We would like to call your attention to a publication by Martin et al. (“Claudin-20 promotes an aggressive phenotype in human breast cancer cells”)1 in Tissue Barriers. The authors show that high claudin-20 expression correlates with the aggressiveness of breast cancer and poor survival rates for affected patients. They discuss a connection between the sensitivity of cancer cells to Gefitinib and claudin-20 content and suggest that claudin-20 might serve as a biomarker for Gefitinib resistance. However, this conclusion is based on a misconception on the part of the authors. They claim that claudin-20 is the same protein as epithelial membrane protein 1 (EMP-1), and to substantiate this claim, they misleadingly refer to a prior publication demonstrating that EMP-1 is a biomarker for Gefitinib-resistance in an adenocarcinoma model.2
However, Claudin-20 and EMP-1 are 2 distinct proteins. EMP-1 (NCBI reference sequence accession number NP_001414.1) has been described as a tight junction protein in the blood-brain barrier3 as well as in the human airway epithelium, where it regulates tight junction formation.4 To our knowledge, the paper of Martin et al.1 is the first publication that presents functional data regarding human claudin-20 (NP_001001346.1). Their paper will be immediately found (and likely cited) by other scientists working with either of the 2 proteins. Therefore, it is crucial to avoid misrepresentations through a careful review of the existing literature.
According to Price et al.5 (also cited by Martin and colleagues1) both claudin-20 and EMP-1 belong to the Pfam00822 superfamily, which includes claudins, peripheral myelin protein 22 (PMP-22), lens intrinsic membrane protein (LIM-2/MP20), EMP-1, −2, −3 and voltage dependent calcium channel gamma subunits (CACNGs). The study places claudin-20 within the claudin family (where it is most closely related to the classic claudins 2 and 14), while EMP-1 is placed in the LIM2/PMP22/EMP family.5 This classification was confirmed in an independent investigation, which came to the same results and confirmed the separate identities of claudin-20 and EMP-1.6
Nevertheless, the phylogeny of the Pfam00822 family has not yet been completely resolved. TMEM114 and TMEM235 were first assigned to the claudin family using PMP-22 as an outgroup.7 Later, these TMEMs were renamed claudin-26 and −27, respectively.8 Subsequent work that also included CACNGs, EMPs and LIM-2 and used clarin-1 as an outgroup revealed that TMEM114 and TMEM235 were more closely related to the CACNG family than with the claudin family.6 Thus, Pfam00822 superfamily nomenclature has not yet been conclusively determined.
The members of this superfamily are tetraspanning membrane proteins and share a WX9–44GLWXXC(X9–26C) signature motif in the first extracellular loop (whereas the second cysteine is not conserved in the LIM2/PMP22/EMP family). To illustrate the differences between EMP-1 and claudin-20, we modeled their structures using I-TASSER.9 The recently published crystal structure of claudin-1510 was applied as a template. Figure 1 shows the topology of the transmembrane regions and the extracellular loops of claudin-15, claudin-20 and EMP-1. The transmembrane domains form a four-helix bundle, while large parts of the extracellular loops reveal a β-sheet propensity as shown in the ribbon models (Fig. 1, left).
Figure 1.
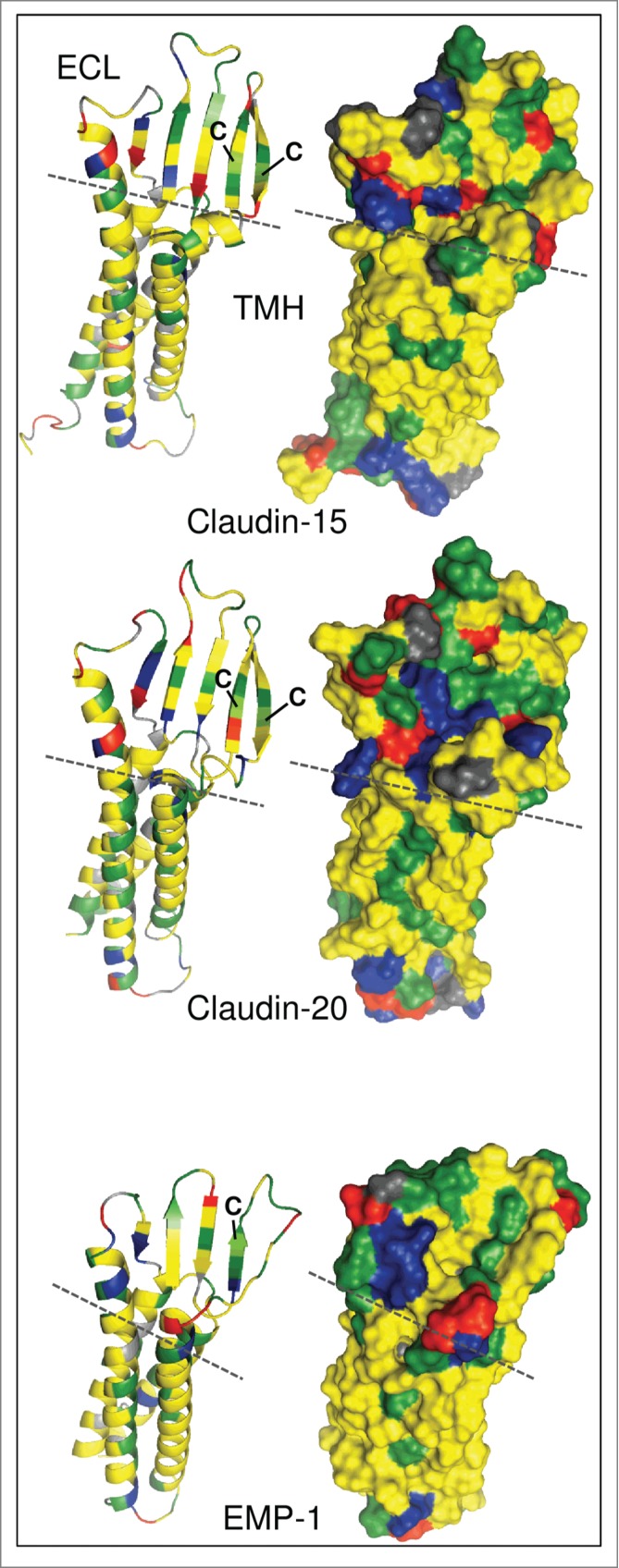
Molecular modeling of human claudin-15, claudin-20 and epithelial membrane protein 1 (EMP-1). Left: Ribbon models showing secondary structural elements. Right: Surface models showing exposed amino acid (aa) residues. Modeling was performed using I-TASSER.9 without additional restraints and excluding C-terminal cytosolic aa. Claudin-20 and EMP-1 models were aligned to claudin-15 using PyMOL (http://www.pymol.org). Claudin-15: aa 1–193, Claudin-20: aa 1–186, EMP-1: aa 1–154. Green, hydrophilic aa; yellow, hydrophobic aa; blue, positively charged aa; red, negatively charged aa; gray, glycine, proline; dashed line, border between transmembrane helices (TMH) and extracellular loops (ECL); C, conserved cysteines in the first ECL.
This reveals clear structural differences in the proteins: the extracellular loops of the claudins are noticeably larger and form 5 β-sheets. In the EMP-1 model only 4 β-sheets are predicted, and they are considerably shorter than in the claudins (Fig. 1, left). The two conserved cysteines in the signature motif of the claudins have been found to form an intramolecular disulfide bond and mediate paracellular tightening.11 In EMP-1 the intramolecular disulfide bond cannot be formed, as only one of these cysteines is conserved (Fig. 1, left). In addition, more charged amino acids are exposed on the surface of claudin-20 whereas lipophilic patches dominate the surface of EMP-1 (Fig. 1, right). These structural differences strongly suggest that the 2 proteins have different functions and certainly cannot – in any case – be considered identical.
In conclusion, the Pfam00822 superfamily contains proteins that share a conserved overall topology and a conserved motif. However, the different protein groups within the family have distinct features, giving them functions that vary considerably, ranging from signaling (as in the case of EMP-1) to adhesion and barrier functions (e.g. claudin-15), or contributions to channel formation. These distinct and unique functions in tissue barriers must not be confused, particularly as we attempt to understand how specific molecules within the family contribute to disease pathologies and responses to drugs.
References
- 1.Martin TA, Lane J, Ozupek H, Jiang WG. Claudin-20 promotes an aggressive phenotype in human breast cancer cells. Tissue Barriers 2013; 1(3):e26518; PMID:24665404; http://dx.doi.org/ 10.4161/tisb.26518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain A, Tindell CA, Laux I, Hunter JB, Curran J, Galkin A, Afar DE, Aronson N, Shak S, Natale RB, Agus DB. Epithelial membrane protein-1 is a biomarker of gefitinib resistance. Proc Nat Acad Sci USA 2005; 102(33):11858-63; PMID:16087880; http://dx.doi.org/ 10.1073/pnas.0502113102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bangsow T, Baumann E, Bangsow C, Jaeger MH, Pelzer B, Gruhn P, Wolf S, von Melchner H, Stanimirovic DB. The epithelial membrane protein 1 is a novel tight junction protein of the blood–brain barrier. J Cereb Blood Flow Metab 2008; 28:1249-60; PMID:18382472; http://dx.doi.org/ 10.1038/jcbfm.2008.19 [DOI] [PubMed] [Google Scholar]
- 4.Durgan J, Tao G, Walters MS, Florey O, Schmidt A, Arbelaez V, Rosen Neal, Crystal Ronald G, Hall A. SOS1 and Ras regulate epithelial tight junction formation in the human airway through EMP1. EMBO Reports 2015; 16:87-96; PMID:25394671; http://dx.doi.org/ 10.15252/embr.201439218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price MG, Davis CF, Deng F, Burgess DL. The α-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate receptor trafficking regulator “stargazin” is related to the claudin family of proteins by its ability to mediate cell-cell adhesion. J Biol Chem 2005; 280(20):19711-20; PMID:15760900; http://dx.doi.org/ 10.1074/jbc.M500623200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maher GJ, Hilton EN, Urquhart JE, Davidson AE, Spencer HL, Black GC, Manson FD, The cataract-associated protein TMEM114, and TMEM235, are glycosylated transmembrane proteins that are distinct from claudin family members. FEBS Letters 2011; 585(14):2187-92; PMID:21689651; http://dx.doi.org/ 10.1016/j.febslet.2011.05.060 [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Helftenbein G, Koslowski M, Sahin U, Tureci Ö. Identification of new claudin family members by a novel PSI‐BLAST based approach with enhanced specificity. Proteins 2006; 65:808-15; PMID:17022085; http://dx.doi.org/ 10.1002/prot.21218 [DOI] [PubMed] [Google Scholar]
- 8.Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, Tsukita S. Predicted expansion of the claudin multigene family. FEBS Letters 2011; 585(4):606-12; PMID:21276448; http://dx.doi.org/ 10.1016/j.febslet.2011.01.028 [DOI] [PubMed] [Google Scholar]
- 9.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols 2010; 5(4):725-38; PMID:20360767; http://dx.doi.org/ 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y. Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 2014; 344(6181):304-7; PMID:24744376; http://dx.doi.org/ 10.1126/science.1248571 [DOI] [PubMed] [Google Scholar]
- 11.Dabrowski S, Staat C, Zwanziger D, Sauer RS, Bellmann C, Günther R, Krause E, Haseloff RF, Rittner H, Blasig IE. Redox-sensitive structure and function of the first extracellular loop of the cell-cell contact protein claudin-1: lessons from molecular structure to animals. Antioxid Redox Signal 2015; 22(1):1-14; PMID:24988310; http://dx.doi.org/ 10.1089/ars.2013.5706 [DOI] [PMC free article] [PubMed] [Google Scholar]


