To the Editor
Multidrug-resistant tuberculosis (MDR-TB) and extensively drug-resistant tuberculosis (XDR-TB) are an increasing public health threat.1 Bedaquiline and delamanid are two drugs that were recently approved by the Food and Drug Administration for treatment of MDR-TB and XDR-TB.2 Here we describe the stepwise amplification of drug resistance in a patient who had emigrated from Tibet to Switzerland in December 2010 and who presented to a Swiss hospital with preextensively drug-resistant tuberculosis at that time.
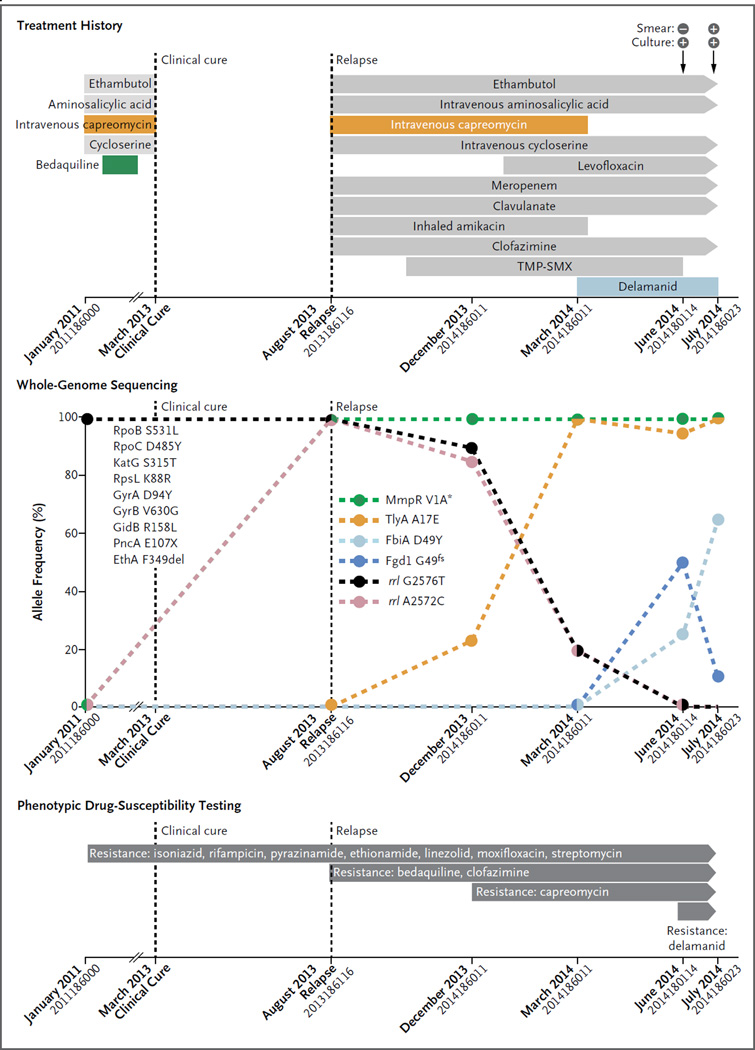
Genome sequencing revealed that the initial Mycobacterium tuberculosis isolate harbored nine mutations in genes associated with resistance to seven drugs (Fig. 1, and the Supplementary Appendix, available with the full text of this letter at NEJM.org). The isolate also showed a compensatory mutation in rpoC,3 indicating a “mature” preextensively drug-resistant strain that had evolved under drug pressure for some time. Given that the patient reported no previous treatment for tuberculosis, he was probably infected with a strain that was already resistant to these drugs.
Figure 1. Clinical Features, Treatment History, Amplification of Drug Resistance, and Phenotypic Drug-Susceptibility Testing in the Patient.
Mutations that confer drug resistance are shown. Resistance to bedaquiline, intravenous capreomycin, and delamanid developed. Percentages on the y axis are based on the number of genome-sequencing reads supporting the corresponding drug resistance–conferring mutation (details are provided in Table S4 in the Supplementary Appendix). Large colored circles indicate measurements, and dashed lines linking these circles indicate the changes in mutation frequency. The loss of the start codon is indicated with an asterisk, and fs indicates frame shift. The mutations listed on the left side of the graph were detected in the first and all subsequent isolates obtained from the patient. All these mutations were seen in 100% of sequencing reads in each isolate. X indicates the stop codon, and del indicates deletion. On the x axis, isolate numbers are listed under the month when the isolate was obtained. TMP-SMX denotes trimethoprim–sulfamethoxazole.
In September 2011, bedaquiline was added to the regimen, which had consisted of four drugs (ethambutol, aminosalicylic acid, intravenous capreomycin, and cycloserine). The patient was considered to be clinically cured in March 2013, but he had a relapse in August 2013. Genome sequencing of five follow-up isolates revealed a mutation in mmpR that was associated with bedaquiline resistance. This mutation persisted even though bedaquiline was discontinued in February 2012; this suggests that it did not cause any clinically significant reduction in the virulence of the infecting bacteria.4
Additional resistance to second-line injectable agents (such as capreomycin) also developed. This was reflected in the emergence of mutations in tlyA and rrs. The latter mutation remained at low frequency and was detected only in drug-containing bacterial cultures (Table S3 in the Supplementary Appendix). All the molecular findings were supported by phenotypic drug-susceptibility testing.
Following this amplification in resistance, delamanid was added to the regimen in March 2014. However, two mutations in fbiA and fgd1 increased in frequency by June 2014, which coincided with the emergence of phenotypic resistance to delamanid. These genes have previously been associated with delamanid resistance. The fgd1 mutation decreased in frequency thereafter, indicating the presence of multiple delamanid-resistant clones in the patient. In August and September 2014, the patient underwent a lobectomy. After surgery, the sputum specimens obtained from the patient were culture-negative, and the patient received treatment on an ambulatory basis.
This case highlights the development of resistance in the context of inadequate MDR-TB and XDR-TB treatment regimens, despite personalized patient care in a well-resourced health care setting. It serves as a warning for the future rollout of new antituberculosis drugs and emphasizes the need for the use of appropriate companion drugs when bedaquiline and delamanid are administered. Our results add to previous findings showing that the development of drug resistance is a dynamic process involving multiple heterogeneous populations of bacteria within individual patients.4,5
Supplementary Material
Acknowledgments
Supported by the Swiss Federal Office of Public Health, the University of Zurich, SystemsX.ch, the Novartis Foundation, and grants from the Swiss National Science Foundation (PP00P3_150750), the National Institutes of Health (AI090928), and the European Research Council (309540-EVODRTB).
Footnotes
Contributor Information
Guido V. Bloemberg, University of Zurich, Zurich, Switzerland
Sebastien Gagneux, Email: sebastien.gagneux@unibas.ch, Swiss Tropical and Public Health Institute, Basel, Switzerland.
Erik C. Böttger, University of Zurich, Zurich, Switzerland
References
- 1.Geneva: World Health Organization; 2014. Global tuberculosis control — surveillance, planning, financing. [Google Scholar]
- 2.Zumla A, Memish ZA, Maeurer M, et al. Emerging novel and antimicrobial-resistant respiratory tract infections: new drug development and therapeutic options. Lancet Infect Dis. 2014;14:1136–1149. doi: 10.1016/S1473-3099(14)70828-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comas I, Borrell S, Roetzer A, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shcherbakov D, Akbergenov R, Matt T, Sander P, Andersson DI, Böttger EC. Directed mutagenesis of Mycobacterium smegmatis 16S rRNA to reconstruct the in vivo evolution of aminoglycoside resistance in Mycobacterium tuberculosis. Mol Microbiol. 2010;77:830–840. doi: 10.1111/j.1365-2958.2010.07218.x. [DOI] [PubMed] [Google Scholar]
- 5.Sun G, Luo T, Yang C, et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J Infect Dis. 2012;206:1724–1733. doi: 10.1093/infdis/jis601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.