order to estimate resolution, we compensated mask effects by performing the high-resolution noise substitution procedure within RELION49 (Extended Data Fig. 1c). The resulting maps were post-processed in RELION by sharpening with an automatically calculated B-factor55 of −136 Å2 and −140 Å2 for class 1 and 2 of apo Pol III and −100 Å2 for elongating Pol III, respectively.
Local resolution was assessed using the tool RESMAP48 (Extended Data Fig. 1c). The elongating Pol III map shows local resolution beyond 3.5 Å in the core and generally a uniform distribution of resolution between 3.5-4.6 Å for all subunits except the C34 WH3 (Extended Data Fig. 1c, 2, Extended Data Table 2). Both apo Pol III maps show stronger resolution differences between the core and peripheral subunits due to mobility of the heterotrimer and the stalk.
Model building and localization of subunits in the elongating Pol III density
The building of initial models into the experimental density map was guided by homology models of Pol III subunits and crystal structures of archaeal RNA polymerase (PDB 4abc), Pol I (PDB 4c3i), Pol II (PDB 1wcm) as well as available crystal structures for the stalk (PDB 2ckz, 3ayh), the C82 homolog hRPC62 (PDB 2xub) and the TFIIF heterodimer (PDB 1f3u). All homology models were built using MODexplorer56, HHsearch57 and Modeller58. Homology models of subunits C160 and C128 were built based on the Pol II structure, C11 based on the A12.2 subunit of Pol I, C82 on hRPC62 (PDB 2xub) and C34 WH3 domain based on an unrelated WH domain of highest sequence similarity (PDB 1ldd). The homology model of the stalk (C17/C25 dimer) was built based on the structures of C17/C25 dimer from S. cerevisiae (PDB 2ckz) and the HRDC (helicase and RNaseD C-terminal) domain of C17 taking the domain orientations from S. pombe C17/C25 structure (PDB 3ayh) that improved the fit with the EM map.
Manual model building and local real-space refinement was performed in COOT59. For initial model building, the homology models were used to assemble a 15-subunit Pol III model consisting of all subunits except C34 and C31 which together with the above-mentioned crystal structures was placed into the elongating Pol III map by rigid body fitting. The fitted structures of individual subunits were used to aid de novo model building and re-building of reference models. Additionally, Xlink Analyzer60 was used to display available lysine-lysine crosslinks and photo-crosslinks16 on the Pol III model, which allowed us to assign several peripheral densities to subunits C31, C34, C37 and C53. Our current models of apo and elongating Pol III account for most features observed in the experimental EM density maps, except for a few short stretches of density where unambiguous assignment to a subunit was not possible and two strong density peaks observed in both apo Pol III reconstructions and located close to the active site. We speculate that these densities might arise from variations in sample preparation and correspond to bound nucleotides and metal ions, although one of them could also result from a conformational switch of the rudder. There are six established Zn2+ binding sites located in the Pol III core subunits C160 (2), C128 (1), C11 (1), ABC10β (1) and ABC10α (1). We accounted for metal binding at these sites where the experimental density sufficiently supported the placement of metal ions.
To identify the position of the C34 WH3 domain, the homology model was fitted into the full density map at 5.5 Å using a systematic global search with the ‘Fit In Map’ tool of UCSF Chimera61. The fitting was performed using 100,000 random initial placements. These fits were clustered (leading to 30,000 unique fits) and scored by the normalized cross-correlation coefficient with the density map. The statistical significance of the scores was assessed as previously described62. Out of the statistically significant fits, the best scoring fit located C34 WH3 into a stretch of density unassigned by any other subunit next to subunit C82. The other significant fits corresponded to regions occupied by WH domains of C82 thus validating the ability of the systematic fitting procedure to locate WH domains in the experimental density. Although the WH1 and WH2 domains of C34 also fitted to this unassigned region, the positioning of WH3 domain was validated by cross-links to C82 and Consurf63 that revealed a conserved surface of the WH3 domain towards C82 in contrast to C34 WH2 and WH1.
Refinement of elongating Pol III
To improve the model after initial model building, we performed automated real-space coordinate refinement against the elongating Pol III density map using a scripted workflow based on CCP4 and cctbx/PHENIX libraries64,65. The sharpened experimental density map of elongating Pol III was used as the refinement target. Due to the resolution differences in the map we initially performed refinement of sub-complexes or individual subunits against map segments filtered at the appropriate resolution, ranging from 3.9 – 4.5 Å (Extended Data Table 1). To better account for the differences in map resolution observed for different parts of the model, we calculated the local resolution distribution using RESMAP48. We then computed a local mean resolution for each subunit by averaging all RESMAP voxels contained in a low-pass filtered volume generated from the model coordinates of this subunit (Extended Data Table 2). Segments comprising all voxels including and extending 3.5 Å outwards of the model coordinates were then carved from the elongating Pol III density map. A uniform isotropic B-factor of 50 Å2 was assigned to all model atoms at the start of the refinement. The individual map segments and the corresponding coordinate models were centered in a cubic box of P1 symmetry with a cell dimension of 260.16 Å (240 × 240 × 240 pixels) to allow uniform grid sampling of model and experimental maps at the experimental pixel size. Coordinate refinement was performed by geometry-restrained real-space refinement based on gradient-driven minimization of a combined map and restraint target as implemented in cctbx/PHENIX up to the computed local mean resolution of each subunit. Grid searches were employed for automated identification of optimal refinement weights to balance the relative contribution of geometry and experimental restraints. Additional restraints were applied between hydrogen-bonded atoms in secondary structure elements. For the DNA/RNA of the transcription bubble, an initial model was obtained from the structure of a Pol II-Mediator complex37 (PDB 4v1n) and restraints were generated with the LibG program distributed with CCP4/REFMAC (Fei Long, unpublished). The geometries of Zn2+ binding sites in C160, C128, C11 and ABC10β and ABC10α were restrained to reference values according to Harding66,67.
Each round of model optimization was evaluated by computing the real-space cross-correlation (RSCC) between experimental map and a map calculated from the model coordinates. To this end, model maps were generated by calculating B-factor-weighted structure factors from the model coordinates (using electron atomic form factors68) and computing the inverse Fourier transform. We refined individual isotropic atomic displacement parameters (ADPs) by optimizing the real-space correlation between model and experimental map. ADPs were obtained by fitting to the computed RSCC profile. The resulting B-factor distributions correlate well with local resolution estimates, with lowest B-factors in the Pol III core and higher B-factors in the peripheral subunits of the stalk (C17/C25) and the C53/C37 and C82/C34/C31 sub-complexes (Extended Data Fig. 4).
Automated real-space refinement cycles were alternated with manual model building in COOT59. Manual model building was aided by experimental maps filtered at lower resolution and experimental maps that were re-sharpened by applying the structure factor amplitudes computed from the current model55,69, which leads to decreased noise levels and increased visibility of structural features in some regions of the map, in particular in the peripheral Pol III subunits. Taking into account the refined B-factor distribution in the model, this map manipulation enhances structural features due to better correction of the Fourier amplitude fall-off resulting from experimental factors, but does not introduce model bias since experimental phases are used.
Following initial refinement of all individual subunits against the corresponding map segments, the entire elongating 17-subunit Pol III complex and the DNA/RNA transcription bubble was assembled and subjected to additional refinement against the full map reconstruction filtered at 3.9 Å resolution to account for inter-subunit interactions. The DNA/RNA model was not further refined at this stage. Since refinement target weights are optimized based on overall map resolution, restraint weighting can be suboptimal in regions of the map with a resolution lower than the average map resolution. To compensate for this effect in the full map refinement, we restrained peripheral subunits by an additional harmonic potential applied to the Cα positions, which was scaled relative to the average local resolution in this map region calculated as described above.
The Fourier shell correlation between model map and half-set 3D reconstructions was used to assess the possibility of overfitting70. Briefly, the atoms in the model were randomly displaced by up to a maximum of 0.5 Å, followed by five cycles of real-space refinement against one of the half maps (work map) using the same protocol as described above. We then computed the FSC between the resulting model and the work map (FSCwork) as well as the cross-validated FSC between the refined model and the other half map not used in refinement (FSCtest). The close agreement between both curves for the elongating Pol III model indicates that no overfitting took place (Extended Data Fig. 4). We attribute the minor discrepancies in particular at low-resolution between FSCwork/FSCtest and the FSC between two independent half sets (Extended Data Fig. 1) to the incomplete model and solvent effects55,71. The very good correlation in the high-resolution region beyond 6 Å provides support that our model accounts well for most of the defined density features observed in the experimental map. The quality of the final model was validated using MOLPROBITY72 and was found to range in the top percentiles for the corresponding resolution range.
Model Building and refinement of apo Pol III (closed/open clamp conformation)
The initial model for apo Pol III in the closed and open clamp conformations was obtained from the model coordinates of the individual subunits refined against the higher resolution map of elongating Pol III. The 10-subunit core, the C53/C37 heterodimer, the C82/C34/C31 heterotrimer and the stalk (C25/C17) were placed by rigid body fitting into the respective density maps. The C-terminal TFIIS-like domain of C11, not visible in the elongating Pol III structure, was added to complete the model. The complete model was then refined against the respective maps filtered at 4.6/4.7 Å resolution. Sharpened maps for both ‘closed’ and ‘open’ clamp conformations of apo Pol III were used as the refinement target. Restraints on secondary structure and metal binding geometries were implemented as described for elongating Pol III. Automatic real-space refinement was performed as described above, with additional harmonic restraints on Cα positions weighted by the average local resolution to account for resolution anisotropy observed in both maps. Manual adjustments to the model were done in COOT; model assessment and validation were performed as described above. A summary of the refinement statistics for all three models can be found in Extended Data Table 1.
Figure Preparation
Graphs and figures were prepared using XMGrace, Pymol73 and UCSF Chimera61.
Extended Data
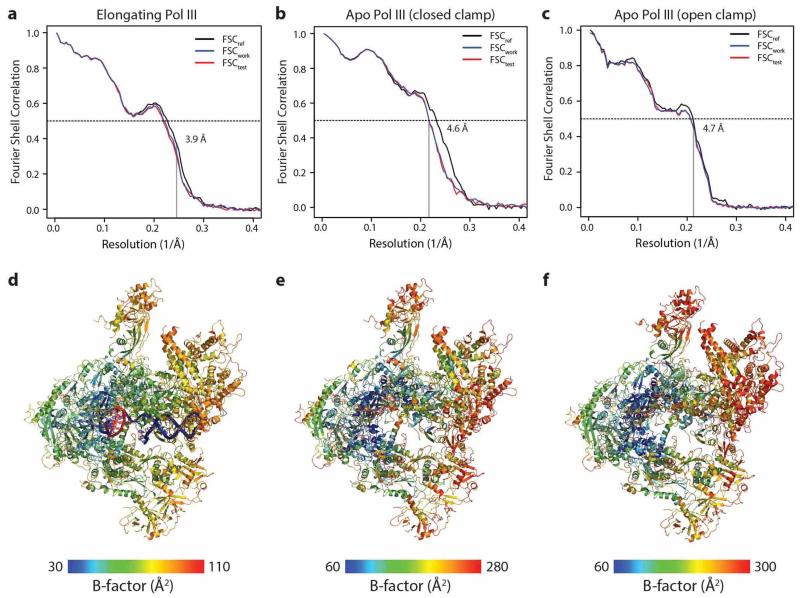
Extended Data Figure 1. Pol III processing pipeline, Fourier-shell correlation (FSC) curves and local resolution assessment.
a, Exemplary micrograph section of elongating Pol III. All micrographs were low-pass filtered for particle picking. b, General processing pipeline. The orange boxes display micrograph number and particles for elongating Pol III (left) and apo Pol III (right). The middle panel shows the general workflow that was followed for both datasets. For elongating Pol III (bottom left), a local classification step yielded one class with 49 k particles (purple) that was subsequently refined and post-processed. For apo Pol III (bottom right), local classification diverged into two classes (purple) with 69 k particles and 52 k particles that were subsequently refined and post-processed. c, Fourier-shell correlation (FSC) and local resolution assessment with RESMAP48. All FSC calculations were performed with two independent half maps using RELION’s masking procedure49. The resolution for the elongating Pol III cryo-EM map (top panel) is 3.9 Å according to the FSC 0.143 criterion, indicated by the black dashed line. The two apo Pol III cryo-EM reconstructions have a resolution of 4.6 Å (closed clamp Pol III, middle panel) and 4.7 Å (open clamp Pol III, bottom panel) according to the FSC 0.143 criterion. Local resolution is displayed on the post-processed full maps (first image column on the right) and a cross-section representation (second image column on the right). In both apo Pol III reconstructions, the peripheral subcomplexes show a strong decay in resolution compared to the core. In the elongating Pol III reconstruction, the resolution is more uniformly distributed indicating stabilization of peripheral subunits.
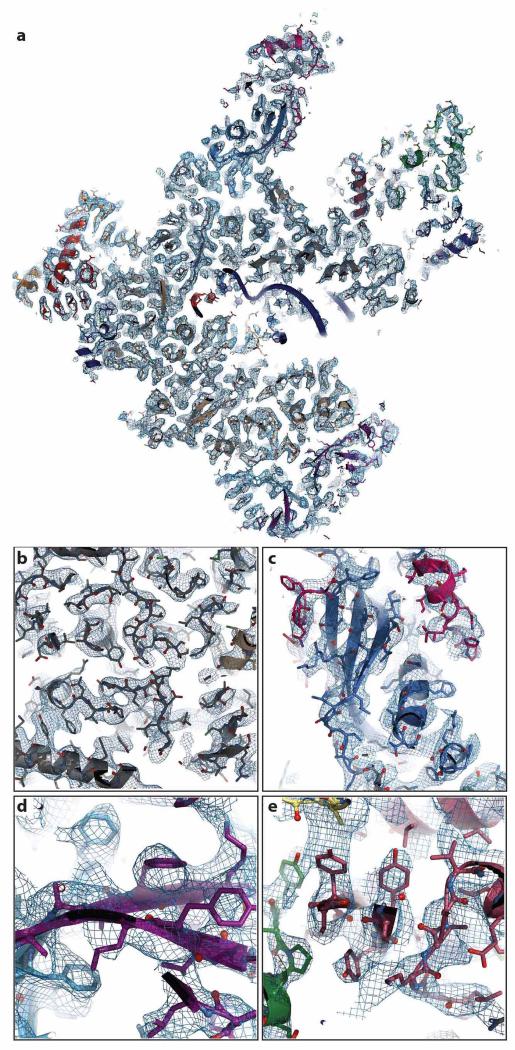
Extended Data Figure 2. Representative sections of the cryo-EM density for elongating Pol III.
a, Cross-section of elongating Pol III in ribbon and stick representation, embedded in the experimental density at 3.9 Å, displayed in tungsten. b, Section displaying the core subunits C160 (grey) and C128 (wheat) shown in stick and ribbon representation. The experimental density of the core (tungsten) is well defined and has been filtered at 3.5 Å resolution for display. c, Section of stalk subunits C25 (blue) and C17 (pink). The estimated local resolution in this part is lower compared to the core (Extended Data Table 2). In panel c, d and e, the cryo-EM density is shown at 3.9 Å resolution. d, Section showing subunits C53 (purple) and C37 (lanthanum). e, Close-up view of C82-WH1 (brown), C82–WH2 (green) and C31 (yellow) interface.
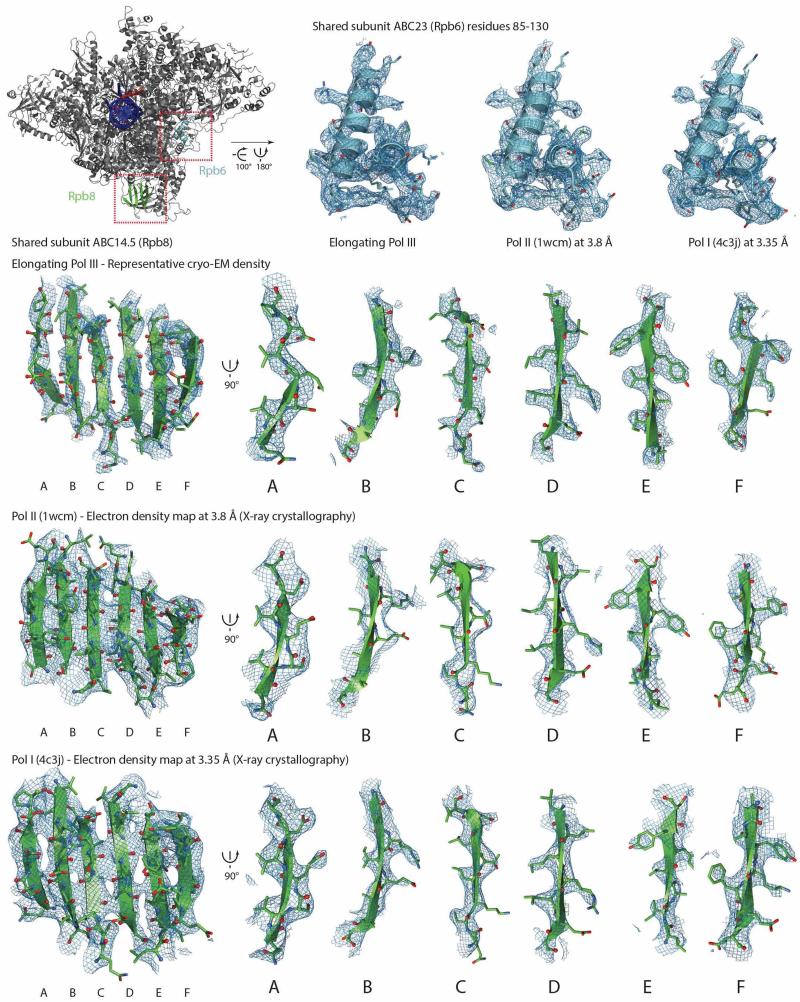
Extended Data Figure 3. Comparison of electron microscopy densities with X-ray electron densities for shared subunits ABC23 (Rpb6) and ABC14.5 (Rpb8).
Top left shows Pol III in front view, a stretch in ABC23 (cyan) and ABC14.5 (green) is colored. The red boxes indicate the regions that are enlarged in the neighboring panels. Corresponding density is displayed in tungsten. Models of Pol II and Pol I at nominally higher resolution are available, but for better comparison models in a similar resolution range are shown. For the 2Fo-Fc electron density maps obtained by X-ray crystallography a threshold of 1σ was used for display. The top right shows three close-up views of the shared subunit ABC23 from elongating Pol III, Pol II (PDB 1wcm) and Pol I (PDB 4c3j). The bottom panels show 6 strands of shared subunit ABC14.5. Front view of the β- sheet and orthogonal views of individual strands in elongating Pol III, Pol II (PDB 1wcm) and Pol I (PDB 4c3j).
Extended Data Figure 4. Model validation and temperature factor distribution of atomic models.
a–c, Fourier shell correlation (FSC) curves calculated between the refined atomic model and the half map used in refinement (FSCwork) are shown in blue, those calculated between the refined atomic model and the second half map not used for refinement (FSCtest) in red. Vertical lines mark the regular FSC 0.143 cutoff and the resolution target used in refinement as shown. Close agreement between FSCwork and FSCtest and the absence of a sharp drop beyond the refinement target resolution indicate that no overfitting took place. The respective FSC between the refined atomic model and the map obtained from 3D reconstruction using the entire dataset (FSCref) is also shown (black). d–e, Atomic B-factor distributions mapped on ribbon representations of elongating and apo Pol III. The overall distribution and relative differences between core and peripheral subunits for the different models correlate well with the distribution of local resolution (Extended Data Fig. 1).
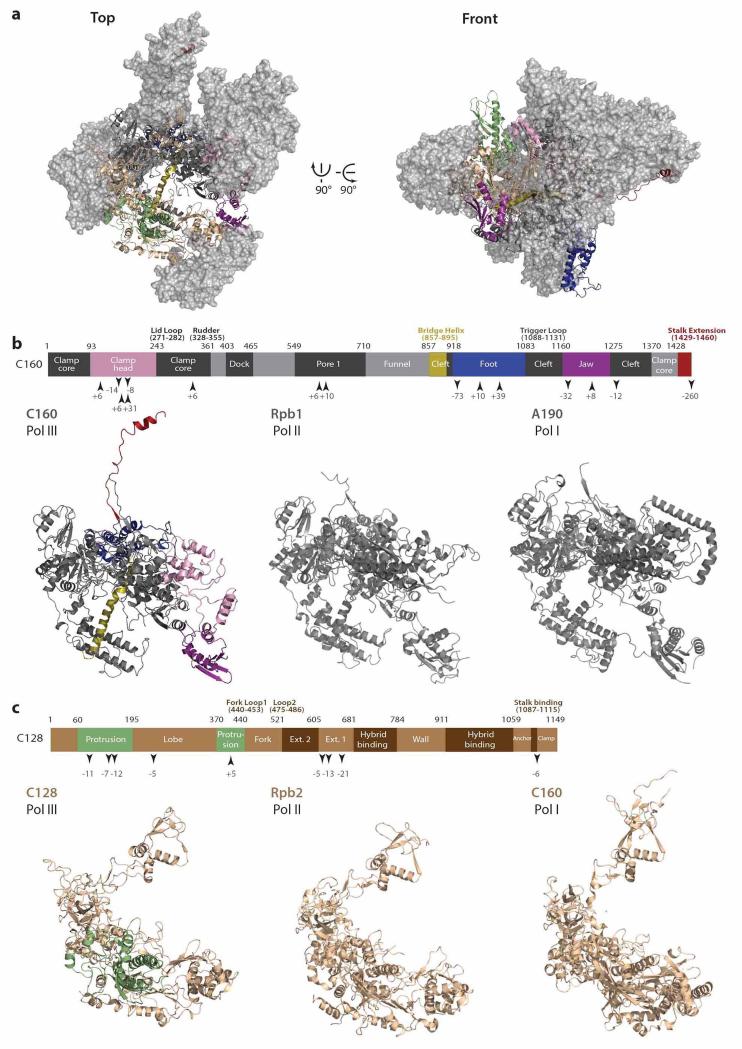
Extended Data Figure 5. Pol III-specific features of subunits C160 and C128 and comparison to the homologous Pol II and Pol I subunits.
a, Top (left) and front (right) view of Pol III, with subunits C160 and C128 displayed in ribbon representation and the other subunits in surface representation (grey). Colored stretches highlight characteristic features denoted in b. b, Bar diagram shows the domain organization of Pol III C160. Arrows and corresponding numbers below the bar diagram indicate insertions and deletions of five or more residues in Pol III relative to Pol II subunit Rpb1 as indicated by structure-based alignment. Colored regions are also shown in Pol III subunit C160 (lower panel, left) and in a. Lower panel middle and right show Pol II Rpb1 and Pol I A190 subunits, respectively. c, Same as in b for Pol III subunit C128. Insertions and deletions compared to Pol II subunit Rpb2 are displayed in the box diagram.
Extended Data Figure 6. Open and closed clamp conformation in Pol III compared to other RNA polymerases.
a, Top view of aligned elongating Pol III and apo Pol III (closed clamp - left panel; open clamp – middle panel) and both ’closed clamp‘ and ’open clamp‘ apo Pol III conformations (right panel). RMSD values (core-heterodimer:all) for elongating Pol III – apo Pol III (closed clamp) (0.43 Å3490 CA:0.43 Å4813 CA), elongating Pol III – apo Pol III (open clamp) (0.71 Å3496 CA:2.73 Å4795 CA) and both apo Pol III open and closed clamp (0.71 Å3540 CA:2.71 Å4829 CA) demonstrate the similarity between ’closed clamp‘ apo Pol III and elongating Pol III conformations. b, Schematic representation of Pol III in top view showing the conformational changes of clamp head, heterotrimer and stalk. The closed clamp conformation (elongating Pol III and ’closed clamp‘ apo Pol III) is displayed in red, the open clamp conformation (‘open clamp‘ apo Pol III) in green. The DNA/RNA duplex is shown in blue, the core and heterodimer in grey. c, Front view on open and closed clamp conformations in other RNA polymerases. The closed clamp state (green) and open clamp state (red) is indicated for archaeal polymerase (left panel, PDB 4ayb and 4qiw), for Pol II (middle panel, PDB 1wcm and 1twf) and for Pol III (right panel). Green and red angles describe the cleft opening in the closed and open clamp conformations. Black arrows and corresponding values indicate the relative distance of the subunits between the two conformations. d, Front view of apo Pol III closed cleft (left panel), apo Pol III closed cleft vs apo Pol II (middle panel, Pol II (PDB 1wcm) in red) and apo Pol III closed cleft vs apo Pol I (right panel, Pol I (PDB 4c3i) in blue). The cleft opening is indicated by a dashed line and the Cα-Cα distance across the cleft (black for Pol III, red for Pol II and blue for Pol I). However, some of the observed differences in cleft width between Pol I, Pol II and Pol III might also reflect differences between conditions of cryo-EM and crystal structures as well as different packing contacts in the crystals.
Extended Data Figure 7. Pol III-specific subunits C82, C31 and C11.
a, Left panel: Overall surface representation of Pol III with the C82/C34/C31 heterotrimer in ribbon representation. Right panels: Two enlarged and orthogonal views of the region marked with a dotted black square. In subunit C82 WH4 inserts in the DNA binding cleft passing through a canyon in the clamp head. WH2 and WH3 extensions reach over the clamp head and are positioned in close proximity to downstream DNA. b, Ribbon model of Pol III fitted into the EM density of the ‘open clamp’ apo Pol III filtered at 6 Å resolution. For C31, additional density is visible in the cavity between the stalk and the heterotrimer, as shown in the top right panel. The described densities are also present in the ‘closed clamp‘ apo Pol III and the elongating Pol III reconstructions. No attempts were made to fit atomic models into these densities. c, EM density of the C11 TFIIS-like domain at 6 Å resolution as observed in the ‘open clamp’ apo Pol III reconstruction. The left panel shows a side view of Pol III, the middle and right panels show close-ups at two different density thresholds.
Extended Data Figure 8. C53/C37 heterodimer and stalk subunits C17/C25.
a, Visualization of the photo-crosslinks between C53/C37 heterodimer and subunit C128. Pol III is shown in surface, C53/C37 and C11 in ribbon representation. In addition, the C128 lobe is shown in cartoon representation (small inset). Purple spheres on the lobe indicate residues that photo-crosslink to C3713, beige spheres on C37 indicate residues that photo-crosslink to C12813. The dashed line marks the tentative path of the non-template DNA strand. The experimental photo-crosslinks fit well to the cryo-EM structure. The C37 loop is disordered between Glu196 and Asn225, although photo-crosslinks indicate that this region is in close proximity to the lobe and the non-template DNA strand. b, Bottom view of Pol III in surface representation, with C53/C37 and C11 shown in ribbon representation. The black dotted square indicates the enlarged area in the center of the image (small inset). The red density (shown at 4.5 Å) was not of sufficient quality to build an atomic model. However, photo-crosslinks from C37 and C128 to C53 (blue spheres on C37 ribbon and C128 surface mark crosslink positions)13 indicate that C53 N-terminal residues are located in this region. c, Stalk anchoring with C160 extensions. Top view (left panel) and bottom view (right panel) of the stalk subunits C17 (magenta), C25 (blue) and the C160 extensions (grey). EM density corresponding to the C160 N- and C-terminal extensions is shown in tungsten blue. Individual entities and subunits are labeled.
Extended Data Table 1. Refinement statistics.
| Elongating Pol III | Apo Pol III (closed clamp) | Apo Pol III (open clamp) | |
|---|---|---|---|
|
| |||
| Model composition | |||
| No. of chains | 17+3 | 17 | 17 |
| Non-hydrogen atoms | 39276 | 38677 | 38427 |
| Protein residues | 4839 | 4882 | 4845 |
| Nucleic acid | 47 | -- | -- |
| Ligand (Zn2+) | 6 | 6 | 6 |
|
| |||
| Refinement | |||
| PDB ID | 5fj8 | 5fj9 | 5fja |
| Resolution (Å) | 260.2-3.9 | 260.2-4.6 | 260.2-4.7 |
| Map sharpening B-factor (Å2) | −100 | −136 | −140 |
| Average B-factor (Å2) | |||
| Protein | 64.9 | 161.8 | 182.0 |
| Nucleic acid | 76.3 | -- | -- |
| Ligand (Zn2+) | 58.6 | 62.4 | 107.9 |
| Molprobity score | 2.6 | 2.58 | 2.48 |
| Clashscore (all atoms) | 14.5 | 13.41 | 13.84 |
| Rotamer outliers (%) | 2.00 | 2.08 | 1.5 |
| Ramachandran statistics | |||
| Favored (%) | 82.46 | 82.52 | 82.58 |
| Disallowed (%) | 1.15 | 1.23 | 1.03 |
| RMS (bonds, Å) | 0.0032 | 0.0032 | 0.0030 |
| RMS (angles, °) | 1.03 | 1.01 | 0.89 |
| Nucleic Acid | |||
| Correct sugar puckers (%) | 91.5 | -- | -- |
| Good backbone conform. (%) | 68.7 | -- | -- |
Extended Data Table 2. Model statistics for elongating Pol III.
| Subunit | Protein | Chain ID | Mw (kDa) | No. of residues | Residues built | Chain breaks | All-atom clashscore | Molprobity score | Average B-factor (Å2) | Local resolution (Å) |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Core | ||||||||||
| RPC1 | C160 | A | 162.3 | 1460 | 1422 (97.4%) | 3 | 10.42 | 2.24 | 56.35 | 3.8 |
| RPC2 | C128 | B | 129.5 | 1149 | 1115 (97.0%) | -- | 12.61 | 2.40 | 60.10 | 3.8 |
| RPC40 | AC40 | C | 37.7 | 335 | 335 (100%) | -- | 10.60 | 2.40 | 60.42 | 3.8 |
| RBP5 | ABC27 | E | 25.1 | 215 | 215 (100%) | -- | 16.07 | 2.81 | 67.97 | 4.1 |
| RBP6 | ABC23 | F | 17.9 | 155 | 83 (53.5%) | -- | 3.67 | 2.34 | 57.97 | 3.6 |
| RBP8 | ABC14.5 | H | 16.5 | 146 | 140 (95.9%) | 1 | 9.05 | 2.15 | 62.64 | 4.2 |
| RPC11 | C11 | I | 12.5 | 110 | 43 (39.1%) | -- | 16.00 | 2.73 | 73.40 | 4.3 |
| RBP10 | ABC10β | J | 8.3 | 70 | 68 (97%) | -- | 5.40 | 2.39 | 56.79 | 3.6 |
| RPC19 | AC19 | K | 16.1 | 142 | 101 (71.1%) | -- | 8.85 | 2.41 | 59.38 | 3.7 |
| RPC10 | ABC10α | L | 7.2 | 70 | 46 (65.7%) | -- | 12.13 | 2.65 | 67.80 | 4.2 |
| Stalk | ||||||||||
| RPC9 | C17 | D | 18.6 | 161 | 119 (73.9%) | 2 | 15.82 | 2.51 | 73.39 | 4.4 |
| RPC8 | C25 | G | 24.3 | 212 | 191 (90.1%) | 2 | 16.86 | 2.67 | 70.03 | 4.3 |
| Heterotrimer | ||||||||||
| RPC3 | C82 | O | 74.0 | 654 | 539 (82.4%) | 2 | 15.99 | 2.86 | 73.08 | 4.5 |
| RPC6 | C34 | P | 36.1 | 317 | 89 (28.1%) | 2 | 26.41 | 3.54 | 75.84 | 5.3 |
| RPC7 | C31 | Q | 27.7 | 251 | 63 (25.1%) | 2 | 10.13 | 2.13 | 76.61 | 4.6 |
| Heterodimer | ||||||||||
| RPC5 | C37 | M | 32.1 | 282 | 164 (58.2%) | 1 | 11.72 | 2.59 | 74.18 | 4.3 |
| RPC4 | C53 | N | 46.7 | 422 | 110 (26.1%) | 1 | 12.10 | 2.98 | 71.51 | 4.2 |
| RNA | RNA | R | 18 | 9 (50.0%) | -- | 75.00 | 3.9 | |||
| DNA | Non-Template | S | 38 | 15 (39.5%) | -- | 75.91 | 3.9 | |||
| DNA | Template | T | 38 | 23 (60.5%) | -- | 78.03 | 3.9 | |||
ACKNOWLEDGEMENTS
We thank Y. Sadian, H. Grötsch and R. Wetzel for help in Pol III purification. We are grateful to F. Thommen and M. Wahlers for setup and maintenance of the high-performance computational environment of RELION. We acknowledge F. Schur, S. Fromm, C. Bertipaglia, M. Beck and G. Hofhaus for helpful advice regarding sample preparation and image processing. We also thank the ‘Fermentation et culture de microorganisms’ (IFR88, CNRS, Marseille). M.M.-M. and A.J.J. were supported by Marie-Sklodowska-Curie fellowships (FP7-PEOPLE-2011IEF301002, PIEF-GA-2012-331285). N.A.H. acknowledges support by the EMBL International PhD program, A.J.J. and J.K. by postdoctoral fellowships from the EMBL Interdisciplinary Postdoc Program (EIPOD) under Marie Curie COFUND actions (PCOFUND-GA-2008-229597) and C.W.M. by an ERC Advanced Grant (ERC-2013-AdG340964-POL1PIC).
Footnotes
AUTHOR INFORMATION
The 3.9 Å cryo-EM map of elongating Pol III and the two maps of apo Pol III have been deposited in the Electron Microscopy Data Bank with accession codes EMD-3178 (elongating Pol III) and EMD-3179 (‘closed clamp’ apo Pol III) EMD-3180 (‘open clamp’ apo Pol III). The coordinates of the corresponding atomic models have been deposited in the Protein Data Bank under accession code 5fj8 (elongating Pol III) and 5fj9 (‘closed clamp’ apo Pol III), 5fja (‘open clamp’ apo Pol III).
The authors declare no competing financial interests.
REFERENCES
- 1.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 2.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 4.Arimbasseri AG, Maraia RJ. Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werner M, Thuriaux P, Soutourina J. Structure-function analysis of RNA polymerases I and III. Curr Opin Struct Biol. 2009;19:740–745. doi: 10.1016/j.sbi.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Jasiak AJ, Armache KJ, Martens B, Jansen RP, Cramer P. Structural biology of RNA polymerase III: subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol Cell. 2006;23:71–81. doi: 10.1016/j.molcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J Biol Chem. 2010;285:2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landrieux E, et al. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter R, Drouin G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol. 2010;27:1035–1043. doi: 10.1093/molbev/msp316. [DOI] [PubMed] [Google Scholar]
- 11.Geiger SR, et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell. 2010;39:583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Tornero C, et al. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol Cell. 2007;25:813–823. doi: 10.1016/j.molcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Wu CC, Lin YC, Chen HT. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol. 2011;31:2715–2728. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane LA, et al. Mass spectrometry reveals stable modules in holo and apo RNA polymerases I and III. Structure. 2011;19:90–100. doi: 10.1016/j.str.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Roeder RG. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes & Development. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 16.Wu CC, et al. RNA polymerase III subunit architecture and implications for open promoter complex formation. Proc Natl Acad Sci U S A. 2012;109:19232–19237. doi: 10.1073/pnas.1211665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thuillier V, Stettler S, Sentenac A, Thuriaux P, Werner M. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995;14:351–359. doi: 10.1002/j.1460-2075.1995.tb07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khoo SK, Wu CC, Lin YC, Lee JC, Chen HT. Mapping the protein interaction network for TFIIB-related factor Brf1 in the RNA polymerase III preinitiation complex. Mol Cell Biol. 2014;34:551–559. doi: 10.1128/MCB.00910-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boguta M. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta. 2013;1829:376–384. doi: 10.1016/j.bbagrm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Vannini A, et al. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Sainsbury S, Bernecky C, Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:129–143. doi: 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- 23.Svetlov V, Nudler E. Basic mechanism of transcription by RNA polymerase II. Biochim Biophys Acta. 2013;1829:20–28. doi: 10.1016/j.bbagrm.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Tornero C, et al. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502:644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- 25.Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Tornero C, et al. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J. 2010;29:3762–3772. doi: 10.1038/emboj.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Morcillo M, et al. Solving the RNA polymerase I structural puzzle. Acta Crystallogr D Biol Crystallogr. 2014;70:2570–2582. doi: 10.1107/S1399004714015788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes CO, et al. Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble. Mol Cell. 2015;59:258–269. doi: 10.1016/j.molcel.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 31.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 32.Lefevre S, et al. Structure-function analysis of hRPC62 provides insights into RNA polymerase III transcription initiation. Nat Struct Mol Biol. 2011;18:352–358. doi: 10.1038/nsmb.1996. [DOI] [PubMed] [Google Scholar]
- 33.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 34.He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 36.Ferri ML, et al. A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol Cell Biol. 2000;20:488–495. doi: 10.1128/mcb.20.2.488-495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plaschka C, et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- 38.Arimbasseri AG, Rijal K, Maraia RJ. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription. 2014;5:e27639. doi: 10.4161/trns.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu HL, Wu CC, Lee JC, Chen HT. A region of Bdp1 necessary for transcription initiation that is located within the RNA polymerase III active site cleft. Mol Cell Biol. 2015 doi: 10.1128/MCB.00263-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rijal K, Maraia RJ. RNA polymerase III mutants in TFIIFalpha-like C37 that cause terminator readthrough with no decrease in transcription output. Nucleic Acids Res. 2013;41:139–155. doi: 10.1093/nar/gks985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaaban SA, Krupp BM, Hall BD. Termination-altering mutations in the second-largest subunit of yeast RNA polymerase III. Mol Cell Biol. 1995;15:1467–1478. doi: 10.1128/mcb.15.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitehall SK, Bardeleben C, Kassavetis GA. Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J Biol Chem. 1994;269:2299–2306. [PubMed] [Google Scholar]
- 44.Iben JR, et al. Point mutations in the Rpb9-homologous domain of Rpc11 that impair transcription termination by RNA polymerase III. Nucleic Acids Res. 2011;39:6100–6113. doi: 10.1093/nar/gkr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehara H, Sekine S, Yokoyama S. Crystal structure of the C17/25 subcomplex from Schizosaccharomyces pombe RNA polymerase III. Protein Sci. 2011;20:1558–1565. doi: 10.1002/pro.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakraborty A, et al. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jun SH, et al. The X-ray crystal structure of the euryarchaeal RNA polymerase in an open-clamp configuration. Nat Commun. 2014;5:5132. doi: 10.1038/ncomms6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 52.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 53.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 55.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Kosinski J, Barbato A, Tramontano A. MODexplorer: an integrated tool for exploring protein sequence, structure and function relationships. Bioinformatics. 2013;29:953–954. doi: 10.1093/bioinformatics/btt062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 58.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Kosinski J, et al. Xlink Analyzer: software for analysis and visualization of cross-linking data in the context of three-dimensional structures. J Struct Biol. 2015;189:177–183. doi: 10.1016/j.jsb.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Bui KH, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 63.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harding MM. Geometry of metal-ligand interactions in proteins. Acta Crystallogr D Biol Crystallogr. 2001;57:401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 67.Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- 68.Colliex C, et al. International Tables of Crystallography C. International Union of Crystallography; 2006. pp. 259–429. 2006. [Google Scholar]
- 69.Sachse C, et al. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J Mol Biol. 2007;371:812–835. doi: 10.1016/j.jmb.2007.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiMaio F, Zhang J, Chiu W, Baker D. Cryo-EM model validation using independent map reconstructions. Protein Sci. 2013;22:865–868. doi: 10.1002/pro.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang JS, Brunger AT. Protein hydration observed by X-ray diffraction. Solvation properties of penicillopepsin and neuraminidase crystal structures. J Mol Biol. 1994;243:100–115. doi: 10.1006/jmbi.1994.1633. [DOI] [PubMed] [Google Scholar]
- 72.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]