Sir,
Tetracycline can still be used for treatment of infections caused by Gram-negative organisms that lack resistance genes. In Acinetobacter baumannii tetracycline resistance is often caused by tetA(A) and tetA(B), which encode efflux pumps,1,2 and occasionally by tet(M), which encodes a ribosomal protection protein.1 In addition, a novel determinant, tet39 (tetA39-tetR39), was found in tetracycline-resistant Acinetobacter strains recovered from freshwater fish farms in Denmark3 and Thailand4 and from a clinical Acinetobacter calcoaceticus/baumannii complex isolate recovered from human urine in 1986.3 The tetA39 gene encodes an efflux pump that was reported to confer resistance to tetracycline but not minocycline.3
In strains belonging to global clone 1 (GC1), the tetA(A) gene is part of the AbaR resistance island5 while in GC2 isolates the tetA(B) gene is found in an AbGRI1 resistance island.6 However, there is little information about the tetracycline resistance determinants and their context in strains that do not belong to the two major global clones. Here, we have examined the cause of tetracycline resistance in RCH52, a clinical multiply antibiotic-resistant A. baumannii strain recovered prior to 2010 in a Queensland hospital.
RCH52 was found to be resistant to ampicillin, ceftazidime, ticarcillin/clavulanic acid, imipenem, meropenem, streptomycin, spectinomycin, sulphonamides, trimethoprim, kanamycin, gentamicin and tetracycline. The genome of RCH52 was sequenced using the Illumina Hiseq platform and assembled as described previously,7 generating 60 contigs. RCH52 was ST729 (Institut Pasteur scheme), a novel single locus variant of ST3 (rpoB4 in ST729 differs from rpoB3 in ST3 by a single base pair) and RCH52 therefore belongs to the European clone III. To the best of our knowledge this is the first report of the European clone III in Australia. ResFinder 2.1 (https://cge.cbs.dtu.dk//services/ResFinder/) was used to identify resistance genes, and RCH52 contains aphA1b, aacC2, aadA1, floR, cmlA1, arr-2, sul1, sul2, dfrA14, oxa10, blaTEM, oxa23 (encoding a variant of OXA-23 differing by four amino acids) and the tet39 determinant, accounting for the resistances observed. The comM gene is uninterrupted and ISAba1 was not found upstream of the ampC gene.
The tet39 determinant was found on an 11 kb contig that was shown to be circular using PCR followed by sequencing. The plasmid carrying tet39, named pRCH52-1 (GenBank accession number KT346360; Figure 1), is 11146 bp. Plasmid DNA was isolated and electroporated into A. baumannii ATCC 17978, which is tetracycline susceptible. Transformants were selected on L-agar supplemented with 20 mg/L tetracycline (transformation frequency = 4.5 × 107 transformants/μg of DNA). None of the transformants grew on L-agar containing other antibiotics that RCH52 was resistant to, indicating that only the tetracycline resistance had transferred into the recipient cells. Primers tet39-F (5′-GCAGCTAATGCCCATACCAT-3′) and tet39-R (5′-GCCTTTTGCGTTGTTACCAT-3′) were designed to amplify a 219 bp internal fragment of the tetA39 gene. This PCR generated the expected product for all of the transformants tested. Although it was reported that tet39 does not confer resistance to minocycline,3 transformants tested here exhibited reduced susceptibility to minocycline, with inhibition zone diameters of 18 mm compared with 26 mm for ATCC 17978.
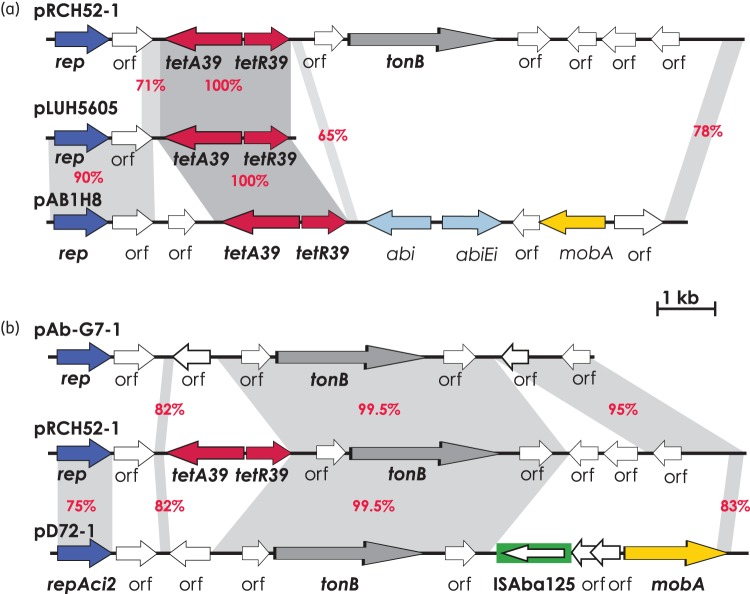
Figure 1.
Linearized map of pRCH52-1 compared with other plasmids. (a) Comparison of the original tet39 region found in pLUH5605 with the corresponding region found in pRCH52-1 and pAB1H8. (b) pRCH52-1 compared with the cryptic plasmids pAb-G7-1 and pD72-1 seen in GC1 and GC2 strains, respectively. Arrows indicate the extent and direction of genes and ORFs. The tetA gene and tetR genes of tet39 are shown in red and rep genes are coloured blue. The tonB gene encodes a TonB-dependent transporter homologue. The green box indicates ISAba125 and the arrow inside represents the transposase gene. The extents of regions with significant DNA identities are shown in grey and red numbers represent DNA identities. A scale bar is also shown. The picture is drawn to scale from the following GenBank entries: pRCH52-1, GenBank accession number KT346360; pAb-G7-1, GenBank accession number KJ586856; pD72-1, GenBank accession number KM051986; pLUH5605, GenBank accession number AY743590; and pAB1H8, GenBank accession number ANNC01000048. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The original study showed that tet39 was located on plasmids but only a 3727 bp fragment of one plasmid (named here pLUH5605) was sequenced (GenBank accession number AY743590). This sequence, the only complete tet39 sequence found in the GenBank non-redundant database, includes tetA39, tetR39 and two ORFs (Figure 1a), one of which encodes a replication initiation protein.3 Only 2360 bp of the pLUH5605 sequence was present in pRCH52-1 (Figure 1a). To explore the distribution of the tet39 determinant in Acinetobacter strains, the whole genome sequence database of NCBI was explored using the sequences of pRCH52-1 and pLUH5605. Twenty-one Acinetobacter strains belonging to different species were found to contain tet39 (Table S1, available as Supplementary data at JAC Online). However, the tet39 region appears to be in different contexts in all but one A. baumannii strain, AB1H8 (GenBank accession number ANNC01000048). AB1H8 appears to include a plasmid that contains a Rep that is 97% identical in terms of amino acids to the pLUH5605 RepA (Figure 1a). Hence, discrete boundaries surrounding tet39 were not found.
pRCH52-1 encodes a replication initiation protein Rep that belongs to the Rep_3 superfamily (pfam01051) and differs from the Rep found in pLUH5605 by 43%. The pRCH52-1 Rep is identical to the Rep protein found in a strain belonging to the A. calcoaceticus/baumannii complex (NCBI Reference Sequence number WP_000845850). Thereafter, the closest Rep is RepAci7 (GenBank accession number GU978996), with 96% amino acid identity.
pRCH52-1 also encodes a TonB-dependent transporter homologue. These outer membrane proteins bind and transport siderophores, vitamin B12, nickel complexes and carbohydrates.8 This gene is also present in several cryptic plasmids of A. baumannii,6,7,9,10 including pAb-G7-1 and pD72-1 from GC1 and GC2 strains, respectively. However, the amino acid sequence of Rep encoded by pRCH52-1 differs from those in pAb-G7-1 (GenBank accession number KJ586856) and pD72-1 (GenBank accession number KM051986) by 24% and 21%, indicating that the tonB gene, which is a potential virulence determinant, is widely distributed.
The tet39 determinant is widespread in Acinetobacter species. It has also been found in other species of Gram-negative and Gram-positive bacteria recovered from a polluted Nigerian river.11 Its location on plasmids in clinical isolates would facilitate the spread of tetracycline resistance amongst Acinetobacter strains, leading to further restriction of treatment options.
Funding
This study was supported by NHMRC Project Grant 1026189 and Wellcome Trust grant number 098051. M. H. was supported by NHMRC Project Grant 1026189. K. E. H. was supported by an NHMRC Fellowship (no. 1061049).
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
We thank Dr Mohammad Katouli for sending us RCH52. We also thank the team of curators of the Institut Pasteur Acinetobacter MLST system for curating the data and making them publicly available at http://pubmlst.org/abaumannii/.
References
- 1.Huys G, Cnockaert M, Vaneechoutte M et al. . Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res Microbiol 2005; 156: 348–55. [DOI] [PubMed] [Google Scholar]
- 2.Mak JK, Kim M-J, Pham J et al. . Antibiotic resistance determinants in nosocomial strains of multidrug-resistant Acinetobacter baumannii. J Antimicrob Chemother 2009; 63: 47–54. [DOI] [PubMed] [Google Scholar]
- 3.Agerso Y, Guardabassi L. Identification of Tet 39, a novel class of tetracycline resistance determinant in Acinetobacter spp. of environmental and clinical origin. J Antimicrob Chemother 2005; 55: 566–9. [DOI] [PubMed] [Google Scholar]
- 4.Agerso Y, Petersen A. The tetracycline resistance determinant Tet 39 and the sulphonamide resistance gene sulII are common among resistant Acinetobacter spp. isolated from integrated fish farms in Thailand. J Antimicrob Chemother 2007; 59: 23–7. [DOI] [PubMed] [Google Scholar]
- 5.Hamidian M, Wynn M, Holt KE et al. . Identification of a marker for two lineages within the GC1 clone of Acinetobacter baumannii. J Antimicrob Chemother 2014; 69: 557–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nigro SJ, Hall RM. Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J Antimicrob Chemother 2012; 67: 1342–6. [DOI] [PubMed] [Google Scholar]
- 7.Hamidian M, Holt KE, Pickard D et al. . A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J Antimicrob Chemother 2014; 69: 955–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noinaj N, Guillier M, Barnard TJ et al. . TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 2010; 64: 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt KE, Hamidian M, Kenyon JJ et al. . Genome sequence of Acinetobacter baumannii strain A1, an early example of antibiotic-resistant global clone 1. Genome Announc 2015; 3: e00032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nigro SJ, Hall RM. Amikacin resistance plasmids in extensively antibiotic-resistant GC2 Acinetobacter baumannii from two Australian hospitals. J Antimicrob Chemother 2014; 69: 3435–7. [DOI] [PubMed] [Google Scholar]
- 11.Adelowo OO, Fagade OE. The tetracycline resistance gene tet39 is present in both Gram-negative and Gram-positive bacteria from a polluted river, Southwestern Nigeria. Lett Appl Microbiol 2009; 48: 167–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.