Abstract
Rapid and accurate diagnosis of influenza is important for patient management and infection control. We determined the performance of the cobas® Influenza A/B assay, a rapid automated nucleic acid assay performed on the cobas® Liat System for qualitative detection of influenza A and influenza B from nasopharyngeal (NP) swab specimens. Retrospective frozen and prospectively collected NP swabs from patients with signs and symptoms of influenza collected in universal transport medium (UTM) were tested at multiple sites including CLIA-waived sites using the cobas® Influenza A/B assay. Results were compared to the Prodesse Pro-Flu+ assay and to viral culture. Compared to the Prodesse ProFlu+ Assay, sensitivities of the cobas® Influenza A/B assay for influenza A and B were 97.7 and 98.6%, respectively; specificity was 99.2 and 99.4%. Compared to viral culture, the cobas® Influenza A/B assay showed sensitivities of 97.5 and 96.9% for influenza virus A and B, respectively; specificities were 97.9% for both viruses. Polymerase chain reaction (PCR)/sequencing showed that the majority of viral culture negative but cobas® Influenza A/B positive results were true positive results, indicating that the cobas® Influenza A/B assay has higher sensitivity compared to viral culture.
In conclusion, the excellent accuracy, rapid time to result, and remarkable ease of use make the cobas® Influenza A/B nucleic acid assay for use on the cobas® Liat System a highly suitable point-of-care solution for the management of patients with suspected influenza A and B infection.
Keywords: influenza, influenza A, influenza B, diagnosis, PCR, viral culture, sensitivity, point of care
Introduction
Influenza is a contagious respiratory illness caused by influenza viruses A and B in humans. Worldwide, three to five million individuals develop severe influenza each year and 250,000 to 500,000 die of influenza-related causes [1]. Even in developed countries, such as the United States, influenza is responsible for more than 200,000 hospitalizations annually and 3000 to 49,000 deaths [2]. While hospitalization costs are important contributors, lost productivity from missed work days and lost lives comprise the bulk of the economic burden of influenza [3]. Moreover, as illustrated by the 2009 H1N1 pandemic that affected hundreds of countries, influenza has the potential to rapidly spread globally.
Early identification of influenza is important for optimal patient management and infection control. However, case definitions of influenza-like illness including cough, sore throat, rhinorrhea, and nasal congestion only have modest sensitivity and specificity [4, 5]. For this reason, physicians rely on the use of laboratory tests to diagnose influenza [6] and initiate prompt administration of antiviral therapy, mainly oseltamivir [7]. Additional benefits of rapid identification are infection control, public health notification and tracking, and prevention of unnecessary use of antibiotics, hospital procedures, and laboratory tests [8, 9].
Current diagnostic techniques for the detection and identification of influenza virus include rapid influenza detection tests (RIDTs), viral culture, and nucleic acid amplification tests (NAAT). Commercially available RIDTs are widely used in clinical practice as point of care tests because they are simple to use and provide results within 15 to 30 min [10–12]. However, their sensitivities vary widely depending on the manufacturer and can be as low as 10%, with specificities ranging from 90 to 100% [10, 13]. Viral culture has increased sensitivity over both RIDTs and DFAs but requires skilled technologists and specialized laboratory settings and has a long turnaround time (2 to 14 days) [14]. NAAT are highly sensitive and are replacing culture as the gold standard, but these tests are generally more expensive, require highly skilled molecular technologists, and have turnaround times of up to 24 h from receipt to results [15–20]. In clinical practice, specimens with negative RIDTs are usually tested subsequently by more sensitive culture or molecular assays. Based on the poor performance of RIDTs, the Food and Drug Administration (FDA) has recently proposed tightening standards for rapid infl uenza tests in order to avoid impeding of diagnosis and treatment of influenza patients (http://www.gpo.gov/fdsys/pkg/FR-2014-05-22/html/2014-11635.htm?source=govdelivery&utm_medium=email&utm_ source=govdelivery). To mitigate the risks of false-negative and (equally detrimental) false-positive results from RIDTs, special measures that identify the minimum acceptable performance criteria, identify appropriate comparators for establishing performance of new assays, and mandate annual analytical reactivity testing of contemporary influenza strains should be implemented.
Polymerase chain reaction (PCR)-based molecular assays have shown excellent clinical utility for the detection and identification of influenza viruses; numerous FDA-cleared commercial devices are now available [17, 21, 22]. More recently, an isothermal amplification-based test with fast turnaround time and simplicity has been described [23, 24].
In the present study, we investigated the performance of the cobas® Influenza A/B assay (Roche Molecular Systems, Pleasanton, CA) for the detection and differentiation of influenza A and influenza B viruses in nasopharyngeal swab (NPS) specimens. We observed very high sensitivity and specificity for the detection of both influenza A and B viruses compared to viral culture and PCR/sequencing. High accuracy combined with the ease of use and rapid time to result should prove useful for the management of patients with suspected influenza.
Materials and methods
Clinical study design
Clinical trials to demonstrate the clinical performance of the cobas® Influenza A/B assay on the cobas® Liat System were performed using two sets of patient samples. First, nasopharyngeal swabs (retrospective samples) obtained in 2013–2014 from TriCore Reference Laboratories (Albuquerque, NM) and Lahey Clinic (Burlington, MA) were archived at −80 °C upon receipt. Samples were distributed to three CLIA waived sites (Advanced Pediatrics, Vienna, VT; Meridian Clinical Research, Bellevue, NE; Med Center Medical Clinic, Fair Oaks, CA) for testing, and results were compared to those obtained in the Prodesse ProFlu+ Assay (Hologic, MA, USA) [22, 25].
During the 2008/9, 2013/14, and 2014/15 flu seasons, 12 CLIA-waived sites in the US prospectively collected nasopharyngeal swabs. Of these, nine sites were primary care offices, and three sites were hospital emergency departments. Thirty-three operators participated in the study, including ten nurses, 14 medical assistants, one administrative assistant, one nurse aide, two personnel with Emergency Medical Technician training, and five other personnel with no formal medical training. Patients were enrolled based on the following inclusion criteria: presence (self-reported) of two or more flu-like clinical signs and symptoms (fever; headache; extreme tiredness; dry cough; sore throat; runny or stuffy nose; muscle aches; and gastrointestinal symptoms, such as nausea, vomiting, and diarrhea) within the past 48 h. Subjects taking antiviral medication at the time of the visit or within 7 days of the visit or who had received a nasal flu vaccine within the last 6 weeks were excluded. Nasopharyngeal swabs collected in universal transport medium (UTM) from each patient were tested using the cobas® Influenza A/B assay at clinical sites, and the Prodesse ProFlu+ Assay (Hologic, MA, USA) [22, 25], and viral culture at reference laboratories. To prevent sampling bias, samples from the same vial used for testing with the cobas® Influenza A/B assay were used for viral culture and the ProFlu+ assay, if available. Bidirectional sequencing was used to investigate discordant results.
cobas® Liat System
The cobas® Liat System is a new point-of-care instrument with sample-to-answer capabilities in which all sample processing steps as well as detection are carried out using a single-use, disposable Liat Tube (Fig. 1A–C). Each test cartridge handles one sample and has a unique barcode identifier that is scanned into the cobas® Liat System to code the sample ID. An NPS sample is loaded directly into a Liat assay tube using a transfer pipette. After the tube is capped, the cobas® Liat System scans the tube barcode, and the tube is inserted into the analyzer. Specimen sampling and handling is controlled using multiple sample processing modules contained within the cobas® Liat System. The sample processing modules are composed of two assemblies, a moving side assembly comprised of multiple sample processing plungers and clamps, and a fixed side assembly. When performing an assay, a Liat tube is inserted into the tube slot of a cobas® Liat System. The plungers and clamps selectively compress the Liat tube segments against the fixed side assembly to release reagents from the segments, move the sample from one segment to another, and control reaction conditions. Test cartridges are single use and part of a closed system. An internal control used in conjunction with procedural checks monitors instrument functionality, performance, fluidics, and result determination based on a predefined decision algorithm. The cobas® Liat System is 510(k) cleared by the US Food and Drug Administration (FDA).
Fig. 1.
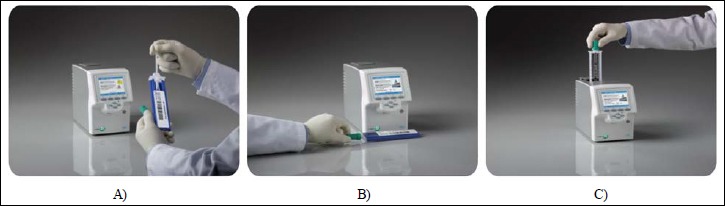
cobas® Influenza A/B assay workflow. A sample is collected directly into a Liat Tube (A). After the tube is capped, the analyzer scans the tube barcode (B), and the tube is inserted in the analyzer (C). Then, the analyzer automatically performs all the nucleic acid extraction and amplification steps and reports results in ~20 min
cobas® Influenza A/B assay
The cobas® Influenza A/B assay uses an established nucleic acid test chemistry and assay protocol for the detection of viral RNA. The sample preparation methodology is based on lysis by a chaotropic agent followed by magnetic particle based nucleic acid purification. First, the NPS sample in UTM is diluted and mixed with an internal process control (IPC). Second, nucleic acids are isolated from lysates through binding to silica magnetic beads in the presence of the chaotropic salt followed by removal of possible inhibitors. Target amplification and detection use TaqMan-probe based real-time polymerase chain reaction (RT-PCR). The influenza A primer and probe set detects matrix RNA from type A influenza virus, and influenza B primer and probe sets detect nonstructural protein RNA from influenza B viruses. An IPC primer and probe set is also included to amplify the target region of the internal control. Dual-labeled fluorogenic hydrolysis (TaqMan) probes anneal to specific target sequences and are degraded causing reporter dyes to separate from the quenchers, thereby generating fluorescent signals and cycle threshold (Ct) values for the specific analytes.
The internal process control (IPC) comprises an encapsulated RNA that is prepacked in each Liat tube. When conducting an assay, it is first mixed with sample and then processed through all the test steps to monitor both the sample preparation and the RT-PCR reaction performance. The IPC RNA is detected in a separate channel by IPC specific primers and probe.
Positive control is provided in the cobas® Influenza A/B assay Quality Control Kit. The positive control comprises of inactivated Influenza A and B virus. Negative control is provided in the cobas® Influenza A/B assay Quality Control Kit. The negative control comprises UTM.
The limit of detection (LOD) of the assay was determined as the lowest log virus concentration that was detected ≥95% of the time (i.e., log concentration at which at least 19 out of 20 replicates tested positive). The LOD for three strains of Influenza A were 10-2 to 10-1 TCID50/ ml, while those for the two strains of Influenza B were 10-3 to 10-1 TCID50/ml. The reactivity of the cobas® Influenza A/B assay has been confirmed for a large number of influenza A and influenza B strains from multiple geographical regions including contemporary strains (cobas® Influenza A/B assay package insert). The assay demonstrated no cross reactivity with a large number of non-influenza respiratory pathogens and other microorganisms which may be found in NPS specimens, nor did the presence of such other microorganisms interfere with the detection of influenza. The cobas® Influenza A/B assay does not show inhibition when nasopharyngeal samples contain relevant concentrations of potentially interfering substances including antiviral (zanamivir, oseltamivir), antibacterial (tobramycin, muporicin) drugs, nasal sprays (ozymetazoline, flutocasone), throat lozenges, anaesthetics, analgesics, or nasal gels.
The workflow of the cobas® Influenza A/B assay is shown in Fig. 1. The entire workflow takes ~1 min of human resource time, and the assay generates results in only 20 min.
The assay is 510(k)-cleared by the FDA, and the FDA has recently granted CLIA (Clinical Laboratory Improvement Amendments) waiver for the cobas® Influenza A/B test for use on the cobas® Liat System.
Performance using fresh vs. frozen samples
The performance of the cobas® Influenza A/B assay was tested comparing results obtained using fresh and frozen specimens. One influenza A strain (A/Brisbane/10/07) and one influenza B strain (B/Malaysia/2506/04) were individually spiked into NPS matrix at different viral loads, including levels near LOD and levels reflecting the clinical range. For each strain, 60 samples were tested immediately while another 60 samples were frozen at −80 °C for 7 days, thawed, and then tested.
Reference testing
Reference testing using viral culture was performed concurrent with the prospective sample collection in 2008 and 2009 by Laboratory Corporation of America at its ViroMed Laboratories (Minnetonka, MN). Reference testing comprised influenza A or B positive shell vial culture and indirect fluorescent antibody (IFA) procedures using Millipore Influenza Mab A, B IFA Reagent. Viral culture for clinical samples collected in the 2013–2014 and the 2014–1015 flu seasons was performed by Focus Diagnostics Inc. (Cypress, CA) using direct fluorescent antibody staining with the FDA-cleared D3 Ultra DFA Respiratory Virus Screening Kit (Diagnostic Hybrids, Athens, OH). Reference testing using the Prodesse ProFlu+ Assay was performed by Cleveland Clinic (Cleveland, OH).
PCR and bidirectional sequencing was used as the reference method for retrospective samples, as well as to investigate discordances in prospective sample results. The protocol utilized was adapted from Ghedin et al. [26, 27] and World Health Organization Sequencing Primers and Protocols (WHO Sequencing primers & protocol, May 12, 2009, WHO Sequencing Primers & Protocol, www.who.int/entity/csr/resources/publications/swineflu/GenomePrimers_20090512.pdf). For influenza A, due to mismatches between the 2009 H1N1 strain and the Inf A primers reported in the WHO Sequencing Primers and Protocols, a 2009 H1N1 specific primer set was derived by modifying the WHO Inf A primer sequence to be complementary to the 2009 H1N1 sequence, and was used to test all Inf A discordances in addition to the WHO Inf A primer set. Briefly, RNA extraction was performed on 200 μl of each sample using the Qiagen Viral RNA Mini Kit following the kit instructions for use. RNA from each sample was eluted in 60 μl of 10 mM Tris Buffer, pH 8.5. Inf A discordant samples were tested with both the Inf A and 2009 H1N1 primer sets. For influenza B, discordant samples were tested using the Inf B primer sets. Twenty microliters (20 μl) of the purified RNA was used as template in a 30 μl RT-PCR reaction using the Qiagen One-Step RT-PCR kit. The first RT-PCR was performed using a thermal protocol comprising: 1) a reverse transcription step of 50 °C for 30 min; 2) an initial denaturation step of 94 °C for 15 min; 3) 50 cycles of denaturing at 94 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 60 s; and 4) a final extension step of 72 °C for 10 min. First RT-PCR products were diluted 1:100 in molecular grade water and used as template in the subsequent second PCR. Two microliters (2 μl) of the diluted first RT-PCR product was combined with 28 μl PCR Mastermix containing the corresponding second (nested) PCR primers. The second PCR was performed using a thermal protocol comprising: 1) an initial denaturation step of 94 °C for 2 min; 2) 30 cycles of denaturing at 94 °C for 15 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s. After amplification, five microliters (5 μl) of second PCR product was loaded onto a 2% agarose gel and electrophoresed at 100 volts using 1× TAE buffer. After electrophoresis, gels were visualized under ultraviolet (UV) light and appropriately sized gel bands were excised. DNA from each gel fragment was purified with the Qiagen MinElute Gel Extraction Kit and eluted in 20 μl of 10 mM Tris buffer, pH 8.5. Two microliters (2 μl) of the gel extract was combined with 0.5 μl of 100 μM sequencing primer and 17.5 μl of ddH2O for a total of 20 μl primer–target mix. The mix was then sent to the Massachusetts General Hospital DNA Core Facility for sequencing. After obtaining the sequencing data, NCBI BLAST search was used against the nucleotide collection (nr/nt) database. Acceptance criteria for sequence analysis were a) sequence contains a minimum of 200 overlapping contiguous bases, b) bases have a Quality Value of 20 or higher as measured by PHRED (probability of an error of 1% or lower), and c) sequence matches the reference or consensus sequence with an expected value (E value) <10-30 for the specific target. If only one direction of the bidirectional sequencing data met the acceptance criteria, the sequencing procedure was repeated by performing PCR using diluted amplicon from the previous PCR as template. The new PCR product was then sent for bidirectional sequencing. If the retest still did not yield results where both directions met the acceptance criteria, the sample was called “indeterminate”.
Positive and negative controls were tested with each batch of samples. Positive controls comprised of NP swab matrix spiked with A/NY/01/2009 H1N1, A/Brisbane/59/07, and B/Malaysia/2506/04, respectively, to give a final concentration of 1.0 TCID50/ml. Negative control comprised blank NP swab matrix.
Statistical analysis
Sensitivity and specificity of the cobas® Influenza A/B assay were determined and compared to reference results including the Prodesse ProFlu+ Assay and viral culture; results obtained by discordant analysis were not used to recalculate assay performance. Ninety-five percent confidence intervals are stated.
Ethical considerations
The study was approved by the Institutional Review Boards with waiver of Health Insurance Portability and Accountability Act of 1996 (HIPAA) authorization. All patients completed the Informed Consent Form. When under the age of 18, parents provided guardian consent; patients aged 6–17 provided the Minor Assent form. Exclusion criteria were the absence of flu-like symptoms and current antiviral medication.
Results
Performance of the cobas® Influenza A/B assay in fresh vs. frozen samples
The cobas® Influenza A/B assay detected 100% of all fresh and frozen samples across all tested levels of viral load including those near LOD. Of interest, Ct values observed in the cobas® Influenza A/B assay when performed on frozen samples compared to fresh samples showed a delay of 0.5 Ct on average for influenza A and 1.2 Ct on average for influenza B. The Ct delay reflects the degradation of virus during freeze-thaw cycles. The concordance of results demonstrates that the cobas® Influenza A/B assay had equivalent performance for fresh and frozen samples.
Comparison of the cobas® Influenza A/B assay with the Prodesse ProFlu+ assay in retrospective and prospective samples
Tables 1a and b show results comparing the cobas® Influenza A/B assay against the Prodesse ProFlu+ assay in 300 retrospective samples. The cobas® Influenza A/B assay demonstrated 98.8% sensitivity and 100% specificity for influenza A, and 100% sensitivity and 100% specificity for influenza B.
Table 1a.
Clinical performance of the cobas® Influenza A/B assay compared to the Prodesse ProFlu+ assay in retrospective samples for the detection of influenza A
| Influenza A | ProFlu+ | ||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Agreement (%) | 95% CI | |||
| cobas Liat | Positive | 79 | 0 | 79 | Positive | 98.8 | 93.3–99.8 |
| Negative | 1* | 220 | 221 | Negative | 100.0 | 98.3–100.0 | |
| Total | 80 | 220 | 300 | ||||
* One Liat negative, ProFlu+ positive specimen was negative by PCR/sequencing
Table 1b.
Clinical performance of the cobas® Influenza A/B assay compared to the Prodesse ProFlu+ assay in retrospective samples for the detection of influenza B
| Influenza B | ProFlu+ | ||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Agreement (%) | 95% CI | |||
| cobas Liat | Positive | 100 | 0 | 100 | Positive | 100.0 | 96.3–100.0 |
| Negative | 0 | 200 | 200 | Negative | 100.0 | 98.1–100.0 | |
| Total | 100 | 200 | 300 | ||||
Tables 2a and 2b show results comparing the cobas® Influenza A/B assay against the Prodesse ProFlu+ assay in 842 prospectively collected samples. The cobas® Influenza A/B assay demonstrated 96.9% sensitivity and 98.9% specificity for influenza A, and 95.5% sensitivity and 99.2% specificity for influenza B. Three and two “false negative” results were detected among prospectively collected samples for influenza A (n = 841) and B (n = 842), respectively, and eight and six “false positives” were reported for Influenza A and B, respectively. PCR and bidirectional sequencing used to investigate discordant results confirmed the positive cobas results in three and four cases; three and two negative cobas results were confirmed in two cases (Tables 2a and b).
Table 2a.
Comparison of the cobas® Influenza A/B assay with Prodesse ProFlu+ Assay on prospective samples for the detection of influenza A
| Influenza A | ProFlu+ | ||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Accuracy (%) | 95% CI | |||
| cobas Liat | Positive | 94 | 8* | 102 | Sensitivity | 96.9 | 91.3–98.9 |
| Negative | 3† | 736 | 739 | Specificity | 98.9 | 97.9–99.5 | |
| Total | 97 | 744 | 841‡ | ||||
* Of eight cobas Liat positive, ProFlu+ negative specimen, three were positive and five were negative by PCR/sequencing. One was positive, and seven were negative by culture
† Of three cobas Liat negative, ProFlu+ positive specimens, three were positive by PCR/sequencing. One was positive, one was negative, and one was not tested by culture
‡ One specimen was indeterminate for Inf A by ProFlu+ due to late Ct. This sample was positive for influenza A by cobas Liat and PCR/sequencing
Table 2b.
Comparison of the cobas® Influenza A/B assay with Prodesse ProFlu+ Assay on prospective samples for the detection of influenza B
| Influenza B | ProFlu+ | ||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Accuracy (%) | 95% CI | |||
| cobas Liat | Positive | 42 | 6* | 48 | Sensitivity | 95.5 | 84.9–98.7 |
| Negative | 72† | 792 | 794 | Specificity | 99.2 | 98.4–99.7 | |
| Total | 44 | 798 | 842 | ||||
* Of six cobas Liat positive, ProFlu+ negative specimens, four were positive and two were negative by PCR/sequencing. All six were negative by culture
† Of two cobas Liat negative, ProFlu+ positive specimens, one was positive and one was negative by PCR/sequencing. One was negative, and one was not tested by culture
Tables 3a and b show the combined performance characteristics of the cobas® Influenza A/B assay in 1,142 retrospective and prospective samples. Overall sensitivity of the cobas® Influenza A/B assay compared to the Prodesse ProFlu+ assay was 97.7% for influenza A and 98.6% for influenza B; specificities were 99.2% and 99.4% for influenza A and B, respectively.
Table 3a.
Comparison of the cobas® Influenza A/B assay with Prodesse ProFlu+ Assay on retrospective and prospective samples for the detection of influenza A
| Influenza A | Viral culture | ||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Accuracy (%) | 95% CI | |||
| cobas Liat | Positive | 173 | 8* | 181 | Sensitivity | 97.7 | 94.3–99.1 |
| Negative | 4† | 956 | 960 | Specificity | 99.2 | 98.4–99.6 | |
| Total | 177 | 964 | 1141‡ | ||||
* Of eight cobas Liat positive, ProFlu+ negative specimens, three were positive and five were negative by PCR/sequencing. One was positive, and seven were negative by viral culture
† Of four cobas Liat negative, ProFlu+ positive specimens, three were positive and one was negative by PCR/sequencing. One was positive, one was negative, and two were not tested by viral culture
‡ One specimen was indeterminate for Inf A by ProFlu+ due to late Ct. This sample was positive for Influenza A by cobas Liat and PCR/sequencing
Table 3b.
Comparison of the cobas® Influenza A/B assay with Prodesse ProFlu+ Assay on retrospective and prospective samples for the detection of influenza B
| Influenza B | Viral culture | ||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Accuracy (%) | 95% CI | |||
| cobas Liat | Positive | 142 | 6* | 148 | Sensitivity | 98.6 | 95.1–99.6 |
| Negative | 2† | 992 | 994 | Specificity | 99.4 | 98.7–99.7 | |
| Total | 144 | 998 | 1142 | ||||
* Of six cobas positive, ProFlu+ negative specimens, four were positive and two were negative by PCR/sequencing. All six were negative by viral culture
† Of two cobas negative, ProFlu+ positive specimens, one was positive and one was negative by PCR/sequencing. One was negative, and one was not tested by viral culture
Positive correlation (linear regression line slope = 0.97 and 0.87 for influenza A and B, respectively) was observed for Ct values between the Liat and ProFlu+ assays. On average, Liat Ct values were lower than those in the Pro-Flu+ assay by 3.2 for influenza A, and 3.7 for influenza B suggestive of higher PCR efficiency and higher assay sensitivity. Furthermore, the percent positive results in the Prodesse ProFlu+ assay correlated with the Ct values in the cobas® Influenza A/B assay. For both influenza A and B, the Prodesse ProFlu+ assay reported positive results for all samples with Liat assay Ct values of ≤ 25 (n = 108/108 for influenza A, n = 108/108 for influenza B). For specimens with 25 < Ct < 30 values in the cobas® Influenza A/B assay, we observed a ~94% detection rate in the Prodesse ProFlu+ assay (n = 45/48 for influenza A, n = 33/35 for influenza B). In samples with cobas® Influenza A/B assay Ct values of >30, the Prodesse ProFlu+ detection rate fell to ~80% for influenza A (n = 21/26) and ~20% for influenza B (n = 1/5). This trend underlines our analytical observations including a lower analytical LOD of the cobas® Influenza A/B assay compared to the Prodesse ProFlu+ assay, lower average Ct values in the cobas® Influenza A/B assay compared to Prodesse ProFlu+, and the assay performance near the cut-off (data not shown).
Performance of the cobas® Influenza A/B assay compared to viral culture in prospectively collected samples
Data from prospective samples were compared against viral culture as the reference method. Tables 4a and b show results comparing the cobas® Influenza A/B assay against viral culture for influenza A and B. Overall, the cobas® Influenza A/B assay demonstrated 97.5% sensitivity and 97.9% specificity for influenza A, and 96.9% sensitivity and 97.9% specificity for influenza B. Two and one “false negative” results were detected among 784 prospectively collected samples for influenza A and B, respectively, and 15 and 16 “false positives” were reported for influenza A and B, respectively. PCR and bidirectional sequencing used to investigate these discordant results confirmed the cobas result in the majority of cases (Tables 4a and b).
Table 4a.
Clinical performance of the cobas® Influenza A/B assay compared to viral culture on prospective samples for the detection of influenza A
| Influenza A | Viral culture | ||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Accuracy (%) | 95% CI | |||
| cobas Liat | Positive | 77 | 15* | 92 | Sensitivity | 97.5 | 91.2–99.3 |
| Negative | 2† | 690 | 692 | Specificity | 97.9 | 96.5–98.7 | |
| Total | 79 | 705 | 784 | ||||
* Of 15 cobas Liat positive, culture negative specimens, nine were positive and six were negative by PCR/sequencing. Seven were positive, one was indeterminate due to high Ct, and seven were negative by PoFlu+
† Of two cobas Liat negative, culture positive specimens, two were positive by PCR/sequencing. One was positive, and one was negative by ProFlu+
Table 4b.
Clinical performance of the cobas® Influenza A/B assay compared to viral culture on prospective samples for the detection of influenza B
| Influenza B | Viral culture | ||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Accuracy (%) | 95% CI | |||
| cobas Liat | Positive | 31 | 16* | 47 | Sensitivity | 96.9 | 84.3–99.4 |
| Negative | 1† | 736 | 737 | Specificity | 97.9 | 96.6–98.7 | |
| Total | 32 | 752 | 784 | ||||
* Of 16 cobas Liat positive, culture negative specimens, 14 were positive and two were negative by PCR/sequencing. Ten were positive, and six were negative by PoFlu+
† One cobas Liat negative, culture positive specimen was positive by PCR/sequencing and was negative by ProFlu+
Discussion
The ideal diagnostic technique for the management of patients with suspected influenza A and B must combine high accuracy, speed, and ease of use. While viral cell culture and molecular diagnostic techniques are considered the “gold standard” for detection of influenza A and B due to their high accuracy, they lack speed and ease of use [6, 10]. Rapid antigen tests are therefore commonly used in outpatient clinics, physicians’ offices, and in some hospitals despite the fact that they have insufficient sensitivity [10]. In the present multicenter study, we evaluated the performance of the cobas® Influenza A/B assay rapid point of care test that has recently been cleared and CLIA-waived by the FDA. Based on the results observed, the cobas® Influenza A/B assay addresses the requirements of accuracy, speed, and ease of use needed to provide management decisions for patients with suspected infection with influenza A and B.
First, using a collection of 300 retrospective nasopharyngeal swabs from U.S. sites, the cobas® Influenza A/B assay demonstrated sensitivities of 98.8 and 100% for the detection of influenza A and B, respectively, compared to the ProdessaFlu+ assay. Discordant testing using PCR/ sequencing supported the results obtained in the cobas® Influenza A/B test. The excellent sensitivity was accompanied by excellent specificities of 100% for both influenza A and B.
Second, we compared the cobas® Influenza A/B assay with the Prodesse ProFlu+ test using a large number of prospectively collected samples. Accuracy of the cobas® Influenza A/B assay was excellent and mirrored the results obtained in the retrospective sample set. Overall, the sensitivity and specificity of the cobas® Influenza A/B assay in retrospective and prospective samples were 97.7 and 98.6% for the detection of influenza A, respectively, and 98.6 and 99.4% for the detection of influenza B. PCR and bidirectional sequencing used to further investigate discordant results showed that the cobas® Influenza A/B assay detected influenza virus in certain specimens that gave negative results in the ProdessaFlu+ assay.
To confirm these findings, we then compared the cobas® Influenza A/B assay to viral culture considered the gold standard by some. The observed sensitivity was 97.5% for influenza A and 96.9% for influenza B. Similarly, this high sensitivity was achieved while maintaining a very high specificity of 97.9% for both influenza A and B. PCR and bidirectional sequencing used to investigate the discordant results confirmed the results obtained in the cobas® Influenza A/B assay in the majority of cases.
Recently, several PCR-based assays have been FDA-cleared for the detection of influenza A and B. The Luminex Xtag and Biofire FilmArray tests cover influenza A and B virus in addition to a number of other respiratory viruses [22, 28, 29] while the Prodesse ProFlu+ test detects influenza A and B and RSV [22] and the Cepheid Xpert Flu assay detects influenza A and B [9, 30]. Sensitivities of these assays for the detection of influenza virus A and B range between 90 and 100%. While showing slightly lower or similar accuracy compared to the cobas® Influenza A/B assay, these PCR-based tests do not offer the necessary speed and ease of use for use in environments other than the (centralized) laboratory.
Since rapid antigen tests are commonly performed as true point-of-care solutions in emergency rooms and physician’s offices, we also compared the cobas® Influenza A/B assay to rapid antigen tests. Compared to viral culture, we observed markedly higher sensitivities of the cobas® Influenza A/B assay vs. the rapid antigen tests (97.5 vs. 55.4% for influenza A, 96.9 vs. 71.4% for influenza B; data not shown). Specificities for the detection of influenza A and B were high for both methods. These results are consistent with recent literature reporting the low sensitivity of rapid antigen tests for the detection of influenza virus [31–33]. While these tests are widely used based on their speed and ease of use, they are characterized by a high rate of false negative results that require confirmatory testing using culture or molecular tests. Furthermore, the low accuracy may result in a significant numbers of misdiagnoses affecting quality and costs of patient management.
The Alere i Influenza A&B assay is isothermal-amplification-based and detects influenza virus A and B and has been compared to other diagnostic tests [23, 24, 34–36]. The reported sensitivity for influenza A compared with viral culture and the Prodesse ProFlu+, FilmArray RP, and Xpert Flu A/B ranged from 70 to 99%, and the specificity ranged from 62 to 100%; for influenza B, the sensitivity ranged from 91.8–100% and the specificity ranged from 53 to 100% reported by others [24, 34, 35]. The sensitivity was markedly lower in another study, particularly for the detection of influenza B [36].
Conversely, the cobas® Liat RT-PCR technology demonstrated high sensitivity and equivalent performance compared other RT-PCR-based tests. Compared to the Simplexa Flu A/B & RSV PCR assay, the cobas® Influenza A/B assay achieved sensitivities of 99.2 and 100% for influenza A and B, respectively; the specificity was 100% for each target resulting in an overall agreement of 99.5% [37]. While both the Alere test and cobas® Influenza A/B assay are considered point-of-care solutions and demonstrate comparable time to result, the user-friendliness and low human resource time of the cobas® Influenza A/B assay are markedly less than the same features using the Alere i Influenza A&B test procedure. Two sequences of manual steps are required for the Alere assay with a three-minute wait time between each sequence. The sample is not added until after the three-minute wait. In contrast, the cobas® Influenza A/B assay (Fig. 1) runs automatically to a result after sample insertion in the setup. Future studies will have to directly compare the accuracy, time to result, human-resource time, and overall clinical value of these molecular point-of-care tests for the detection of influenza A and B.
Comparing currently available techniques for the diagnosis of infection with influenza virus A and B, the cobas® Influenza A/B assay – to our knowledge – is the first test that combines the high accuracy of molecular techniques or viral culture with the speed and ease of use of rapid antigen tests.
In regard to speed and ease of use, the total time to result was 20 min, and the total hands-on time to start the test was ~1 min. Thus, the time to result is only slightly longer than the one observed using rapid antigen tests but still well within the time frame needed to impact the management of patients with suspected influenza in outpatient clinics, emergency room, or on regular and intensive care wards. The ease of use of the cobas® Liat System is further underlined by the fact that testing of samples in this trial was conducted by personnel having received only minimal training at the initiation of the study. The ease of use and minimal human resource time also lends itself to expand testing to other settings such as pharmacies and practitioner’s offices. The small footprint of the cobas® Liat System adds to the value of this technology, i.e., in a true point-of-care setting.
The ease-of-use is further underlined by the fact that the prospective collection of NPS was performed by operators who had limited or no training or hands-on experience in conducting laboratory tests. Operators included nurses, medical assistants, administrative assistants, nurse aides, personnel with Emergency Medical Technician training, or other personnel with no formal medical training. Operators were only provided with labeling and product materials that are included with the test kit (Liat Influenza A/B Assay Package Insert, Quick Reference Instructions, and the Liat Analyzer Quick Start Guide and User Manual) but did not receive additional instructions on the operation of the Liat system. In a questionnaire, operators rated the overall ease of use as 4.2/5 (including the instructions to test specimens, load samples, start the assay, read and understand the test results, data not shown). The cobas® Liat System has been CLIA-waived in conjunction with the cobas® Strep A test; the cobas® Influenza A/B assay recently was granted CLIA-waiver by the FDA.
The diagnostic accuracy and speed of the new rapid PCR-based molecular and near-patient diagnostic tests for influenza have strong potential to translate into improved clinical management. Recently, Hansen et al. (G. Hansen, Clinical Decision Making in the Emergency Department During Influenza Season; Can Test Results Influence Practice and the Role of the Cobas Liat Influenza A/B Assay, 2015 AACC Annual Meeting and Clinical Lab Expo, Atlanta, Georgia, July 26–30, 2015, manuscript in preparation) reported the excellent accuracy of the cobas® Liat assay compared to the standard of care (GenMark RVP assay with provider judgment) in the emergency room. Introduction of the cobas® Liat assay had a significant impact on clinical management as changes in patient management occurred in 86 (57%) of 150 patients studied in the emergency room. Patient management changes included changes to antiviral–antimicrobial stewardship (53%), changes in admission/discharge orders (17%), and changes to procedures/lab orders (19%). Of interest, 61% of patients in whom management was changed had a negative cobas® Influenza A/B result.
Previous studies have found that the mean length of stay in the hospital for inpatients with respiratory viral isolates was reduced by almost 50%, and mean variable costs for these patients were reduced by 2/3 after introduction of rapid testing [38] Thus, the introduction of highly accurate and PCR-based rapid tests is likely to have significant clinical benefits ranging from decreases in turnaround time, mortality, length of stay, and overall costs; at the same time, these benefits will also result in improved antibiotic stewardship.
PCR testing offers significant improvements in accuracy over rapid antigen testing and/or clinical judgment [39]. The trade-off is that molecular assays are more expensive than rapid antigen tests. All new rapid molecular systems should be compared to both rapid antigen testing and PCR to validate their accuracy, ease of use, and degree of benefit offered. Rapid antigen tests have demonstrated striking differences in accuracy and, the same may prove to be true for POC molecular tests based on PCR and other molecular techniques. Additional studies and health economic analysis are needed to assess the performance benefits, cost, and reimbursement of new molecular testing.
Our study has limitations. First, performance characteristics for influenza A were prospectively established at influenza seasons when particular influenza strains including A/H1 and A/H3 were the predominant influenza A viruses in circulation. However, 180 other influenza A and B strains obtained from the CDC were successfully identified by the cobas® Influenza A/B assay and analytical studies also included a large number of influenza strains of temporal and geographic diversity (data not shown). Furthermore, data generated to demonstrate changes in patient management were generated from a limited cohort of patients with suspected influenza. Therefore, future studies will have to demonstrate the overall outcome benefits of the cobas® Influenza A/B assay for the management of these patients.
In conclusion, the cobas® Influenza A/B assay demonstrated excellent sensitivity and specificity compared to viral culture and a comparator PCR-based molecular test; performance compared to standard rapid antigen tests was markedly superior. Results were generated within 20 min, and the assay could be performed by personnel untrained in laboratory procedures. Results of the present study strongly support the use of the cobas® Influenza A/B assay for the detection of influenza A and B in nasopharyngeal specimens obtained from patients with suspected influenza.
Acknowledgements
The authors wish to thank all the staff involved in this study at the collection and testing sites. We thank Ruchika Mohan and Joanna Sickler for expert help during study execution and article preparation.
All authors are employees of Roche Molecular Systems.
References
- 1.Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, et al. : Influenza-associated hospitalizations in the United States. Jama 292, 1333–1340 (2004) [DOI] [PubMed] [Google Scholar]
- 2.Thompson WW Shay DK Weintraub E Brammer L Cox N et al.: Mortality associated with influenza and respiratory syncytial virus in the United States. Jama 289, 179–186 (2003) [DOI] [PubMed] [Google Scholar]
- 3.Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, et al. : The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25, 5086–5096 (2007) [DOI] [PubMed] [Google Scholar]
- 4.Call SA, Vollenweider MA, Hornung CA, Simel DL, McKinney WP: Does this patient have influenza? Jama 293, 987–997 (2005) [DOI] [PubMed] [Google Scholar]
- 5.Petrozzino JJ, Smith C, Atkinson MJ: Rapid diagnostic testing for seasonal influenza: an evidence-based review and comparison with unaided clinical diagnosis. J Emergency Med 39, 476–490 e471 (2010) [DOI] [PubMed] [Google Scholar]
- 6.Chartrand C, Leeflang MM, Minion J, Brewer T, Pai M: Accuracy of rapid influenza diagnostic tests: a meta-analysis. Ann Intern Med 156, 500–511 (2012) [DOI] [PubMed] [Google Scholar]
- 7.Aoki FY, Macleod MD, Paggiaro P, Carewicz O, El Sawy A, et al. : Early administration of oral oseltamivir increases the benefits of influenza treatment. J Antimicrob Chemother 51, 123–129 (2003) [DOI] [PubMed] [Google Scholar]
- 8.Singh BK, Savill NJ, Ferguson NM, Robertson C, Wool-house ME: Rapid detection of pandemic influenza in the presence of seasonal influenza. BMC Public Health 10, 726 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novak-Weekley SM, Marlowe EM, Poulter M, Dwyer D, Speers D, et al. : Evaluation of the Cepheid Xpert Flu Assay for rapid identification and differentiation of influenza A, influenza A 2009 H1N1, and influenza B viruses. J Clin Microbiol 50, 1704–1710 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chartrand C, Pai M: How accurate are rapid influenza diagnostic tests? Expert Rev Anti-Infect Ther 10, 615–617 (2012) [DOI] [PubMed] [Google Scholar]
- 11.van Doorn HR Kinh N Tuan HM Tuan TA Minh NN et al.: Clinical validation of a point-of-care multiplexed in vitro immunoassay using monoclonal antibodies (the MSD influenza test) in four hospitals in Vietnam. J Clin Microbiol 50, 1621–1625 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale SE, Mayer C, Mayer MC, Menegus MA: Analytical and clinical sensitivity of the 3M rapid detection influenza A+B assay. J Clin Microbiol 46, 3804–3807 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurt AC, Alexander R, Hibbert J, Deed N, Barr IG: Performance of six influenza rapid tests in detecting human influenza in clinical specimens. J Clin Virol 39, 132–135 (2007) [DOI] [PubMed] [Google Scholar]
- 14.Lee GC Jeon ES Kim WS Le DT Yoo JH et al.: Evaluation of a rapid diagnostic test, NanoSign(R) Influenza A/B Antigen, for detection of the 2009 pandemic influenza A/ H1N1 viruses. Virol J 7, 244 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierce VM, Elkan M, Leet M, McGowan KL, Hodinka RL: Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J Clin Microbiol 50, 364–371 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginocchio CC: Strengths and weaknesses of FDA-approved/cleared diagnostic devices for the molecular detection of respiratory pathogens. Clin Infect Dis 52 Suppl 4, S312–325 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teo J, Di Pietro P, San Biagio F, Capozzoli M, Deng YM, et al. : VereFlu: an integrated multiplex RT-PCR and microarray assay for rapid detection and identification of human influenza A and B viruses using lab-on-chip technology. Arch Virol 156, 1371–1378 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babady NE, Mead P, Stiles J, Brennan C, Li H, et al. : Comparison of the Luminex xTAG RVP Fast assay and the Idaho Technology FilmArray RP assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J Clin Microbiol 50, 2282–2288 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chidlow G, Harnett G, Williams S, Levy A, Speers D, et al. : Duplex real-time reverse transcriptase PCR assays for rapid detection and identification of pandemic (H1N1) 2009 and seasonal influenza A/H1, A/H3, and B viruses. J Clin Microbiol 48, 862–866 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang YW, Lowery KS, Valsamakis A, Schaefer VC, Chappell JD, et al. : Clinical accuracy of a PLEX-ID flu device for simultaneous detection and identification of influenza viruses A and B. J Clin Microbiol 51, 40–45 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, Bose ME, Beck ET, Fan J, Tiwari S, et al. : Rapid multiplex reverse transcription-PCR typing of influenza A and B virus, and subtyping of influenza A virus into H1, 2, 3, 5, 7, 9, N1 (human), N1 (animal), N2, and N7, including typing of novel swine origin influenza A (H1N1) virus, during the 2009 outbreak in Milwaukee, Wisconsin. J Clin Microbiol 47, 2772–2778 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeffelholz MJ, Pong DL, Pyles RB, Xiong Y, Miller AL, et al. : Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens. J Clin Microbiol 49, 4083–4088 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie S, Roth RB, Stiles J, Mikhlina A, Lu X, et al. : Evaluation of Alere i Influenza A&B for rapid detection of influenza viruses A and B. J Clin Microbiol 52, 3339–3344 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell J, Bonner A, Cohen DM, Birkhahn R, Yogev R, et al. : Multicenter clinical evaluation of the novel Alere i Influenza A&B isothermal nucleic acid amplification test. J Clin Virol 61, 81–86 (2014) [DOI] [PubMed] [Google Scholar]
- 25.Legoff J, Kara R, Moulin F, Si-Mohamed A, Krivine A, et al. : Evaluation of the one-step multiplex real-time reverse transcription-PCR ProFlu-1 assay for detection of influenza A and influenza B viruses and respiratory syncytial viruses in children. J Clin Microbiol 46, 789–791 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, et al. : Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 437, 1162–1166 (2005) [DOI] [PubMed] [Google Scholar]
- 27.Ghedin E, Fitch A, Boyne A, Griesemer S, DePasse J, et al. : Mixed infection and the genesis of influenza virus diversity. J Virol 83, 8832–8841 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang SM, Lim MS, Han M, Hong YJ, Kim TS, et al. : Comparison of xTAG respiratory virus panel and verigene respiratory virus plus for detecting influenza virus and respiratory syncytial virus. J Clin Lab Anal 29(2), 116–121 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popowitch EB, O’Neill SS, Miller MB: Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP fast multiplex assays for detection of respiratory viruses. J Clin Microbiol 51, 1528–1533 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiMaio MA, Sahoo MK, Waggoner J, Pinsky BA: Comparison of Xpert Flu rapid nucleic acid testing with rapid antigen testing for the diagnosis of influenza A and B. J Virol Methods 186, 137–140 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruest A, Michaud S, Deslandes S, Frost EH: Comparison of the Directigen flu A+B test, the QuickVue influenza test, and clinical case definition to viral culture and reverse transcription-PCR for rapid diagnosis of influenza virus infection. J Clin Microbiol 41, 3487–3493 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petric M, Comanor L, Petti CA: Role of the laboratory in diagnosis of influenza during seasonal epidemics and potential pandemics. J Infect Dis 194 Suppl 2, S98–110 (2006) [DOI] [PubMed] [Google Scholar]
- 33.Rouleau I, Charest H, Douville-Fradet M, Skowronski DM, De Serres G: Field performance of a rapid diagnostic test for influenza in an ambulatory setting. J Clin Microbiol 47, 2699–2703 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell JJ, Selvarangan R: Evaluation of the Alere I influenza A&B nucleic acid amplification test by use of respiratory specimens collected in viral transport medium. J Clin Microbiol 52, 3992–3995 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapin KC, Flores-Cortez EJ: Performance of the molecular Alere I influenza A&B test compared to that of the xpert flu A/B assay. J Clin Microbiol 53, 706–709 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jokela P, Vuorinen T, Waris M, Manninen R: Performance of the Alere i influenza A&B assay and mariPOC test for the rapid detection of influenza A and B viruses. J Clin Virol 70, 72–76 (2015) [DOI] [PubMed] [Google Scholar]
- 37.Binnicker MJ, Espy MJ, Irish CL, Vetter EA: Direct Detection of influenza A and B viruses in less than 20 minutes using a commercially available rapid PCR assay. J Clin Microbiol 53, 2353–2354 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barenfanger J, Drake C, Leon N, Mueller T, Troutt T: Clinical and financial benefits of rapid detection of respiratory viruses: an outcomes study. J Clin Microbiol 38, 2824–2828 (2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller MR, Peters TR, Suerken CK, Snively BM, Poehling KA: Predictors of influenza diagnosis among patients with laboratory-confirmed influenza. J Infect Dis (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]


