Abstract
Epithelial sheets, a synapomorphy of all metazoans but porifers, are present as 2 layers in cnidarians, ectoderm and endoderm, joined at their basal side by an extra-cellular matrix named mesoglea. In the Hydra polyp, epithelial cells of the body column are unipotent stem cells that continuously self-renew and concomitantly express their epitheliomuscular features. These multifunctional contractile cells maintain homeostasis by providing a protective physical barrier, by digesting nutrients, by selecting a stable microbiota, and by rapidly closing wounds. In addition, epithelial cells are highly plastic, supporting the adaptation of Hydra to physiological and environmental changes, such as long starvation periods where survival relies on a highly dynamic autophagy flux. Epithelial cells also play key roles in developmental processes as evidenced by the organizer activity they develop to promote budding and regeneration. We propose here an integrative view of the homeostatic and developmental aspects of epithelial plasticity in Hydra.
Keywords: autophagy, epithelial plasticity, evolution, Hydra epitheliomuscular layers, injury-induced response, neuromuscular transmission, regeneration and organizer activity
Hydra, a Classical Model for Studying the Multiple Functions of Epithelial Layers
Eumetazoans, defined as the large cohort of “true” animals formed by cnidarians and bilaterians (Fig. 1A), are multicellular organisms whose organization relies on epithelial cells. Epithelial cells are characterized by a typical apical to basal polarity and by a variety of junction and adhesive properties that allow them to form epithelial sheets. All cnidarians share a bi-layered body wall made of an external layer named ectoderm, and an internal layer named endoderm, which are tightly connected through an extracellular matrix called mesoglea (Fig. 1B-D). The ectoderm provides a protective function analogous to the one of epidermis whereas the endoderm, also named gastrodermis as it lines the surface of the gastric cavity, is involved in food uptake and digestion. Hydra makes use of a third stem cell population, the multipotent interstitial stem cells (i-cells) that are predominantly distributed in the central body column, intermingled between the ectodermal epithelial cells (see in1). These i-cells provide migratory progenitors that after one or several rounds of divisions differentiate into nerve cells, nematocytes (mechano-sensory cells) and gland cells. Indeed some of these interstitial progenitors traverse the mesoglea to reach the gastrodermis where they differentiate as secretory gland cells. In summary, the endodermal layer contains myoepithelial digestive cells, gland cells, and a few neurons. In contrast, the ectodermal layer contains a different population of myoepithelial cells, a large fraction of proliferating stem cells and progenitors of the i-cell lineage, which differentiate into neurons and nematocytes in asexual animals.
Figure 1.
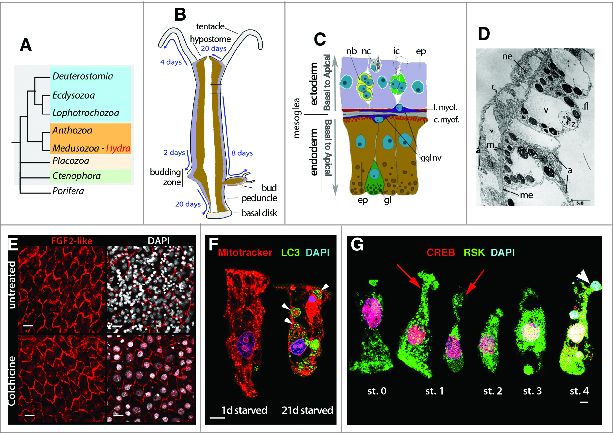
Hydra epithelial cells in homeostatic and stressed conditions. (A) Phylogenetic position of Hydra among metazoans. Note the sister group position of cnidarians that include anthozoans and medusozoans (orange background) to bilaterians (blue background). Among the early-diverged metazoan phyla (Porifera, Placozoa, Ctenophora), only Porifera do not differentiate epithelia. (B) Anatomy and tissue dynamics in Hydra. Hydra polyps have a cylindrical tube shape, terminated at the oral pole by a dome named hypostome and a single opening, the mouth, encircled by tentacles. At the basal pole, the basal disk or foot secretes mucus that helps animals to attach to substrates. Upon regular feeding, polyps reproduce asexually through budding, however when the environment becomes critical for survival, the animals shift to gametogenesis and sexual reproduction (not shown). Epithelial and interstitial stem cells continuously cycle along the body column. Arrows indicate the displacement in time of the epithelial cells toward the bud and the extremities.90 When reaching the poles, epithelial cells stop cycling to undergo terminal differentiation as head- or foot-specific cells (gray zones). (C) Schematic view of the bilayered tissue organization (framed region in B) with endodermal (brown) and ectodermal (mauve) epithelial cells (ep), gland cells (gl), ganglia nerve cell (ggl), a pair of interstitial stem cells (ic), nematoblasts (nb), nematocytes (nc). (D) Low magnification electron micrograph of a segment of body wall of Chlorohydra viridissima reproduced from4 (Fig. 1). Note the acellular mesoglea (me) that separates the thinner epidermis on the left from the gastrodermis, which, in this species, contains intracellular symbiotic green algae (z); the myofibrils (m) in the epidermis (cross-section) and in the gastrodermis (longitudinal section); in the gut lumen the flagellae (fl) of endodermal epithelial cells; the intracellular vacuoles (v) in both layers; the thin cuticle (c) covering the epidermis; a nematocyst within a nematocyte (ne); regions of increased density (a), which correspond to the attachment areas. Scale bar: 5 μm. (E) Immunodetection of the ectodermal epithelial cell membranes with the anti-FGF2 antibody (Santa Cruz sc7911) in untreated animals and Colchicine-treated animals fixed 10 days after an 8 hour colchicine exposure. Note the elimination of the interstitial cells and their derivatives as evidenced by the absence of small DAPI-stained nuclei in colchicine-treated animals. Scale bar: 20 μm. (F) Starvation induces autophagy in Hydra epithelial cells as evidenced here by the dramatic increase in autophagosomes (arrowhead) immunodetected after 21 days of starvation with the anti-LC3 antibody (Novus Biological NB100-2220, green).55,56 Note the presence of numerous mitochondria inside the autophagic vacuoles detected with Mitotracker (red, arrowheads). Scale bar: 10 μm. (G) Engulfment of apoptotic bodies and loss of epithelial polarity in head-regenerating tips (ref. 62, Supplt S2). Efferocytosis by the epithelial endodermal cells (digestive cells) is detected here with Hoechst staining (blue) and anti-CREB (red) and anti-RSK (green) immunodetection. At stage 0 cells display the usual apical to basal hourglass morphology; at stage 1 their apical part gradually detaches (red arrows); at stage 2 they shape ovoid and come into contact with apoptotic bodies, thus named “early engulfing cells;" at stage 3, the “mature engulfing cells” include phagosomes that are large vesicles containing strongly condensed DNA surrounded by a rim of RSK-positive cytoplasm; at stage 4 cells contain phagosomes (blue, arrowheads) but have regained their epithelial cell shape.
The freshwater Hydra cnidarian polyps, a classical model system in cell and developmental biology over the past centuries,2 greatly contributed to the identification of the typical features of epithelia. The behavior of the ciliated endodermal cells during digestive processes was described in Hydra in the late XIXe century.3 Seventy years later, the discovery and visualization of septate junctions (SJs) in Hydra epithelia by electronic microscopy provided the basis to apprehend cell-cell communication,4 completed a few years later by the comparative analysis of SJs and gap junctions (GJs) in the same animal.5 More recently, the analysis of the Hydra genome indicated that the molecular toolkit for establishing apical basal polarity, for differentiating SJs, GJs but also adherens junctions (AJs) and hemidesmosome-like structures is shared between cnidarians and bilaterians.6
Beside the analysis of the Hydra genome, efforts were made over the last decades to systematically identify the molecular signatures of the different Hydra cell types, first through peptidomic approaches that led to the discovery of epitheliopeptides and neuropeptides,7,8 then through cDNA microarrays,9 and more recently through strategies that combine transgenesis, cell sorting and RNA-seq.10 Hydra transgenesis was established in 200611 and led to the production of transgenic strains that constitutively express eGFP in one or the other cell lineage, offering the possibility to FACS-sort GFP expressing cells and to analyze their cell-type specific transcriptomes.10 To complement the transcriptomic profiles of stem cells in Hydra, we recently applied this latter approach. We dissected the central body column of animals from AEP transgenic strains produced by the Bosch laboratory (which constitutively express GFP, either in the endodermal epithelial cells11 or in the ectodermal epithelial cells,12 or in the interstitial stem cells10), dissociated the tissues to sort the GFP-expressing cells by flow cytometry,13 and quantified the level of expression of each gene by RNA-seq (Fig. 2A) (for details, see14). Hence, detailed expression levels of transcripts in endodermal and/or ectodermal epithelial cells were obtained (Table 1).
Figure 2.
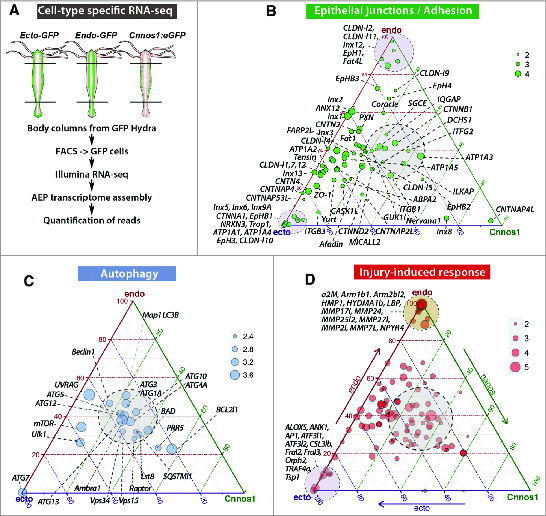
Molecular patterns of the ectodermal and endodermal epithelial cells as deduced from RNA-seq transcriptomic analyses. (A). Scheme depicting the procedure to produce RNAs from each stem cell population by dissecting the body columns of 3 transgenic AEP strains that constitutively express GFP either in the ectodermal epithelial cells (ECTO actin::eGFP12), or in the endodermal epithelial cells (ENDO actin::eGFP11), or in the interstitial stem cells.39 The quantitative RNA-seq analysis was performed on FACS-sorted cells.13,14 (B–D). Ternary plots showing the cellular distribution of gene transcripts encoding epithelial junction - cell adhesion proteins (B), injury-induced immune proteins (C) and autophagy proteins (D). Each dot represents the expression of a unique gene as the computation of the median values of 4 biological replicates in each cell type. Maximal endodermal expression is at the top (endo), ectodermal at the bottom left (ecto) and interstitial at the bottom right (cnnos1). The position of each dot results from the relative transcript abundance in these 3 cell types, with genes similarly expressed in the 3 cell types located in the gray central zone. The dot size is proportional to the number of log10(reads) reads as indicated on the scale.
Table 1.
Cell-type specific expression of epithelial cell markers in Hydra (for protein sequences and expression levels see Supplemental Data). Table showing the relative level of expression of several classes of epithelial markers in the ectodermal epithelial cells (ecto), endodermal epithelial cells (endo) or interstitial cells (i-cells) of the body column of AEP Hydra as deduced from quantitative RNA-seq applied to GFP-sorted cells (see Fig. 2A). “Expressing cells” column: >>> or <<< indicate a minimal 10× difference, >> or << a minimal 2× difference, uppercase writing indicates over 1'000 reads. Hydra protein sequences are available on Uniprot.org either as individual sequences or as sequences from RNA-seq transcriptomes designed to identify cell-type specific proteins,10 Hydra vulgaris/human orthologs,91,92 GO-annotated immune proteins,82 neuromuscular transmission proteins,14 epithelial markers (this work, all annotated protein sequences are accessible from UniProt release 2015_10. For the nomenclature of claudin-like proteins, see Ganot et al. 2015 (ref. 21).)
| Predicted FUNCTIONS | PROTEIN NAMES Gene families | EXPRESSION in GFP Hv-AEP CELLS | Protein ACCESSION (Hv-Basel, Zürich, Jussy) |
|---|---|---|---|
| Sub-apical complex | INADL InaD-like protein (PatJ) | ECTO > Endo >> i-cells | T2M9I3_HYDVU |
| LIN7C Protein lin-7 homolog C | Ecto, Endo > i-cells | T2M567_HYDVU | |
| MPP5 MAGUK p55 (Stardust, Pals1) | Ecto > Endo >> i-cells | T2M9J3_HYDVU | |
| Notch2 (Crumbs-like) | ENDO >>> Endo >> i-cells | T2MDK9_HYDVU | |
| Apico-lateral complex | CDC42 Cell division control protein 42 homolog | ENDO > ECTO > I-CELLS | T2MEG1_HYDVU |
| PARD3 Partitioning defective 3 homolog | ECTO > ENDO > i-cells | T2M994_HYDVU | |
| PARD6G Partitioning defective 6 homolog gamma | Ecto > Endo >> i-cells | T2M6J3_HYDVU | |
| PRKCI Protein kinase C | ECTO > Endo >> i-cells | T2MGA0_HYDVU | |
| Lateral complex | DLG1 Disks large homolog 1 | ECTO > ENDO >> I-CELLS | T2ME64_HYDVU |
| DLG5 Disks large homolog 5 | Ecto >> Endo, i-cells | T2M8B5_HYDVU | |
| LLGL1 Lethal(2) giant larvae prot. homolog 1 | ECTO > ENDO > I-CELLS | T2MCV2_HYDVU | |
| SCRIB Protein scribble homolog | ECTO > ENDO > I-CELLS | T2MDC2_HYDVU | |
| Structural | CLDN-l1, 7, 12 Claudin-like 1,7, 12 | ECTO > ENDO >> i-cells | CRX73236, CRX73250, CRX73241 |
| Septate Junctions | CLDN-l10 Claudin-like 10 | ECTO >> Endo >> i-cells | CRX73238, |
| (St SJs) | CLDN-l2, 9, 11 Claudin-like 2,9,11 | Ecto << Endo >> i-cells | CRX73242, CRX73253, CRX73239 |
| CLDN-l3, CLDN-l4, Claudin-like 3, 4 | Ecto, Endo | CRX73247, T2MFM9_HYDVU | |
| CLDN-l5 Claudin-like 5 | Ecto > Endo >> i-cells | T2MBI9_HDYVU | |
| CLDN-l6, 8, 14, 15 Claudin-like 6,8,14,15 | No or very low expression | CRX73249, CRX73252, CRX73244, CRX73246 | |
| CNTN2 Contactin 2 | ENDO > ECTO >> i-cells | T2MEK3_HYDVU | |
| CNTN4 Contactin 4 | ECTO >> ENDO >> i-cells | CRX73254 | |
| CNTNAP2 Contactin assoc. prot 2 | ECTO > ENDO >> i-cells | T2M432_HYDVU | |
| CNTNAP2l Contactin assoc. prot 2like | Ecto > i-cells > Endo | CRX73256 | |
| CNTNAP4 Contactin assoc. prot 4 | Ecto >> Endo | CRX73257 | |
| CNTNAP4l Contactin assoc. prot 4like | Ecto, Endo << I-CELLS | CRX73258 | |
| CNTNAP5 Contactin assoc. prot.-like 5 | ECTO > ENDO >> i-cells | T2M8X1_HYDVU | |
| CNTNAP53l Contactin assoc. prot. like 5-3 | Ecto >> endo | CRX73259 | |
| DSCAM Down syndrome cell adhesion mol. | Endo < Ecto < i-cells | T2MIF2_HYDVU | |
| NRXN1 Neurexin-1a like | Apical expression only | CRX73281 | |
| NRXN3 Neurexin-3a like | ECTO >> Endo > i-cells | T2M365_HYDVU | |
| Scaffold | ATP1A1 NaK ATPase-α1 | ECTO >>> Endo, i-cells | CRX73229 |
| Septate Junctions | ATP1A2 NaK ATPase-α2 | ECTO >> Endo >> i-cells | CRX73230 |
| (Sc SJs) | ATP1A3 / AT1A NaK ATPase-α3 | ECTO < ENDO < I-CELLS | AT1A_HYDVU, T2MGY6_HYDVU |
| ATP1A4 NaK ATPase-α4 | Ecto | CRX73232 | |
| ATP1A5 NaK ATPase-α5 | Ecto > Endo > i-cells | CRX73233 | |
| ATP1B1 NaK ATPase-β2 (NRV Nervana) | ECTO > I-CELLS > ENDO | T2MHY2_HYDVU | |
| EPB41L4A Band 4.1 l4 (Coracle) | Endo > Ecto > i-cells | T2M572_HYDVU | |
| EPB41L5 Band 4.1 l5 (Yurt) | ECTO >> Endo >> i-cells | T2M5L9_HYDVU | |
| ZO-1 Zonula Occludens 1 (TJP1) | ECTO >> ENDO >> i-cells | T2MDH6_HYDVU | |
| Adherens Junctions | ACTN1 α-actinin | ECTO > ENDO >> i-cells | T2MHI5_HYDVU |
| (AJs) | CDH Classical cadherin | ECTO >> i-cells >> Endo | CRX73223 |
| CELSR2 Cadherin EGF LAG 7pass | ECTO > Endo > i-cells | T2M506_HYDVU | |
| CTNNA1 α-catenin | ECTO >> i-cells, Endo | T2M3Z5_HYDVU | |
| CTNNB1 β-catenin | ENDO > I-CELLS> ECTO, | T2MGP6_HYDVU | |
| CTNND2 δ-catenin | Ecto > i-cells > Endo | T2M3M0_HYDVU | |
| DAG1 Dystroglycan | Ecto | T2MDZ1_HYDVU | |
| DCHS1 Protocadherin 16 | Ecto > Endo> i-cells | T2M7D2_HYDVU | |
| FAT1 Protocadherin 1 | ENDO > ECTO >> i-cells | T2MDR8_HYDVU | |
| FAT4l Protocadherin Fat4-like | ENDO >> ECTO > i-cells | CRX73260 | |
| MICALl2 MICAL like protein 2 | ECTO > Endo > i-cells | T2MAH1_HYDVU | |
| MLLT4 (Afadin) | ECTO >> ENDO >> i-cells | T2MF28_HYDVU | |
| SGCE Sarcoglycan | Endo > Ecto > i-cells | T2MJ55_HYDVU | |
| VCL Vinculin | ECTO > ENDO >> I-CELLS | T2MH95_HYDVU | |
| Gap junctions (GJs) | Inx1, Innexin 1 | ENDO > ECTO >>> i-cells | Q2EMV6_HYDVU, |
| Inx2, Inx9, Inx10, Inx11, Inx14, Inx15 | No or very low expression in body column | seq57378, seq46622 (pending), CRX73266, seq79106, seq05316, seq64623 (pending) | |
| Inx3, Inx13 Innexin 3, 13 | ECTO > ENDO >> i-cells | CRX73271, CRX73269 | |
| Inx4, Inx5, Inx6, Inx7, | ECTO or Ecto | CRX73272, CRX73275, CRX73274, CRX73277, | |
| Inx8 Innexin 8 | I-CELLS >> Ecto >> Endo | Seq55322 (pending) | |
| Inx12 Innexin12 | Endo >> Ecto >>> i-cells | CRX73268 | |
| Hemi-desmosomes | ADAM10 | ECTO > ENDO >> i-cells | T2MJ41_HYDVU |
| (HDs) | ADAM12 | Endo > Ecto > i-cells | T2MIA5_HYDVU |
| ADAM17 | ECTO < ENDO < I-CELLS | T2MEE2_HYDVU | |
| ADAM33 | Ecto < Endo < i-cells | T2M6H9_HYDVU | |
| ADAMTS9 Disintegrin MP thrombospondin | Endo > i-cells > Ecto | T2M4C5_HYDVU | |
| CIB1 Calcium and integrin-binding protein 1 | Endo > Ecto >> i-cells | T2M774_Hydvu | |
| FAK1 Focal adhesion kinase | ECTO, ENDO >> i-cells | T2MDJ8_HYDVU | |
| ILK Integrin linked kinase | ECTO, ENDO >> i-cells | T2ME09_HYDVU | |
| ILKAP ILK-associated protein | ECTO < ENDO < I-CELLS | T2M6A7_HYDVU | |
| ITFG2 Integrin-a FG-GAP | Endo, i-cells > Ecto | T2M8F8_HYDVU | |
| ITGA4 integrin-alpha4 | ECTO > ENDO >> i-cells | CRX73278 | |
| ITGA8 Integrin-alpha8 | ENDO > ECTO > I-CELLS | T2MFQ0_HYDVU | |
| ITGA9 Integrin-alpha9 | ECTO > ENDO >>i-cells | T2ME15_HYDVU | |
| ITGB1 Integrin-beta1 | ECTO >> ENDO >> i-cells | T2MHW4_HYDVU | |
| ITGB2 Integrin-beta2 | ECTO > ENDO >> I-CELLS | T2MGW7_HYDVU | |
| ITGB3 Integrin-beta3 | Ecto >>> Endo < i-cells | CRX73280 | |
| PXN Paxillin | ENDO > ECTO >> I-CELLS | T2MG05_HYDVU | |
| TLN2 Talin2 | ECTO > ENDO > I-CELLS | T2M2W2_HYDVU | |
| TNS1 Tensin1 | ECTO > Endo >>> i-cells | T2M5L6_HYDVU | |
| Cell adhesion | ANX12 Annexin XII / Annexin –B12 | ENDO > ECTO >>> i-cells | P26256_HYDVU |
| Scaffolding proteins | ANXA7 Annexin | ECTO > ENDO >> i-cells | T2MGP1_HYDVU |
| CASK Peripheral plasma mbne protein CASK | Ecto >> Endo, i-cells | CRX73235 | |
| DSCAM Down syndrome cell adhesion mol | Endo < Ecto < i-cells | T2MIF2_HYDVU | |
| EpH1 Ephrin receptor 1 | ENDO >>> i-cells, Ecto | AGO06063.1 | |
| EpH2 / EPHA7 Ephrin receptor 2/7 | ECTO, Endo >> i-cells | AGO06064.1, T2MDF6_HYDVU | |
| EpH3 / EPHA5 Ephrin receptor 3/5 | ECTO >>> i-cells, endo | AGO06066.1, T2MF36_HYDVU | |
| EpH4 / EPHA4 Ephrin receptor 4 | Endo >> Ecto > i-cells | AGO06065.1, T2MEM7_HYDVU | |
| EpHB1 Ephrin ligand B1 | Ecto >>> i-cells, Endo | AGO06067.1, R9WY58_HYDVU | |
| EpHB2 Ephrin ligand B2 | Ecto, Endo << i-cells | AGO06068.1, R9WWC9_HYDVU | |
| EpHB3 Ephrin ligand B3 | ENDO >> Ecto >> i-cells | AGO06069.1, R9X0X4_HYDVU | |
| FARP2 l FERM RhoGEF pleckstrin domain | Ecto > Endo >> i-cells | T2MID3_HYDVU | |
| GUK1 like Guanylate Kinase 1 | Ecto, Endo > i-cells | T2MD66_HYDVU | |
| IQGAP / IQGAP1 GTPase-activating like prot | ENDO > ECTO > I-CELLS | Q9XZE9_HYDVU, T2MFN7_HYDVU | |
| LRIG3 Leu Rich Repeats Ig-like prot 3 | Ecto > Endo > i-cells | T2MAL0_HYDVU | |
| Trop1 Tropomyosin | ECTO >>> Endo | TPM1_HYDVU | |
| Cuticle structure | Sweet Tooth proteins | 22 proteins | See Böttger et al. 2012 (ref. 16) |
| PPOD1 Putative Peroxidase 1 | ECTO >> ENDO >>> i-cells | Q2FBK4_HYDVU, Q2FBK7_HYDVU | |
| PPOD2 Putative Peroxidase 2 | No PPOD2 in Hv-AEP | Q962G1_HYDVU, Q2FBK2_HYDVU | |
| PPOD2-like Putative Peroxidase 2-like | No PPOD2l in Hv-AEP | Q2FBJ9_HYDVU | |
| Extra-Cellular Matrix | ANKFN1 Ankyrin repeat fibronectin III | Ecto > Endo >> i-cells | T2M9C4_HYDVU |
| (ECM) | COL4A1 / COL4A5 collagen-alpha5 (IV) | ENDO >>> Endo, i-cells | Q9GQB1, T2MFW7_HYDVU |
| FARM1 secreted astacin | Endo >>> Ecto | Q9U4X9_HYDVU | |
| FiCol fibrillar collagen | ENDO >>>> Ecto, i-cells | Q8MUF5_HYDVU | |
| FNDC3B FN type III containing protein 3A | ECTO, ENDO >> i-cells | T2MCC9_HYDVU | |
| HMCN1l1 Hemicentin1 like1 | ENDO >>> i-cells > Ecto | CRX73261 | |
| HMCN1l2 Hemicentin1 like2 | ENDO >> i-cells > Ecto | CRX73263 | |
| HMCN2l1 Hemicentin2 like1 | ECTO >>> i-cells, Endo | CRX73264 | |
| HMP1 Metalloendopeptidase | ENDO >> Ecto > i-cells | Q25174_HYDVU | |
| HSPG2 basement membrane-specific heparan sulfate proteoglycan protein | ENDO >> i-cells > Ecto | T2MDT4_HYDVU | |
| LAMA5 Laminin subunit alpha-5 | ENDO >> Ecto > i-cells | T2MIW4_HYDVU | |
| LAMB1 Laminin subunit beta-1 | ENDO >>> i-cells > Ecto | LAMB1_HYDVU | |
| MMP matrix metalloproteinase | ENDO >>>> Ecto > i-cells | Q9U9P0_HYDVU | |
| MP2 Metalloendopeptidase (meprin-like) | Endo >> i-cells > Ecto | Q9XZG0_HYDVU | |
| Stem Cell Behavior | Ets1 / ERG | ECTO > ENDO >>> i-cells | I3V7X0_HYDVU, T2MHK5_HYDVU |
| & Stemness | Ets2 / GABPA | Endo > Ecto >> i-cells | T2MDI3_HYDVU |
| FoxO | ECTO > I-CELLS > ENDO | J7HWF0_HYDVU | |
| Klf1 Krueppel like factor 1 | ECTO > ENDO >>> i-cells | T2MDQ7_Hydvu | |
| Klf3 Krueppel like factor 3 | ECTO > Endo | I3V7X3_HYDVU | |
| Klf7 Krueppel like factor 7 | ECTO > ENDO >> i-cells | T2MIK5_Hydvu | |
| Klf8 Krueppel like factor 8 | ECTO > ENDO | I3V7V7_HYDVU | |
| Klf11 Krueppel like factor 11 | ECTO > ENDO >> i-cells | I3V7X4_HYDVU, T2MJ10_HYDVU | |
| Klf13 Krueppel like factor 13 | Endo < Ecto << I-CELLS | I3V7W6_HYDVU, T2M360_HYDVU | |
| MAX | ECTO < ENDO < I-CELLS | D0EM50_HYDVU | |
| Myc-1 | Endo < Ecto << i-cells | D0EM49_HYDVU | |
| Myc-2 | Ecto < ENDO < I-CELLS | D2KBP8_HYDVU, T2MH01_HYDVU | |
| Myc-3 | Endo < Ecto <<< i-cells | CRX73227 | |
| PIWIL1 /Hywi /Cniwi Piwi-like protein 1 | ENDO < ECTO < I-CELLS | T2M7W7, J7HWM3, T2HRA5 | |
| PIWIL2 / Hyli Piwi-like protein 2 | ENDO < ECTO << I-CELLS | T2M9F7, U5XHW4, T2HRQ9 | |
| PL10 | ENDO < ECTO < I-CELLS | Q9GV14_HYDVU | |
| POU4F2 | Endo << Ecto < i-cells | T2MDR7_HYDVU | |
| SOX2 | ECTO > Endo, i-cells | T2MFM3_HYDVU | |
| TCF Ternary Complex Factor | ENDO > I-CELLS > ECTO | Q9GTK1_HYDVU | |
| TCTP (p23) | ENDO > ECTO > I-CELLS | TCTP_HYDVU | |
| TP53BP2 | ECTO >> ENDO > i-cells | T2MDM1_HYDVU | |
| TP73 | Endo >> Ecto > i-cells | T2MIU9_HYDVU | |
| Vasa1 / Cnvas1 | ECTO < ENDO < I-CELLS | Q9GV13_HYDVU | |
| Vasa2 / Cnvas2 | ECTO < ENDO < I-CELLS | Q9GV12_HYDVU |
In this review, we highlight the recent progress made in our understanding of the multiple functions carried out by Hydra epithelia, such as protection to the environment, nutrient adsorption, cell-cell communication, contractility, resistance to starvation, resistance to pathogens, wound healing, reactivation of developmental programs. Given the evolutionary conservation of epithelial functions among eumetazoans, we assume that tracing back in Hydra epithelia the mechanisms that support these functions will provide new concepts and possibly new tools to face the physiological and pathological consequences of epithelial alterations in mammals.
The Cuticle Provides a Protective Physical Barrier to the Environment
In Hydra, the ectodermal epithelial layer, which delimits the outlines of the animal protects the animal from constant environmental challenges: physical interactions, osmotic pressure or invading pathogens. Similarly to the mammalian epidermis, the ectoderm synthesizes a fibrous assembly called cuticle, which resembles the glycocalix that surrounds many epithelial cells and shields the external surface of the animal (Fig. 1D). Although carefully observed in electron-microscopic studies in the 60s,4,15 the fine structure and the components of the Hydra glycocalyx were only recently identified.16 This fibrous cuticle, up to 1.5 μm thick, is formed of 5 distinct layers that contain 3 main components: (i) glycosaminoglycans, namely unsulfated chondroitin and chondroitin-6-sulfate disaccharides, (ii) several SWT “sweet tooth” proteins, and (iii) 3 distinct PPOD (Putative PerOxiDase) proteins (Table 1). These proteins, stored in vesicles close to the apical side, are secreted by the ectodermal cells. Thanks to their β-trefoil structure and their haemagglutinin activity, these proteins can bind to chondroitin sulfate and thus contribute to the cuticle organization. Interestingly, the family of PPODs found in Hydra seems to be absent in plant or animal species, suggesting that this Hydra specificity was acquired by horizontal gene transfer from bacteria.16,17,18
Epithelial Polarity and Epithelial Junctions
Hydra epithelial cells exhibit a typical apico-basal polarity, possibly resulting from the activity of the 3 complexes that set up the epithelial polarity in bilaterians19: the sub-apical Crumbs complex (Crbs, MPP5/Pals1, InaD/PatJ), the apico-lateral Par complex (Par3, Par6, aPKC, cdc42) and the lateral Scribble complex (Scrbl, Lgl, DLG). Whether the function of the Hydra Crumbs-like protein in the sub-apical complex is conserved remains to be tested. As expected, epithelial cells also express a full set of proteins that establish permeability barriers, the septate junctions (SJs), the anchoring junctions as baso-lateral adherens junctions (AJs) and the basal hemi-desmosome-like structures (see Table 1). Important components of the AJs are the classical cadherins. These are present in Nematostella vectensis20; in Hydra we found a single classical cadherin protein, which encodes a series of cadherin tandem repeat domains and 2 laminin domains as extra-cellular domains, as well as a conserved cadherin cytoplasmic domain (see Table 1).
SJs are shared by all metazoans, but vertebrates also evolved tight junctions (TJs), characterized by the presence of “stricto sensu” claudin proteins, which are not found in invertebrates. Those rather express claudin-like proteins.21 Hydra expresses 14 claudin-like (CLDN-l) genes: 3 exclusively in the endodermal epithelial cells (CLDNl2, CLDN-l9, CLDN-l11), 3 at similar levels in both epithelial layers (CLDN-l3, CLDN-l5), and 4 in both layers although at higher levels in the ectoderm (CLDN-l1, CLDN-l7, CLDN-l10, CLDN-l12) (Fig. 2B, Table 1). Finally, 4 are not detected in the body column or at very low levels (CLDN-l6, CLDN-l8, CLDN-l14, CLDN-l15).
Gap junctions (GJs) play a major role in cell-cell communication in Hydra and epithelial cells communicate by electric conduction through GJs.22 GJs in deuterostomes (including vertebrates) are formed by connexins/pannexins, whereas in protostomes, GJs are formed by proteins from the innexin (Inx) family, similarly to what is observed in Hydra.6,23 Hydra innexins can be expressed either at similar levels in the 2 epithelial layers (Inx1, Inx3, Inx13), or predominantly in the ectoderm (Inx4, Inx5, Inx6, Inx7, Inx10) or in the endoderm (Inx12)14 (Table 1). Surprisingly, innexins were not found so far in other cnidarian species.
Beside the general conservation of the epithelial toolkit in the ectodermal and the endodermal epithelial cells, this analysis also shows that the 2 epithelial cell layers are structurally different as for example, Contactin 4 (CNTN4), CNTNAP53l, Neurexin-3a like, Zonula Occludens 1 (ZO-1), α-catenin (CTNNA1), Inx4, Inx5, Inx6, Inx7, Inx10 genes that are strictly or predominantly expressed in the ectodermal cells, whereas Crumbs-like, Claudin-like 2, 9, 11, Protocadherin Fat4-like, Inx12 are strictly or predominantly expressed in the endodermal ones (Fig. 2B, Table 1). If confirmed at the protein level, this implies that the epithelial organization is largely similar in the epidermis and the gastrodermis although not identical. This difference, previously noted by Hemmrich et al.,10 is not so surprising as the corresponding epithelial cell types have different anatomies, carry functions specific to the layer they belong to, and cannot replace each other.
Extracellular Matrix Production and Regulation of Developmental Processes
The extracellular matrix (ECM) deposit named mesoglea, which separates the 2 epithelial layers in Hydra, contributes to the adhesion and the anchoring of epithelial cells, keeping the 2 layers tightly connected. The mesoglea consists in fine fibrils of different diameters organized as 2 basal lamina matrix with a central fibrous area (see in24). Ultrastructural, histochemical and biochemical studies showed that the structural components of Hydra ECM are highly similar to those found in the basement membrane of vertebrates i.e. type IV and fibrillar collagens, laminins, fibronectin and proteoglycan-like molecules, as well as several types of fibrillar collagens, and confirmed the lax and porous structure of the mesoglea, with pores of 0.5-1 μm in diameter, which facilitate the communication between ectoderm and endoderm. In situ hybridization and cell type specific transcriptomes showed that both epithelial layers produce the ECM components although with specific roles, the ectodermal cells synthesising fibronectin and the α/β integrins, and the endodermal cells synthesising all types of collagens, the laminins (α1, β1) and the matrix metalloproteinases (HMP1, HMP2, HMMP) (see in Table 1, refs10,24). All these components, assembled together in the extracellular space, also play an important role in morphogenetic processes as regeneration and budding.24,25 As an example, the strength of the adhesion of the epithelial cells to the ECM varies with morphogenetic displacements along the body column and in the region where the bud develops.26
Epithelial Cells in the Hydra Body Column are Both Differentiated and Stem Cells
All epithelial cells in Hydra are epithelial-muscular cells that, in the central body column, continuously proliferate and self-renew, displaying thus stem cell properties and differentiated features concomitantly.27 Both ectodermal and endodermal populations exhibit a rather unusual cycling pattern, characterized by the lack of G1-phase and an extended G2-phase, which is reminiscent of the cell cycle properties of embryonic stem cells.28,29,30 A recent flow cytometry analysis confirmed that 85% epithelial stem cells distribute between the S and G2 phases.13 Given the fixed S phase length (about 12 hours), the total length of the epithelial cell cycle is imposed by the length of the G2 phase, which varies according to the feeding regime: An epithelial cell cycle takes 3-4 days to complete in well-fed animals versus up to 10-12 days in starving animals.13,29 Hydra epithelial cells are not migratory, but as a result of their rapid proliferation in the body column, they get progressively displaced laterally into newly developing buds or pushed toward the extremities of the animal (Fig. 1B). When reaching extremities, epithelial cells stop cycling and terminally differentiate in G2 phase, giving rise to foot-, head-, or tentacle-specific cells.13,30
So far, our knowledge concerning the genetic circuitry regulating stemness in Hydra is limited (see in Table 1). The famous “Yamanaka OKSN factors” are not well conserved in cnidarians,31 either completely missing as Nanog (N), or distantly related as Sox2 (S) and Oct4 (O). However, in Hydractinia the Oct4-like transcription factor named “Polynem” promotes self-renewal32 and in Hydra, the related POU4F2 transcription factor, predominantly expressed in ectodermal and interstitial stem cells, might play a similar role. Several Krüppel-like factors (Klf) are expressed in Hydra, 2 of them exclusively in the epithelial cells (KLF3, KLF8) and a third one, KLF11, predominantly but not exclusively in the epithelial cells. Although not a clear vertebrate Sox2 ortholog, the Hydra Sox-2 like gene is a potential regulator of self-renewal.10 As additional stem cell transcription factors, the proto-oncogene Myc is present as 4 copies in the Hydra genome33; HyMyc1 and HyMyc2 contain a typical bHLH-ZIP DNA-binding box and several Myc domains, whereas HyMyc3 and HyMyc4 contain only the DNA-binding domain.34,35 HyMyc1 is predominantly expressed in the interstitial stem cells, likely controlling their proliferation.36 By contrast, hyMyc2 is expressed at high levels in all 3 stem cell populations, suggesting that paralogs of an ancestral Myc gene also control epithelial proliferation.35 Among candidate regulators of stem cells, one also finds the Ets transcription factors that in vertebrates regulate proliferation, inhibit apoptosis and promote neuronal specification.37 Two of them (Ets1, Ets2) are specifically expressed in the epithelial cells.10 The role of all these genes on the behavior of epithelial stem cells remains to be tested in Hydra.
The FoxO gene that encodes a forkhead transcription factor, was initially identified for its role in stress response.38 Subsequently, it was selected together with Tcf, PIWI and vasa1 for its high level of expression in the 3 stem cell populations, providing thus candidate regulators of stem cell behavior in Hydra.10 Indeed FoxO down-regulation in epithelial cells leads to a reduced growth and to an enhanced differentiation of foot and head epithelial cells, supporting a role for FoxO in the control of stem cells.39 Surprisingly, FoxO silencing also affects the innate immune response, enhancing the expression of antimicrobial peptides, suggesting a role in host defense mechanisms.
Hydra expresses 2 PIWI genes, PIWIL1 named Hywi or Cniwi, and PIWIL2 named Hyli, both expressed in the 3 stem cell populations.40,41 The mapping of piRNAs on cell-type specific transcriptomes revealed non-transposon putative PIWI targets in epithelial cells, pointing to adhesion and ECM protein genes in the ectoderm, and to proteolytic and ECM genes in the endoderm.40 The role of PIWI proteins in epithelial cells is largely supported by Hyli, as shown in hyli-RNAi transgenic lines where the epithelial integrity of F1 hatchlings is altered, leading to tissue disintegration and death. In i-cells, the PIWI-piRNA pathway is associated with transposon silencing.41
Pacemaker Contractile Activity of the Epitheliomuscular Cells
The two distinct epithelial cell lineages that build up the body wall of Hydra are actually myoepithelial, i.e. contain at their basal side myofibrils, oriented perpendicular to each other, i.e., circular in the endoderm, longitudinal in the ectoderm, acting thus as circular or longitudinal muscles4,42 (Fig. 1C). Electrophysiological studies have shown that well-fed animals contract on average once every 5 to 10 minutes with periodic bursts of contractions, each layer exhibiting an autonomous pacemaker activity.43,44 Indeed these myoepithelial pacemakers function autonomously as their activity persists, although at a slower pace, in nerve-free animals.45,46 This autonomous contractile behavior possibly reflects the proto-neuronal status of the epithelial cells.47 It occurs thanks to electrical synapses such as gap junctions, which connect epithelial cells48 via innexins23 (Table 1). In fully-equipped animals, neurons control this activity through Inx2: Inx2 is expressed in a small subgroup of nerve cells in the peduncle of the animal, and initiates the epithelial pacemaker activity in this region.49 By contrast the complex feeding response that involves tentacle swirling and mouth opening requires a coordinated neuronal network.50 At the base of the tentacles, the myoepithelial cells express sodium channel receptors (NaC) that are directly activated by the RFamide neuropeptides, implying that peptide-gated ion channels are involved in neuromuscular transmission in Hydra.51 Thus cnidarians, and so far only cnidarians, have independently recruited peptides as fast transmitters for neuromuscular transmission.
Digestive Functions
An important function of the gastrodermis is to digest nutrients and to perform exchanges with the content of the lumen. In its natural environment, i.e. wild ponds, Hydra eat small swimming crustaceans (Daphnia nauplii), whereas in laboratory, the animals feed on desalted Artemia nauplii (brine shrimps larvae). Polyps paralyze preys thanks to a touch-induced discharge of venom contained in the capsules (named nematocysts or cnidocysts) embedded in their nematocytes.52 Then, preys are progressively introduced through the mouth opening inside the gastric cavity by coordinated tentacle movements. Once inside the gastric cavity, the food is partially degraded by the proteolytic enzymes released by the gland cells, and absorption by digestive cells occurs through phagocytosis and pinocytosis. The whole digestive process is highly dynamic, with peristalsis, segmentation movements and defecation reflex, the latter ejecting feces through the mouth opening 6 to 9 hours after feeding.46
The epithelial endodermal cells display a typical columnar shape with short processes at the basal pole, extending microvilli and flagella into the gastric cavity. Early electron-microscopic studies of digestive cells evidenced a very heterogeneous cytoplasmic content, with diverse vesicle types, lipid droplets and glycogen granules that serve as nutrients for the surrounding cells.42,53 Based on precise ultrastructural and immuno-histochemical criteria (Lysotracker red-LTR, MitoFluor 589, LBPA, DAPI, LC3), three distinct types of vacuoles were identified in the digestive cells: digestive vacuoles, autophagic vacuoles and apoptotic bodies.54-56 This diversity of vesicles actually reflects the multiple functions of epithelial cells, which, besides their digestive role, contribute to the elimination of cell debris, or can activate cyto-protective or pro-survival mechanisms.
Autophagy and Maintenance of Fitness
Hydra polyps readily adjust to caloric restriction by activating the autophagy process.55,56 This evolutionarily conserved survival strategy affects both epithelial cell populations that display autophagic vacuoles already 3 days after the onset of starvation.56 After 3 weeks of starvation, epithelial cells contain numerous autophagosomes that can be easily immunodetected with the universal autophagy marker LC3/ATG8 (Fig. 1F). In fact, autophagy activation was first recorded in endodermal epithelial cells of animals knocked-down for Kazal1, a gene that encodes a serine protease inhibitor (SPINK) expressed by the gland cells.57 The phenotype, which mimics the SPINK1/SPINK3 mammalian phenotype, consists in a progressive autophagy of all endodermal cells linked to a progressive loss of fitness, a parallel loss of budding, and in head-regenerating tips, an immediate excessive autophagy after bisection, which in few hours leads to cell death. Hence, autophagy has a double role in Hydra: survival in case of starvation, and cytoprotection in stressed or damaged tissues.58
Orthologs of most components of the autophagy and TOR pathways were identified in Hydra and Nematostella, indicating that the machinery is well conserved in cnidarians.56 As anticipated the drugs rapamycin, wortmannin and bafilomycin similarly modulate autophagy in Hydra and mammals, as the mTOR inhibitor rapamycin that enhances autophagy in all Hydra epithelial cells.55,56 A cell-type specific RNA-seq analysis shows that all members of the autophagy pathway examined here but ATG7, are expressed in epithelial as well as in i-cells (Fig. 2C). However the Ubiquitin-like modifier-activating enzyme ATG7 is almost exclusively expressed in the ectodermal epithelial cells. In addition the mTOR kinase that acts as a central regulator of cellular metabolism, the kinase Ulk1 that responds to starvation, the positive regulator of autophagy UVRAG are predominantly expressed in epithelial cells, likely reflecting the distinct regulations of autophagy between epithelial and interstitial cell types.
Resistance to Cell Death and Efferocytosis
Epithelial cells are extremely resistant to cell death59 and are in charge of engulfing the apoptotic bodies, a process named efferocytosis. Epithelial efferocytosis was first reported by Campbell who observed apoptotic bodies in both the ectodermal and the endodermal epithelial cells of polyps exposed to colchicine.60 Since then, numerous studies confirmed the active role of the epithelial cells in apoptotic cell clearance by engulfment, whatever the pro-apoptotic agent, pharmacological, heat-shock, starvation, gametogenesis, wounding, head regeneration or histocompatibility reaction (see in59). The epithelial cells recognize the dying cells, which in most circumstances are of interstitial origin, probably thanks to “eat-me” signals present on apoptotic membranes. In mammalian cells, phosphatidylserine translocation to the outer cellular membrane provides a typical signal for engulfment, and this classical marker of apoptosis was also identified in Hydra.61,62 However the phagocytic receptors recognizing eat-me signals in Hydra have not been identified yet, but similarly to bilaterian cells, receptor tyrosine kinases expressed in epithelial cells might play an important role in this recognition process.9,10
In case of head regeneration, an immediate and massive wave of efferocytosis can be observed in the endodermal epithelial cells located below the bisection plane.62 Interestingly these cells transiently lose their apico-basal polarity during the first hours (Fig. 1G). A similar transient loss of the polarity of the endodermal epithelial cells was previously observed during early reaggregation.63 Both observations suggest that the maintenance of the endoderm as an epithelial layer requires dynamic interactions with the sus-jacent ectodermal layer. The impact of efferocytosis in head-regenerating tips on the regenerative process was not tested so far, it might be limited to a scavenging function, but it might also trigger the developmental function of the endodermal cells, which at that time start developing an organizer activity.
Antimicrobial Host Defense Role of Hydra Epithelium
As an aquatic species living in an open environment and thus exposed to a multitude of potential pathogens such as protists, bacteria or viruses, Hydra developed host defense strategies that integrate innate immunity tools located in the epithelial layers.64 These immune responses, also present in porifers, were deeply dissected in Hydra, which makes use of Toll-like receptors (TLR), NOD-like pattern recognition receptors (NLR) and the cytoplasmic cascades that mediate the production of antimicrobial peptides (AMPs).65-68 TLR signaling in Hydra was revealed by silencing the universal transducer protein Myd88.67 Unlike Nematostella,69 where the TLR function is achieved by receptors that contain both the LRR (leucine rich repeat) and the TIR (Toll/Interleukin-1 receptor) domains, Hydra possesses 2 distinct proteins that functionally interact, one harboring the LRR, the other the TIR domains.65 The activation of the TLR transduction pathway elicits an antimicrobial response, as the production of the periculin peptide by the endodermal epithelial cells and the interstitial cells.
The second line of defense includes the surprisingly complex inventory of NLR family receptors. Although the function of this family of receptors in the innate defense is well established, the interacting partners and the members of signaling pathway are not completely understood in Hydra. So far, in vitro studies identified one caspase containing a DEATH domain that interacts with hyNLR type 1 protein, suggesting that NLR induction triggers caspase activation.66 As output, 3 classes of AMPs are synthetized by the endodermal epithelial cells, periculin, hydramacin and arminins, which show efficient bactericidal activity.65,70,71 As a third line of defense, the gland cells produce serine protease inhibitors, among them Kazal2 that exhibits a powerful activity against Staphylococcus aureus.72 Moreover, under a massive pathogenic aggression, ectodermal cells are able to emit pseudopods and engulf bacteria, providing another protective defense response.65 Thus, both epithelial layers are well equipped with potent defense molecules and mechanisms showing the adaptability of this simple animal to develop defending strategies against external attacks, but also against internal invasion by ingestion of bacteria into the gastric cavity.
Microbiota Formation and Epithelial Cells - Bacteria Colonization
Like in most animal species, the interactions with commensal bacterial populations that form the microbiota are important for Hydra homeostasis. In fact, polyps cultured in sterile conditions cease to reproduce asexually through budding.73 More recent systematic studies reveal that different Hydra species develop particular preferences for certain bacterial phylotypes.74 This process encompass several steps: the initial colonization of juvenile animals with highly variable groups of bacteria, then the transient selection and extension of a bacterial type that will become the principal species of the colonizing group.68 The severe reduction in variability is thus associated with a stable species-specific microbiota interaction: bacteroidetes and β-proteobacteria are predominant in H. vulgaris, α-proteobacteria (rickettsiales) and endosymbionts in H. oligactis.74 Hence the bacterial community is modeled by continuous interactions between the host epithelial cells and the microbial populations, with host-related components playing a crucial role. Ultimately these interactions are beneficial for the host as the microbiota protects it from pathogens.75
These interactions imply several levels of regulation. The analysis of the colonization process in arminin-deficient Hydra showed that these animals do not select properly their bacterial partners, implying that AMPs control the selection of bacterial phylotypes populating the microbiota.68 Also “epithelial” Hydra lacking nerve and gland cells, show a different composition of their colonizing microbiota.76 However the elimination of the interstitial cells is not sufficient to alter the microbiota, indicating that nerve cells and gland cells play an important role in setting the microbiota.70,72,77 Hence in Hydra, the highly dynamic host-microbiota interactions are modulated by the cellular composition of the epithelial layers.
Immune Response of Epithelial Cells to Stress and Injury
A series of studies investigating the events taking place in head-regenerating tips after bisection, point to an essential role of the MAPK/CREB pathway.62,78,79,80 Immediately after mid-gastric amputation, a massive wave of cell death is observed at the head-regenerating edge, affecting interstitial progenitors and interstitial derivatives. The resulting apoptotic bodies are engulfed by the endodermal epithelial cells, which transiently change their columnar phenotype, loose their apical to basal polarity and become spherical (Fig. 1G). Dying cells release Wnt3, which promotes the division of the surrounding progenitors and is necessary for a later Wnt3 up-regulation in the endodermal epithelial cells62,81. By contrast, cell death remains limited and cell proliferation does not increase in foot-regenerating tips, indicating that head and foot regeneration processes are immediately different.62,80
In an attempt to characterize the genes immediately up-regulated upon injury, we recently applied a transcriptomic approach, which led to the identification of 43 immune-associated genes similarly regulated whatever the regenerative context (Fig. 2D).82 Among them, we identified components of the ROS signaling pathway, TNFR and TLR signaling related transcription factors like jun, fos, ATF1/CREB, SIK2, all possibly modulating the NF-kB pathway. This study suggests that the response to injury involves the innate immune system, and raises the question of the developmental impact of this stress-induced immune response on the regenerative processes, and on the potential of epithelial cells to set up an organizer activity.
Developmental Functions of Epithelial Cells
Thanks to its remarkable competence for regeneration and asexual reproduction through budding, the Hydra polyp provides a unique model for deciphering the mechanisms leading to the reactivation of developmental processes in an adult organism. Except extremities, each piece of Hydra tissue is able to undergo a perfect regeneration process and give rise to a complete animal within few days. Transplantation experiments performed at various time-points after bisection showed that the head- or foot-regenerating tips acquire organizer activity in few hours i.e., become able to instruct and recruit host tissues to rebuild the missing head and/or foot regions.83,84 For head regeneration, activation of the MAPK/CREB pathway and induction of the canonical Wnt pathway play essential roles.62,80,81,85
In this developmental transition, the epithelial cells play the key role, as first, chimeric animals resulting from recombination of strains with different morphologic properties, preserve the morphogenic properties of the parental epithelial cells and not that of the interstitial cells (see in 86). Second, Hydra depleted of their interstitial cells, the so-called “epithelial” Hydra (Fig. 1E), are able to regenerate, although at a slower pace. If manually fed, they can also reproduce asexually through budding, which indicates that the interstitial cells can be dispensable for developmental processes. In fact, the genetic circuitry launched upon amputation is sequentially activated and relies preponderantly on epithelial specific genes in the immediate and immediate-early phase.82,87,88
We view the plasticity of Hydra epithelial cells as an intrinsic property that has multiple facets, quite distinct when regulated in acute or chronic contexts. In fully-equipped animals, signals received from the interstitial cells immediately after amputation (as signals released by the dying cells – see above) speed up the transition phase whereby epithelial cells quickly adopt a developmental role, which is absent before amputation.89 In epithelial animals, we suspect that epithelial cells adapt to the loss of interstitial cells by “slowly” reprogramming a large series of genetic programs already in homeostatic conditions, i.e. in the absence of injury signals (ref. 14 and unpublished). Our hypothesis is that in such “reprogrammed” Hydra, the response to injury is still efficient, although different from that observed in fully equipped Hydra. Nevertheless the reprogramming potential of the epithelial cells remains limited as epithelial cells never transform into cells of the interstitial lineage. In summary, the ability of the epithelial cells to adapt to the loss of the nervous system and the potential of digestive cells to develop at any time an organizer activity are amazing, reflecting distinct roles, to control tissue homeostasis, and to maintain fitness of the organism through repair and regeneration.
Conclusions and Perspectives
As reported above, multiple properties characterize the epithelial cells of the Hydra body column, with some significant quantitative and qualitative differences between the epithelial cells of the outer layer, which form an epidermis, and the epithelial cells of the inner layer, which form a gastrodermis (Fig. 3). However, the cells of a given layer do not express the full repertoire of their properties at the same time. Rather, they provide the animal with the abilities to react and to adapt to stress, infection, starvation, amputation, so that homeostasis is reestablished and maintained over weeks, months and, in favorable environment, over years. Therefore, Hydra offers a unique model system to test the multiple facets of cellular plasticity. Our view of the molecular signaling supporting epithelial plasticity in Hydra is currently limited, but available data point to evolutionarily-conserved signaling pathways, such as (i) a ROS signaling pathway for the immediate response to stress, heatshock and injury, which efficiently contributes to the wound healing process, (ii) a highly diversified innate immune system for a sustained response to stress, infection and injury, (iii) autophagy and TOR signaling pathways to efficiently respond to starvation and thus support animal survival for weeks, (iv) evolutionarily-conserved developmental pathways involving Wnts, FGF, BMP, Notch and Nodal signaling for the full reactivation of developmental processes in an adult organism.
Figure 3.
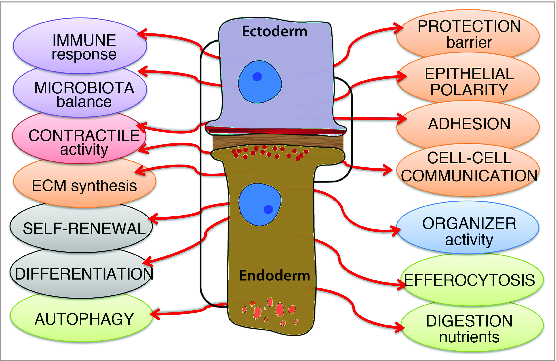
Summary scheme depicting the multiple functions of endodermal and ectodermal epithelial cells in Hydra. Note the functions that are common to both epithelial cell types (brackets).
A series of puzzling questions remain pending: Which of these pathways respond to taxon-specific signals such as epitheliopeptides that are numerous in Hydra? How do these pathways cross-talk? How do the epithelial cells prioritize the different tasks they have to execute? Can we establish hierarchies in the meta-signaling network linking the specific environmental contexts and thus identify master components of environmental-dependent regulators of plasticity? Deciphering the molecular networks supporting epithelial plasticity in Hydra, should highlight the mechanisms that support specific biological competences as the maintenance of fitness to face stressful environmental conditions, the ability to repair tissues and appendages, the ability to reproduce asexually and thus bypass the costs of sexual reproduction, and the ability to resist to aging. No doubt that the most robust molecular regulators of these competences in Hydra should be tested in mammalian contexts, potentially offering new tools for regenerative medicines.
Funding
This work was supported by the State of Geneva, the Swiss National Fonds for Research (snf-31003A-149630), the NIH (grant 7RO1AG037962), the HFSP (grant RHP0016-2010) and the Claraz donation.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. David CN. Interstitial stem cells in hydra: multipotency and decision-making. Int J Dev Biol 2012; 56:489-97; PMID:22689367; http://dx.doi.org/ 10.1387/ijdb.113476cd [DOI] [PubMed] [Google Scholar]
- 2. Galliot B. Hydra, a fruitful model system for 270 years. Int J Dev Biol 2012; 56:411-23; PMID:22855328; http://dx.doi.org/ 10.1387/ijdb.120086bg [DOI] [PubMed] [Google Scholar]
- 3. Greenwood M. On digestion in hydra, with some observations on the structure of the endoderm. J Physiol 1888; 9:317-i316; PMID:16991502; http://dx.doi.org/ 10.1113/jphysiol.1888.sp000290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wood RL. Intercellular attachment in the epithelium of hydra as revealed by electron microscopy. J Biophys Biochem Cytol 1959; 6:343-52; PMID:13845833; http://dx.doi.org/ 10.1083/jcb.6.3.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hand AR, Gobel S. The structural organization of the septate and gap junctions of hydra. J Cell Biol 1972; 52:397-408; PMID:4109925; http://dx.doi.org/ 10.1083/jcb.52.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. The dynamic genome of hydra. Nature 2010; 464:592-6; PMID:20228792; http://dx.doi.org/ 10.1038/nature08830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takahashi T, Muneoka Y, Lohmann J, Lopez de Haro MS, Solleder G, Bosch TC, David CN, Bode HR, Koizumi O, Shimizu H, et al. Systematic isolation of peptide signal molecules regulating development in hydra: LWamide and PW families. Proc Natl Acad Sci U S A 1997; 94:1241-6; PMID:9037037; http://dx.doi.org/ 10.1073/pnas.94.4.1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takahashi T. Neuropeptides and epitheliopeptides: structural and functional diversity in an ancestral metazoan hydra. Protein Pept Lett 2013; 20:671-80; PMID:23030717; http://dx.doi.org/ 10.2174/0929866511320060006 [DOI] [PubMed] [Google Scholar]
- 9. Hwang JS, Ohyanagi H, Hayakawa S, Osato N, Nishimiya-Fujisawa C, Ikeo K, David CN, Fujisawa T, Gojobori T. The evolutionary emergence of cell type-specific genes inferred from the gene expression analysis of hydra. Proc Natl Acad Sci U S A 2007; 104:14735-40; PMID:17766437; http://dx.doi.org/ 10.1073/pnas.0703331104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hemmrich G, Khalturin K, Boehm AM, Puchert M, Anton-Erxleben F, Wittlieb J, Klostermeier UC, Rosenstiel P, Oberg HH, Domazet-Loso T, et al. Molecular signatures of the three stem cell lineages in hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol 2012; 29:3267-80; PMID:22595987; http://dx.doi.org/ 10.1093/molbev/mss134 [DOI] [PubMed] [Google Scholar]
- 11. Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci U S A 2006; 103:6208-11; PMID:16556723; http://dx.doi.org/ 10.1073/pnas.0510163103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anton-Erxleben F, Thomas A, Wittlieb J, Fraune S, Bosch TC. Plasticity of epithelial cell shape in response to upstream signals: a whole-organism study using transgenic hydra. Zoology (Jena) 2009; 112:185-94; PMID:19201587; http://dx.doi.org/ 10.1016/j.zool.2008.09.002 [DOI] [PubMed] [Google Scholar]
- 13. Buzgariu W, Crescenzi M, Galliot B. Robust G2 pausing of adult stem cells in hydra. Differentiation 2014; 87:83-99; PMID:24703763; http://dx.doi.org/ 10.1016/j.diff.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 14. Wenger Y, Buzgariu W, Galliot B. Systematic analysis of gene regulations linked to the loss of neurogenesis in hydra. Philos Trans R Soc Lond B Biol Sci (in revision). 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lentz TL. 1966. Fine structure of the epidermis and mesoglea, The cell biology of hydra. North-Holland Publishing Company, Amsterdam, pp. 38-50 [Google Scholar]
- 16. Böttger A, Doxey AC, Hess MW, Pfaller K, Salvenmoser W, Deutzmann R, Geissner A, Pauly B, Altstätter J, Münder S, et al. Horizontal gene transfer contributed to the evolution of extracellular surface structures: the freshwater polyp hydra is covered by a complex fibrous cuticle containing glycosaminoglycans and proteins of the ppod and swt (sweet tooth) families. PLoS One 2012; 7:e52278; PMID:23300632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoffmeister-Ullerich SA, Herrmann D, Kielholz J, Schweizer M, Schaller HC. Isolation of a putative peroxidase, a target for factors controlling foot-formation in the coelenterate hydra. Eur J Biochem 2002; 269:4597-606; PMID:12230572; http://dx.doi.org/ 10.1046/j.1432-1033.2002.03159.x [DOI] [PubMed] [Google Scholar]
- 18. Thomsen S, Bosch TC. Foot differentiation and genomic plasticity in hydra: lessons from the ppod gene family. Dev Genes Evol 2006; 216:57-68; PMID:16402271; http://dx.doi.org/ 10.1007/s00427-005-0032-9 [DOI] [PubMed] [Google Scholar]
- 19. Martin-Belmonte F, Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nat Rev Cancer 2012; 12:23-38 PMID: 22169974; http://dx.doi.org/ 10.1038/nrc3169 [DOI] [PubMed] [Google Scholar]
- 20. Hulpiau P, van Roy F. New insights into the evolution of metazoan cadherins. Mol Biol Evol 2011; 28:647-57; PMID:20817718; http://dx.doi.org/ 10.1093/molbev/msq233 [DOI] [PubMed] [Google Scholar]
- 21. Ganot P, Zoccola D, Tambutte E, Voolstra CR, Aranda M, Allemand D, Tambutte S. Structural molecular components of septate junctions in cnidarians point to the origin of epithelial junctions in eukaryotes. Mol Biol Evol 2015; 32:44-62; PMID:25246700; http://dx.doi.org/ 10.1093/molbev/msu265 [DOI] [PubMed] [Google Scholar]
- 22. Fraser SE, Green CR, Bode HR, Gilula NB. Selective disruption of gap junctional communication interferes with a patterning process in hydra. Science 1987; 237:49-55; PMID:3037697; http://dx.doi.org/ 10.1126/science.3037697 [DOI] [PubMed] [Google Scholar]
- 23. Alexopoulos H, Böttger A, Fischer S, Levin A, Wolf A, Fujisawa T, Hayakawa S, Gojobori T, Davies JA, David CN, et al. Evolution of gap junctions: the missing link? Curr Biol 2004; 14:R879-80; PMID:15498476; http://dx.doi.org/ 10.1016/j.cub.2004.09.067 [DOI] [PubMed] [Google Scholar]
- 24. Sarras MPJ. Components, structure, biogenesis and function of the hydra extracellular matrix in regeneration, pattern formation and cell differentiation. Int J Dev Biol 2012; 56:567-76; PMID:22689358; http://dx.doi.org/ 10.1387/ijdb.113445ms [DOI] [PubMed] [Google Scholar]
- 25. Shimizu H, Zhang X, Zhang J, Leontovich A, Fei K, Yan L, Sarras MPJ. Epithelial morphogenesis in hydra requires de novo expression of extracellular matrix components and matrix metalloproteinases. Development 2002; 129:1521-32; PMID:11880360 [DOI] [PubMed] [Google Scholar]
- 26. Aufschnaiter R, Zamir EA, Little CD, Özbek S, Münder S, David CN, Li L, Sarras MP, Jr, Zhang X. In vivo imaging of basement membrane movement: ECM patterning shapes hydra polyps. J Cell Sci 2011; 124:4027-38; PMID:22194305; http://dx.doi.org/ 10.1242/jcs.087239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. David CN, Campbell RD. Cell cycle kinetics and development of hydra attenuata. I. Epithelial cells. J Cell Sci 1972; 11:557-68; PMID:5076361 [DOI] [PubMed] [Google Scholar]
- 28. Fluckiger AC, Marcy G, Marchand M, Négre D, Cosset FL, Mitalipov S, Wolf D, Savatier P, Dehay C. Cell cycle features of primate embryonic stem cells. Stem Cells 2006; 24:547-56; PMID:16239321; http://dx.doi.org/ 10.1634/stemcells.2005-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bosch TC, David CN. Growth regulation in hydra: relationship between epithelial cell cycle length and growth rate. Dev Biol 1984; 104:161-71; PMID:6734933; http://dx.doi.org/ 10.1016/0012-1606(84)90045-9 [DOI] [PubMed] [Google Scholar]
- 30. Dubel S, Schaller HC. Terminal differentiation of ectodermal epithelial stem cells of hydra can occur in g2 without requiring mitosis or s phase. J Cell Biol 1990; 110:939-45; PMID:2108971; http://dx.doi.org/ 10.1083/jcb.110.4.939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Okita K, Yamanaka S. Induction of pluripotency by defined factors. Exp Cell Res 2010; 316:2565-70; PMID:20420827; http://dx.doi.org/ 10.1016/j.yexcr.2010.04.023 [DOI] [PubMed] [Google Scholar]
- 32. Millane RC, Kanska J, Duffy DJ, Seoighe C, Cunningham S, Plickert G, Frank U. Induced stem cell neoplasia in a cnidarian by ectopic expression of a pou domain transcription factor. Development 2011; 138:2429-39; PMID:21610024; http://dx.doi.org/ 10.1242/dev.064931 [DOI] [PubMed] [Google Scholar]
- 33. Hobmayer B, Jenewein M, Eder D, Eder MK, Glasauer S, Gufler S, Hartl M, Salvenmoser W. Stemness in hydra - a current perspective. Int J Dev Biol 2012; 56:509-17; PMID:22689357; http://dx.doi.org/ 10.1387/ijdb.113426bh [DOI] [PubMed] [Google Scholar]
- 34. Hartl M, Mitterstiller AM, Valovka T, Breuker K, Hobmayer B, Bister K. Stem cell-specific activation of an ancestral myc protooncogene with conserved basic functions in the early metazoan hydra. Proc Natl Acad Sci U S A 2010; 107:4051-6; PMID:20142507; http://dx.doi.org/ 10.1073/pnas.0911060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hartl M, Glasauer S, Valovka T, Breuker K, Hobmayer B, Bister K. Hydra myc2, a unique pre-bilaterian member of the myc gene family, is activated in cell proliferation and gametogenesis. Biol Open 2014; 3:397-407; PMID:24771621; http://dx.doi.org/ 10.1242/bio.20147005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ambrosone A, Marchesano V, Tino A, Hobmayer B, Tortiglione C. Hymyc1 downregulation promotes stem cell proliferation in hydra vulgaris. PLoS One 2012; 7:e30660; PMID:22292012; http://dx.doi.org/ 10.1371/journal.pone.0030660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharrocks AD. The ets-domain transcription factor family. Nat Rev Mol Cell Biol 2001; 2:827-37; PMID:11715049; http://dx.doi.org/ 10.1038/35099076 [DOI] [PubMed] [Google Scholar]
- 38. Bridge D, Theofiles AG, Holler RL, Marcinkevicius E, Steele RE, Martinez DE. FoxO and stress responses in the cnidarian hydra vulgaris. PLoS One 2010; 5:e11686; PMID:20657733; http://dx.doi.org/ 10.1371/journal.pone.0011686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boehm AM, Khalturin K, Anton-Erxleben F, Hemmrich G, Klostermeier UC, Lopez-Quintero JA, Oberg HH, Puchert M, Rosenstiel P, Wittlieb J, et al. Foxo is a critical regulator of stem cell maintenance in immortal hydra. Proc Natl Acad Sci U S A 2012; 109:19697-702; PMID:23150562; http://dx.doi.org/ 10.1073/pnas.1209714109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Juliano CE, Reich A, Liu N, Götzfried J, Zhong M, Uman S, Reenan RA, Wessel GM, Steele RE, Lin H. Piwi proteins and piwi-interacting rnas function in hydra somatic stem cells. Proc Natl Acad Sci U S A 2014; 111:337-42; PMID:24367095; http://dx.doi.org/ 10.1073/pnas.1320965111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lim RS, Anand A, Nishimiya-Fujisawa C, Kobayashi S, Kai T. Analysis of hydra piwi proteins and pirnas uncover early evolutionary origins of the pirna pathway. Dev Biol 2014; 386:237-51; PMID:24355748; http://dx.doi.org/ 10.1016/j.ydbio.2013.12.007 [DOI] [PubMed] [Google Scholar]
- 42. Lentz TL. 1966; Cell biology of hydra. North Holland Pub. Co, Amsterdam [Google Scholar]
- 43. Passano LM, McCullough CB. Co-ordinating systems and behaviour in hydra. I. Pacemaker system of the periodic contractions. J Exp Biol 1964; 41:643-64 [DOI] [PubMed] [Google Scholar]
- 44. Passano LM, McCullough CB. Co-ordinating systems and behaviour in hydra. II. The rhythmic potential system. J Exp Biol 1965; 42:205-31; PMID:14328679 [DOI] [PubMed] [Google Scholar]
- 45. Campbell RD, Josephson RK, Schwab WE, Rushforth NB. Excitability of nerve-free hydra. Nature 1976; 262:388-90; PMID:958390; http://dx.doi.org/ 10.1038/262388a0 [DOI] [PubMed] [Google Scholar]
- 46. Shimizu H, Koizumi O, Fujisawa T. Three digestive movements in hydra regulated by the diffuse nerve net in the body column. J Comp Physiol A 2004; 190:623-30; http://dx.doi.org/ 10.1007/s00359-004-0518-3 [DOI] [PubMed] [Google Scholar]
- 47. Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, Chera S. Origins of neurogenesis, a cnidarian view. Dev Biol 2009; 332:2-24; PMID:19465018; http://dx.doi.org/ 10.1016/j.ydbio.2009.05.563 [DOI] [PubMed] [Google Scholar]
- 48. McDowall AW, Grimmelikhuijzen CJ. Intercellular junctions in nerve-free hydra. Cell Tissue Res 1980; 209:217-24; PMID:7397766 [DOI] [PubMed] [Google Scholar]
- 49. Takaku Y, Hwang JS, Wolf A, Böttger A, Shimizu H, David CN, Gojobori T. Innexin gap junctions in nerve cells coordinate spontaneous contractile behavior in hydra polyps. Sci Rep 2014; 4:3573; PMID:24394722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pierobon P. Coordinated modulation of cellular signaling through ligand-gated ion channels in hydra vulgaris (cnidaria, hydrozoa). Int J Dev Biol 2012; 56:551-65; PMID:22689363; http://dx.doi.org/ 10.1387/ijdb.113464pp [DOI] [PubMed] [Google Scholar]
- 51. Grunder S, Assmann M. Peptide-gated ion channels and the simple nervous system of hydra. J Exp Biol 2015; 218:551-61; PMID:25696818; http://dx.doi.org/ 10.1242/jeb.111666 [DOI] [PubMed] [Google Scholar]
- 52. Beckmann A, Ozbek S. The nematocyst: a molecular map of the cnidarian stinging organelle. Int J Dev Biol 2012; 56:577-82; PMID:22689365; http://dx.doi.org/ 10.1387/ijdb.113472ab [DOI] [PubMed] [Google Scholar]
- 53. Davis LE. Histological and ultrastructural studies of the basal disk of hydra. Iii. The gastrodermis and the mesoglea. Cell Tissue Res 1975; 162:107-18; PMID:1182023; http://dx.doi.org/ 10.1007/BF00223266 [DOI] [PubMed] [Google Scholar]
- 54. Böttger A, Alexandrova O. Programmed cell death in hydra. Semin Cancer Biol 2007; 17:134-46; PMID:17197196; http://dx.doi.org/ 10.1016/j.semcancer.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 55. Buzgariu W, Chera S, Galliot B. Methods to investigate autophagy during starvation and regeneration in hydra. Methods Enzymol 2008; 451:409-37; PMID:19185734; http://dx.doi.org/ 10.1016/S0076-6879(08)03226-6 [DOI] [PubMed] [Google Scholar]
- 56. Chera S, Buzgariu W, Ghila L, Galliot B. Autophagy in hydra: a response to starvation and stress in early animal evolution. Biochim Biophys Acta 2009a; 1793:1432-43; http://dx.doi.org/ 10.1016/j.bbamcr.2009.03.010 [DOI] [PubMed] [Google Scholar]
- 57. Chera S, de Rosa R, Miljkovic-Licina M, Dobretz K, Ghila L, Kaloulis K, Galliot B. Silencing of the hydra serine protease inhibitor kazal1 gene mimics the human spink1 pancreatic phenotype. J Cell Sci 2006; 119:846-57; PMID:16478786; http://dx.doi.org/ 10.1242/jcs.02807 [DOI] [PubMed] [Google Scholar]
- 58. Galliot B. Autophagy and self-preservation: a step ahead from cell plasticity? Autophagy 2006; 2:231-33; PMID:16874084; http://dx.doi.org/ 10.4161/auto.2706 [DOI] [PubMed] [Google Scholar]
- 59. Reiter S, Crescenzi M, Galliot B, Buzgariu W. Hydra, a versatile model to study the homeostatic and developmental functions of cell death. Int J Dev Biol 2012; 56:593-604; PMID:22689371; http://dx.doi.org/ 10.1387/ijdb.123499sr [DOI] [PubMed] [Google Scholar]
- 60. Campbell RD. Elimination by hydra interstitial and nerve cells by means of colchicine. J Cell Sci 1976; 21:1-13; PMID:932105 [DOI] [PubMed] [Google Scholar]
- 61. Cikala M, Wilm B, Hobmayer E, Böttger A, David CN. Identification of caspases and apoptosis in the simple metazoan hydra. Curr Biol 1999; 9:959-62; PMID:10508589; http://dx.doi.org/ 10.1016/S0960-9822(99)80423-0 [DOI] [PubMed] [Google Scholar]
- 62. Chera S, Ghila L, Dobretz K, Wenger Y, Bauer C, Buzgariu W, Martinou JC, Galliot B. Apoptotic cells provide an unexpected source of wnt3 signaling to drive hydra head regeneration. Dev Cell 2009b; 17:279-89; http://dx.doi.org/ 10.1016/j.devcel.2009.07.014 [DOI] [PubMed] [Google Scholar]
- 63. Murate M, Kishimoto Y, Sugiyama T, Fujisawa T, Takahashi-Iwanaga H, Iwanaga T. Hydra regeneration from recombined ectodermal and endodermal tissue. II. Differential stability in the ectodermal and endodermal epithelial organization. J Cell Sci 1997; 110 (Pt 16):1919-34; PMID:9296391 [DOI] [PubMed] [Google Scholar]
- 64. Bosch TC. Rethinking the role of immunity: lessons from hydra. Trends Immunol 2014; 35:495-502; PMID:25174994; http://dx.doi.org/ 10.1016/j.it.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 65. Bosch TC, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, Rosenstiel P, Jacobs G, Schreiber S, Leippe M, et al. Uncovering the evolutionary history of innate immunity: the simple metazoan hydra uses epithelial cells for host defence. Dev Comp Immunol 2009; 33:559-69; PMID:19013190; http://dx.doi.org/ 10.1016/j.dci.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 66. Lange C, Hemmrich G, Klostermeier UC, López-Quintero JA, Miller DJ, Rahn T, Weiss Y, Bosch TC, Rosenstiel P. Defining the origins of the nod-like receptor system at the base of animal evolution. Mol Biol Evol 2011; 28:1687-702; PMID:21183612; http://dx.doi.org/ 10.1093/molbev/msq349 [DOI] [PubMed] [Google Scholar]
- 67. Franzenburg S, Fraune S, Kunzel S, Baines JF, Domazet-Loso T, Bosch TC. Myd88-deficient hydra reveal an ancient function of tlr signaling in sensing bacterial colonizers. Proc Natl Acad Sci U S A 2012; 109:19374-9; PMID:23112184; http://dx.doi.org/ 10.1073/pnas.1213110109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Franzenburg S, Walter J, Kunzel S, Wang J, Baines JF, Bosch TC, Fraune S. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci U S A 2013; 110:e3730-8; PMID:24003149; http://dx.doi.org/ 10.1073/pnas.1304960110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, Agata K, Bosch TC. The innate immune repertoire in cnidaria–ancestral complexity and stochastic gene loss. Genome Biol 2007; 8:R59; PMID:17437634; http://dx.doi.org/ 10.1186/gb-2007-8-4-r59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Augustin R, Anton-Erxleben F, Jungnickel S, Hemmrich G, Spudy B, Podschun R, Bosch TC. Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob Agents Chemother 2009a; 53:5245-50; http://dx.doi.org/ 10.1128/AAC.00826-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jung S, Dingley AJ, Augustin R, Anton-Erxleben F, Stanisak M, Gelhaus C, Gutsmann T, Hammer MU, Podschun R, Bonvin AM, et al. Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan hydra. J Biol Chem 2009; 284:1896-905; PMID:19019828; http://dx.doi.org/ 10.1074/jbc.M804713200 [DOI] [PubMed] [Google Scholar]
- 72. Augustin R, Siebert S, Bosch TC. Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of hydra's innate immune system. Dev Comp Immunol 2009b; 33:830-7; http://dx.doi.org/ 10.1016/j.dci.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 73. Rahat M, Dimentman C. Cultivation of bacteria-free hydra viridis: missing budding factor in nonsymbiotic hydra. Science 1982; 216:67-8; PMID:7063873; http://dx.doi.org/ 10.1126/science.7063873 [DOI] [PubMed] [Google Scholar]
- 74. Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan hydra. Proc Natl Acad Sci U S A 2007; 104:13146-51; PMID:17664430; http://dx.doi.org/ 10.1073/pnas.0703375104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fraune S, Anton-Erxleben F, Augustin R, Franzenburg S, Knop M, Schroder K, Willoweit-Ohl D, Bosch TC. Bacteria-bacteria interactions within the microbiota of the ancestral metazoan hydra contribute to fungal resistance. ISME J 2014; 9(7):1543-56; PMID:25514534; http://dx.doi.org/ 10.1038/ismej.2014.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kasahara S, Bosch TC. Enhanced antibacterial activity in hydra polyps lacking nerve cells. Dev Comp Immunol 2003; 27:79-85; PMID:12543122; http://dx.doi.org/ 10.1016/S0145-305X(02)00073-3 [DOI] [PubMed] [Google Scholar]
- 77. Fraune S, Abe Y, Bosch TC. Disturbing epithelial homeostasis in the metazoan hydra leads to drastic changes in associated microbiota. Environ Microbiol 2009; 11:2361-9; PMID:19508335; http://dx.doi.org/ 10.1111/j.1462-2920.2009.01963.x [DOI] [PubMed] [Google Scholar]
- 78. Galliot B, Welschof M, Schuckert O, Hoffmeister S, Schaller HC. The cAMP response element binding protein is involved in hydra regeneration. Development 1995; 121:1205-16; PMID:7743932 [DOI] [PubMed] [Google Scholar]
- 79. Kaloulis K, Chera S, Hassel M, Gauchat D, Galliot B. Reactivation of developmental programs: the cAMP-response element-binding protein pathway is involved in hydra head regeneration. Proc Natl Acad Sci U S A 2004; 101:2363-8; PMID:14983015; http://dx.doi.org/ 10.1073/pnas.0306512101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chera S, Ghila L, Wenger Y, Galliot B. Injury-induced activation of the mapk/CREB pathway triggers apoptosis-induced compensatory proliferation in hydra head regeneration. Dev Growth Differ 2011; 53:186-201; PMID:21338345; http://dx.doi.org/ 10.1111/j.1440-169X.2011.01250.x [DOI] [PubMed] [Google Scholar]
- 81. Nakamura Y, Tsiairis CD, Ozbek S, Holstein TW. Autoregulatory and repressive inputs localize hydra wnt3 to the head organizer. Proc Natl Acad Sci U S A 2011; 108:9137-42; PMID:21576458; http://dx.doi.org/ 10.1073/pnas.1018109108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wenger Y, Buzgariu W, Reiter S, Galliot B. Injury-induced immune responses in hydra. Semin Immunol 2014; 26:277-94; PMID:25086685; http://dx.doi.org/ 10.1016/j.smim.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 83. Browne EN. The production of new hydranths in hydra by the insertion of small grafts. J Exp Zool 1909; 7:1-37; http://dx.doi.org/ 10.1002/jez.1400070102 [DOI] [Google Scholar]
- 84. MacWilliams HK. Hydra transplantation phenomena and the mechanism of hydra head regeneration. Ii. Properties of the head activation. Dev Biol 1983; 96:239-57; PMID:6825956; http://dx.doi.org/ 10.1016/0012-1606(83)90325-1 [DOI] [PubMed] [Google Scholar]
- 85. Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H, Holstein TW. Multiple wnts are involved in hydra organizer formation and regeneration. Dev Biol 2009; 330:186-99; PMID:19217898; http://dx.doi.org/ 10.1016/j.ydbio.2009.02.004 [DOI] [PubMed] [Google Scholar]
- 86. Shimizu H. Transplantation analysis of developmental mechanisms in hydra. Int J Dev Biol 2012; 56:463-72; PMID:22689370; http://dx.doi.org/ 10.1387/ijdb.123498hs [DOI] [PubMed] [Google Scholar]
- 87. Galliot B, Miljkovic-Licina M, de Rosa R, Chera S. Hydra, a niche for cell and developmental plasticity. Semin Cell Dev Biol 2006; 17:492-502; PMID:16807002; http://dx.doi.org/ 10.1016/j.semcdb.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 88. Petersen HO, Höger SK, Looso M, Lengfeld T, Kuhn A, Warnken U, Nishimiya-Fujisawa C, Schnölzer M, Krüger M, Özbek S, et al. A comprehensive transcriptomic and proteomic analysis of hydra head regeneration. Mol Biol Evol 2015; PMID:25841488; http://dx.doi.org/ 10.1093/molbev/msv079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gierer A. The Hydra model - a model for what? Int J Dev Biol 2012; 56:437-45; PMID:22451043; http://dx.doi.org/ 10.1387/ijdb.113458ag [DOI] [PubMed] [Google Scholar]
- 90. Campbell RD. Tissue dynamics of steady state growth in hydra littoralis. Ii. Patterns of tissue movement. J Morphol 1967; 121:19-28; PMID:4166265; http://dx.doi.org/ 10.1002/jmor.1051210103 [DOI] [PubMed] [Google Scholar]
- 91. Wenger Y, Galliot B. Punctuated emergences of genetic and phenotypic innovations in eumetazoan, bilaterian, euteleostome, and hominidae ancestors. Genome Biol Evol 2013a; 5:1949-68; http://dx.doi.org/ 10.1093/gbe/evt142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wenger Y, Galliot B. RNAseq versus genome-predicted transcriptomes: a large population of novel transcripts identified in an illumina-454 hydra transcriptome. BMC Genomics 2013b; 14:204; http://dx.doi.org/ 10.1186/1471-2164-14-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


