Abstract
Objectives
The objective of this study was to investigate the silver gull as an indicator of environmental contamination by salmonellae and carbapenemase-producing Enterobacteriaceae (CPE) in south-east Australia.
Methods
A total of 504 cloacal samples were collected from gull chicks at three nesting colonies in New South Wales, Australia [White Bay (n = 144), Five Islands (n = 200) and Montague Island (n = 160)] and were examined for salmonellae and CPE. Isolates were tested for carbapenemase genes and susceptibility to 14 antibiotics. Clonality was determined by PFGE and MLST. Genetic context and conjugative transfer of the carbapenemase gene were determined.
Results
A total of 120 CPE of 10 species, mainly Escherichia coli (n = 85), carrying the gene blaIMP-4, blaIMP-38 or blaIMP-26 were obtained from 80 (40%) gulls from Five Islands. Thirty percent of birds from this colony were colonized by salmonellae. Most isolates contained the gene within a class 1 integron showing a blaIMP-4-qacG-aacA4-catB3 array. The blaIMP gene was carried by conjugative plasmids of variable sizes (80–400 kb) and diverse replicons, including HI2-N (n = 30), HI2 (11), A/C (17), A/C-Y (2), L/M (5), I1 (1) and non-typeable (6). Despite the overall high genetic variability, common clones and plasmid types were shared by different birds and bacterial isolates, respectively.
Conclusions
Our data demonstrate a large-scale transmission of carbapenemase-producing bacteria into wildlife, likely as a result of the feeding habits of the birds at a local waste depot. The isolates from gulls showed significant similarities with clinical isolates from Australia, suggesting the human origin of the isolates. The sources of CPE for gulls on Five Islands should be explored and proper measures applied to stop the transmission into the environment.
Introduction
Carbapenems are one of the most important antibiotics in human medicine and are regarded as last-line drugs to treat infections caused by MDR Gram-negative bacteria. Clinical efficacy of these antibiotics is threatened by carbapenemases, β-lactamases capable of rapid degradation of carbapenems. Metallo-β-lactamases, mainly VIM, NDM and IMP, are among the most disseminated carbapenemases worldwide.1 IMP-type β-lactamases are found in various clinically important Gram-negative bacteria, such as Pseudomonas spp., Acinetobacter spp. and Enterobacteriaceae, all round the world,2 endemically in Japan, Taiwan, China, Korea and the Philippines.3–6 Currently, IMP-4 is the most commonly reported carbapenemase in clinical isolates of Enterobacteriaceae, associated with outbreaks in healthcare settings in Australia,7 but its wide dissemination has been documented also in China.4
Transmission of carbapenem-resistant bacteria to food-producing animals and the environment is of great concern. So far, there are scarce reports of carbapenemase-producing Enterobacteriaceae (CPE) from farm animals and wildlife.8 Salmonella is an important zoonotic pathogen causing infections in humans and domestic animals. Wild birds, such as gulls, influenced by various anthropogenic activities, are important in the epidemiology of human and livestock salmonellosis.9 In Australia, the prevalence of Salmonella, including antibiotic-resistant strains in wild birds, is largely unknown.10
In this work, we studied the silver gull (Chroicocephalus novaehollandiae) as an indicator of environmental contamination by Salmonella and CPE in south-east Australia in respect of the current epidemiological situation in this country. The silver gull feeds on fish, marine and terrestrial invertebrates, but also on human refuse collected at landfills.11,12 This study is the first known report of a large-scale transmission of CPE into wildlife.
Materials and methods
Birds and sampling
The silver gull is widespread throughout Australia, New Zealand and New Caledonia. There has been marked expansion of the population of the silver gull in Australia in the last century, with most increases in numbers occurring near to and within major cities. Although silver gulls do occur near inland wetlands, they are predominantly a coastal species. In south-east Australia they breed mostly on marine islands, the highest densities occurring on islands close to major urban areas, particularly Newcastle, Sydney and Wollongong. Seventy percent of coastal breeding in New South Wales (NSW) occurs at Five Islands off the coast of Wollongong.11,12
A total of 504 cloacal samples were taken from silver gull chicks within the three nesting colonies on the south-eastern coast of NSW, Australia—White Bay (n = 144), Five Islands (n = 200) and Montague Island (n = 160)—in October 2012. The samples were collected using a sterile cotton swab and placed in an Amies transport medium (Coban, Italy).
The small colony at White Bay near Glebe (33°86′ S, 151°18′ E) nest on a diffused industrial wharf within Sydney Harbour, ∼70 km north of Five Islands. In October 2012, the colony contained ∼1500 birds. These birds feed at the nearby fish markets and in other suburban areas surrounding Sydney Harbour,11,12 which provides natural prey enriched by refuse carried by urban runoff.
Big Island, in the Five Islands group (34°29′ S, 150°56′ E), is a 19 ha nature reserve ∼500 m offshore from Port Kembla, 60 km south of Sydney. Big Island represents the largest silver gull breeding colony in NSW, with historical estimates of up to 50 000 pairs,11 although numbers have declined recently for unknown reasons. During our sampling in the 2012 breeding season we estimated only 3854 pairs. Studies have shown that the gulls breeding on Big Island feed predominantly on human refuse.12 As many as 6000 gulls per hour have been recorded leaving Whyte Gully landfill (34°26′ S, 150°48′ E), the main waste depot, located on the mainland 12 km from the nesting colony. Regurgitations from gulls trapped on Big Island contained solely human refuse (85% of samples), solely natural food (13%) or a mixture of both (2%). Meat constituted 63% of the human refuse.12
Montague Island (36°15′ S, 150°14′ E) lies 6 km offshore, 10 km south-west of Narooma, NSW. The entire island is a natural reserve of 82 ha and supports a silver gull population of 1500–2000 pairs, which feed predominantly on a natural diet of worms, fish, crustaceans and insects (N. Carlile, unpublished results).
Isolation of carbapenemase-producing isolates and Salmonella
Individual cloacal samples were enriched overnight in Buffered Peptone Water (Oxoid, UK) and then cultured for carbapenemase-producing Enterobacteriaceae on CHROMagar™ KPC (CHROMagar, Paris, France) and MacConkey agar supplemented with meropenem (0.125 mg/L) and ZnSO4 (100 mg/L) (MCAmer) and for Salmonella using semi-solid Rappaport–Vassiliadis medium (Oxoid) and X.L.D. agar (Oxoid). One colony for each morphological type growing on CHROMagar KPC or MCAmer was selected and identified by MALDI-TOF MS (Microflex LT, Bruker Daltonics, Bremen, Germany). Salmonella isolates were subjected to serotyping using the Kauffmann–White–LeMinor scheme13 and phage typing according to the HPA Colindale protocol.14,15
Antibiotic resistance profiling
Susceptibility to 14 antibiotics, including amoxicillin/clavulanic acid (30 μg), ampicillin (10 μg), cefalotin (30 μg), ceftazidime (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), imipenem (10 μg), meropenem (10 μg), nalidixic acid (30 μg), streptomycin (10 μg), sulfamethoxazole/trimethoprim (25 μg), compound sulphonamides (300 μg) and tetracycline (30 μg) (Oxoid, UK), was tested in Salmonella and blaIMP-positive isolates using a disc-diffusion method.16 Isolates negative for the tested carbapenemase genes were screened for metallo-β-lactamase, AmpC and ESBL production by phenotypic assays.16–18
MICs of meropenem for IMP- and AmpC-producing isolates were determined using the agar dilution method.16
Statistical significance of the data was analysed using the χ2 test. P value ≤0.05 was considered to be statistically significant.
Molecular typing of CPE isolates
Enterobacteriaceae isolates growing on CHROMagar KPC or MCAmer and all Salmonella isolates were screened for carbapenemase genes (blaIMP, blaNDM, blaKPC, blaVIM and blaOXA-48-type) by PCR (Table S1, available as Supplementary data at JAC Online). Isolates positive for blaIMP were tested for the gene blaIMP-4 using PCR followed by sequencing of the amplicons (Table S1). Clonality of blaIMP-positive isolates was determined by XbaI-PFGE. Cluster analysis using Dice similarity indices was done in BioNumerics 6.6 software (Applied Maths, Ghent, Belgium) to generate a dendrogram describing the relationships among PFGE profiles. Isolates with ≥85% similarity of PFGE profiles were assigned to the same cluster (designated as 1, 2, 3, etc. for Escherichia coli and A, B, C, etc. for other species). MLST of representative isolates of E. coli and Klebsiella pneumoniae showing unique PFGE profiles was performed using Achtman's19 and Institut Pasteur's schemes,20 respectively. A PCR-based scheme was used to categorize E. coli isolates into phylogenetic groups.21
Genetic context and transferability of the blaIMP gene
At least one randomly selected isolate for each PFGE pattern/bacterial species was further analysed for context of the blaIMP-4 gene. A reference sequence of typical Australian class 1 integron array blaIMP-4-qacG-aacA4-catB3 (Accession number JX101693) was used to design the PCR mapping.22 The presence of the blaIMP gene inside a class 1 integron was tested using primers specific for the 5′-conserved region of a class 1 integron and blaIMP. The 3′ parts of the integron were analysed in three reactions using forward primer IMP-F in combination with reverse primers targeting the aminoglycoside resistance gene aacA4, the chloramphenicol resistance gene catB3 or the 3′-conserved region of a class 1 integron (for list of primers see Table S1). Selected amplicons were sequenced. Conjugative transfer of blaIMP was tested in all 120 isolates at 37°C using the plasmid-free, rifampicin- and sodium-azide-resistant E. coli MT102 recipient strain. Since IncHI plasmids have a thermosensitive mode for conjugation,23 the experiment was performed in parallel at 25 and 37°C for all isolates harbouring IncHI2 plasmids. Transconjugants were selected on media supplemented with cefotaxime (2 mg/L), sodium azide (100 mg/L) and rifampicin (25 mg/L) and transfer of blaIMP was confirmed by PCR. Plasmids carrying blaIMP were assigned to incompatibility groups by PCR-based replicon typing (PBRT)24,25 and their sizes were determined using PFGE of the total DNA digested with S1 nuclease. Where transconjugants with a single blaIMP-carrying plasmid were obtained, plasmid contents were confirmed in the corresponding donor strains using the same procedure.
Results
Salmonella
A total of 66 (13%) Salmonella enterica isolates were obtained from 504 silver gull cloacal samples from all three nesting colonies. The highest prevalence of salmonellae was found in gulls from Five Islands (56/200; 28%), while the other two locations showed significantly lower prevalences of 0.6% and 6% (Table 1). Salmonellae of 17 different serotypes, with dominance of Salmonella Typhimurium (33/66; 50%), were identified (Table 1). Salmonella Typhimurium DT2 and DT12 were the most common phage types, found in 16 and 8 gulls, respectively. Antibiotic resistance was demonstrated only in 3/66 (4.5%) Salmonella isolates (Table 1).
Table 1.
S. enterica isolates from the silver gull in three nesting colonies in Australia
| Serotype, phage type | Nesting colony (no. of samples) |
||
|---|---|---|---|
| WB (144) | FI (200) | MI (160) | |
| Bovismorbificans | 2 | ||
| Charity | 1 | ||
| Chester | 1 | ||
| Heidelberg | 1 | ||
| Infantis | 1 | ||
| Mbandaka | 1 | ||
| Muenchen | 1 | ||
| Oranienburg | 1 | ||
| Paratyphi B | 1 | ||
| Rissen | 4a | ||
| 3,10:r:– | 1a | ||
| Saintpaul | 5 | ||
| Senftenberg | 4 | ||
| Stanleyville | 4 | ||
| Typhimurium DT2 | 1 | 14a | 1 |
| Typhimurium DT8 | 1 | ||
| Typhimurium DT12 | 8 | ||
| Typhimurium DT13 | 1 | ||
| Typhimurium DT120 | 3 | ||
| Typhimurium RDNC | 4 | ||
| Virchow | 4 | ||
| Wangata | 1 | ||
| Total (prevalence) | 9 (6%) | 56 (28%) | 1 (0.6%) |
WB, White Bay; FI, Five Islands; MI, Montague Island.
aThree isolates showing resistance to tested antibiotics: Salmonella 3,10:r:– (strain no. 1620), resistant to ampicillin and amoxicillin/clavulanic acid; Salmonella Typhimurium DT2 (strain no. 1718), resistant to ampicillin, amoxicillin/clavulanic acid, cefalotin and ceftazidime; and Salmonella Rissen (strain no. 1786), resistant to ampicillin, amoxicillin/clavulanic acid, streptomycin, compound sulphonamides, sulfamethoxazole/trimethoprim and tetracycline.
Isolation of Enterobacteriaceae with the blaIMP gene
A total of 167 Enterobacteriaceae isolates were obtained by culture on selective media, 150 and 17 on MCAmer and CHROMagar KPC, respectively. The only carbapenemase gene detected was blaIMP, found in 120 (72%, n = 167) isolates. All blaIMP-positive isolates originated from 80 of 200 (40%) gulls sampled on Five Islands. No carbapenemase-producing isolates were found in the other two locations, White Bay and Montague Island. Thirty-seven out of 80 (46%) blaIMP-positive gulls carried more than one isolate; 33 and 4 gulls were colonized by two and three different isolates, respectively. The isolates were identified as E. coli (85 isolates), Escherichia fergusonii (10), K. pneumoniae (9), Enterobacter aerogenes (4), Proteus mirabilis (4), Kluyvera georgiana (2), Citrobacter freundii (2), Enterobacter cloacae (2), Proteus penneri (1) and Citrobacter braakii (1). Isolates negative for the carbapenemase genes (n = 36) included P. mirabilis (14), Hafnia alvei (6), E. coli (4), Providencia spp. (4), E. aerogenes (3), E. cloacae (2), Morganella morganii (2) and Proteus vulgaris (1) and were susceptible to third-generation cephalosporins (data not shown). Twelve of them (33%) produced inducible AmpC β-lactamase while neither carbapenemase nor ESBL production was found in the other 24 (67%) isolates. AmpC producers showed resistance to ampicillin, amoxicillin/clavulanic acid, cefalotin and cefoxitin and MICs to meropenem varied from 0.06 to 1 mg/L; they were identified as H. alvei (5), E. aerogenes (3), E. cloacae (2), Providencia spp. (1) and M. morganii (1).
Antibiotic resistance profiles of blaIMP-positive isolates
All blaIMP-positive isolates showed resistance to at least two antibiotic groups. Resistance to sulphonamides was the most prevalent (100%), followed by resistance to ampicillin (99%), amoxicillin/clavulanic acid (99%), ceftazidime (98%), gentamicin (93%), sulfamethoxazole/trimethoprim (93%), chloramphenicol (73%), tetracycline (69%), nalidixic acid (67%), streptomycin (53%), ciprofloxacin (46%), meropenem (25%) and imipenem (6%). According to CLSI criteria, 57% and 18% showed intermediate resistance and susceptibility to meropenem, respectively. Intermediate resistance and susceptibility to imipenem was observed in 33% and 61% of isolates, respectively. A significantly higher number of non-E. coli isolates showed susceptibility to meropenem compared with E. coli isolates (P < 0.01). Such differences were not observed for imipenem. According to CLSI criteria, the majority (79%) of the isolates were susceptible to meropenem, with MICs ≤4 mg/L. MICs were distributed as follows: ≤0.125 mg/L (6 isolates); 0.25 mg/L (7); 0.5 mg/L (39); 1 mg/L (26); 2 mg/L (27); 4 mg/L (12); and 8 mg/L (3).
Clonality of blaIMP-positive isolates
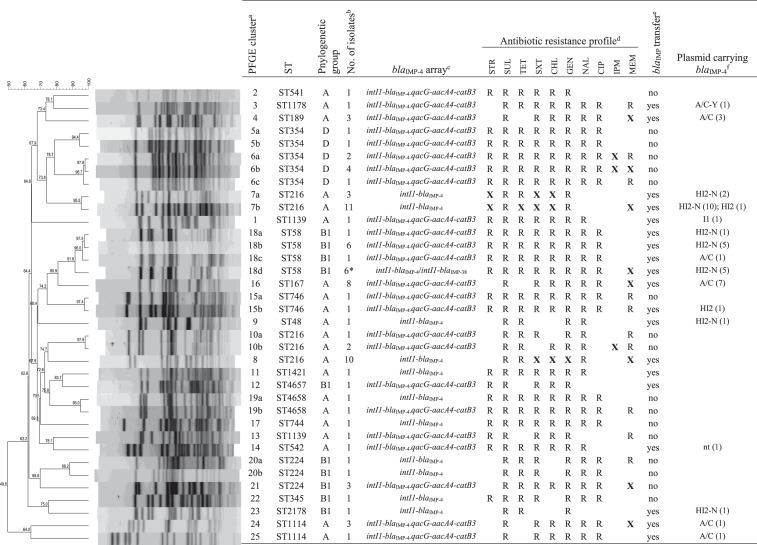
PFGE and MLST analysis demonstrated significant genetic diversity of blaIMP-positive isolates (Figure 1), but also the carriage of identical isolates by different birds (Figure S1). Eighty-five E. coli isolates showed 36 unique PFGE profiles and were divided into 25 clusters based on the criteria of 85% similarity of PFGE patterns. MLST analysis assigned the isolates to 19 different STs (Figure 1). The most prevalent clonal lineages included ST216 (n = 27), ST58 (n = 14), ST354 (n = 9), ST167 (n = 8) and ST224 (n = 5) and constituted 68% of all E. coli isolates. Two novel STs (ST4657 and ST4658) were identified among three isolates. Eleven birds carried two E. coli isolates of different STs. Twenty-four birds were colonized by two or three different species of the Enterobacteriaceae family. Ten E. fergusonii isolates belonged to a single cluster, H, with overall similarity of 95% (Table 2). Nine K. pneumoniae isolates were divided into seven distinct PFGE clusters and belonged to seven different STs, including five novel types (ST1734–ST1738). Two different E. coli strains sharing an identical blaIMP-4-integron array and plasmid replicon type were identified only in one bird, which may suggest a limited role of in vivo plasmid conjugative transfer inside the gull intestine.
Figure 1.
Characteristics of E. coli isolates with the blaIMP-4 gene from the silver gull on Five Islands. aIsolates with ≥85% similarity of PFGE profiles are assigned to the same cluster (designated as 1, 2, 3, etc.). Isolates from the same cluster with <100% identity of PFGE patters are indicated by letters (e.g. 5a, 5b, 6a, 6b, 6c, etc.). bFive of the six E. coli ST58 isolates (indicated by an asterisk) carry both blaIMP-4 and blaIMP-38. cIntegrons with intI1-blaIMP-4 arrays are missing the 3′ parts of the integron conserved region. dSTR, streptomycin; SUL, compound sulphonamides; TET, tetracycline; SXT, sulfamethoxazole/trimethoprim; CHL, chloramphenicol; GEN, gentamicin; NAL, nalidixic acid; CIP, ciprofloxacin; IPM, imipenem; MEM, meropenem. All isolates showed resistance to ampicillin, amoxicillin/clavulanic acid, cefalotin and ceftazidime. Resistance is indicated by ‘R’. Resistance patterns of isolates from the same cluster that show variable profiles are indicated by ‘X’. eTransfer of the gene by conjugation to recipient strains of E. coli. All isolates of E. coli ST58 carrying two blaIMP variants transferred only the gene blaIMP-4. fnt, plasmid non-typeable by PBRT.
Table 2.
Characteristics of non-E. coli isolates with the blaIMP gene from the silver gull on Five Islands
| Species | PFGE clustera | ST | No. of isolates | blaIMP integron | Antibiotic resistance profileb |
blaIMP transferc | Plasmid carrying blaIMPd | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STR | SUL | TET | SXT | CHL | GEN | NAL | CIP | IPM | MEM | |||||||
| Escherichia fergusonii | H1 | 2 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | yes | HI2 (1) | |||||||
| H2 | 7 | intI1-blaIMP-4-qacG-aacA4-catB3 | X | R | X | R | X | X | yes | HI2 (5) | ||||||
| H3 | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | yes | HI2 (1) | ||||||||
| Klebsiella pneumoniae | A | 1735 | 2 | intI1-blaIMP-26-qacG-aacA4-catB3 | R | R | R | R | R | R | R | yes | A/C (2) | |||
| B | 1736 | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | yes | nt (1) | |||||||
| C | 1734 | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | yes | A/C-Y (1) | ||||||||
| D | 394 | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | R | R | no | ||||||
| E | 1737 | 2 | intI1-blaIMP-4-qacG-aacA4-catB3 | X | R | X | X | X | R | X | X | X | yes | HI2-N (1) | ||
| F | 1738 | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | no | |||||||||
| G | 584 | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | yes | L/M (1) | |||||||||
| Kluyvera georgiana | I | 2 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | yes | |||||||||
| Enterobacter aerogenes | J1 | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | yes | L/M (1) | |||||||
| J2 | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | yes | L/M (1) | |||||||||
| J3 | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | yes | nt (1) | ||||||||
| L | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | R | R | yes | L/M (1) | ||||||
| Enterobacter cloacae | O | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | R | R | R | no | |||||
| P | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | yes | L/M (1) | ||||||||
| Citrobacter freundii | M | 1 | intI1-blaIMP-38-qacG-aacA4-catB3 | R | R | R | yes | A/C (1) | ||||||||
| N | 1 | intI1-blaIMP-38-qacG-aacA4-catB3 | R | R | R | R | R | R | yes | A/C (1) | ||||||
| Citrobacter braakii | V | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | R | R | R | R | no | ||||
| Proteus mirabilis | Q | 1 | intI1-blaIMP-4-qacG-aacA4 | R | R | R | R | R | R | R | yes | nt (1) | ||||
| R | 1 | intI1-blaIMP-4-qacG-aacA4 | R | R | R | R | R | R | R | no | ||||||
| S | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | R | yes | ||||||||
| T | 1 | intI1-blaIMP-4-qacG-aacA4-catB3 | R | R | R | R | R | R | yes | nt (1) | ||||||
| Proteus penneri | U | 1 | intI1-blaIMP-4-qacG-aacA4 | R | R | R | R | R | R | R | R | yes | nt (1) | |||
aIsolates with ≥85% similarity of PFGE profiles are assigned to the same cluster (designated as A, B, C, etc.). Isolates from the same cluster with <100% identity of PFGE patters are indicated by numbers (e.g. H1, H2, etc.).
bSTR, streptomycin; SUL, compound sulphonamides; TET, tetracycline; SXT, sulfamethoxazole/trimethoprim; CHL, chloramphenicol; GEN, gentamicin; NAL, nalidixic acid; CIP, ciprofloxacin; IPM, imipenem; MEM, meropenem. All isolates showed resistance to ampicillin, amoxicillin/clavulanic acid, cefalotin and ceftazidime except for two isolates of P. mirabilis (isolate of PFGE type S susceptible to ampicillin, amoxicillin/clavulanic acid and ceftazidime, and isolate of PFGE type T, susceptible to ceftazidime). Resistance is indicated by ‘R’. Resistance patterns of isolates from the same cluster that show variable profiles are indicated by ‘X’.
cTransfer of the gene by conjugation to recipient strains of E. coli.
dnt, plasmid non-typeable by PBRT.
Sequence analysis and genetic environment of the blaIMP gene
The sequence analysis of blaIMP amplicons revealed the presence of blaIMP-4 in 116 (97%) isolates. Single-locus variants of blaIMP-4, the genes blaIMP-26 and blaIMP-38, were found in two (2%) and seven (6%) isolates, respectively. The gene blaIMP-26 was identified in two isolates of K. pneumoniae ST1735 while blaIMP-38 was found in two C. freundii isolates (Table 2) and five E. coli ST58 that were also positive for the variant blaIMP-4 (Figure 1). Seventy randomly selected isolates representing each PFGE cluster/ST/bacterial species, including 40 E. coli and 30 non-E. coli, were tested for the genetic context of the blaIMP gene (Figure S1). The gene blaIMP was present in the class 1 integron as the first gene cassette in all isolates. PCR mapping revealed the blaIMP-4-qacG-aacA4-catB3 cassette array in 50 (71%) isolates (65% E. coli, 80% non-E. coli). The same integron arrangement was also demonstrated for blaIMP-26 and blaIMP-38 from K. pneumoniae and C. freundii isolates, respectively. An additional blaIMP-4-qacG-aacA4 array was identified in three Proteus spp. PCR mapping of the 3′ parts of the integron carrying blaIMP-4 or blaIMP-38 failed in 14 E. coli (21%), suggesting the existence of a structure different from the typical Australian integron used as a reference in this study.
Plasmids carrying the blaIMP gene
All isolates were tested for transfer of blaIMP by conjugation. In 57 (67%) and 22 (63%) E. coli and non-E. coli isolates, respectively, the gene was transferred at 37°C. The experiment was repeated at 25°C in 41 isolates that did not transfer the gene by conjugation at 37°C, revealing another seven positive isolates. Overall, the conjugative transfer of blaIMP was successful in 86 (72%) isolates. Transconjugants carrying blaIMP on a single plasmid were obtained from 66 isolates, including E. coli of 11 different STs, E. fergusonii, K. pneumoniae, E. aerogenes, E. cloacae, C. freundii, P. mirabilis and P. penneri. Furthermore, five isolates produced two or three transconjugants that showed a difference in size of blaIMP-carrying plasmids (Figure S1); therefore, they were all included in the further analysis. Testing of 72 transconjugants demonstrated the association of the gene with plasmids of variable sizes (80–400 kb) and diverse replicons: HI2 (n = 41), N (30), A/C (19), Y (2), L/M (5) and I1 (1). Thirty-five plasmids contained multi-replicons HI2-N (30) and A/C-Y (2). Six plasmids were not typeable by PBRT.
IncA/C plasmids ranged from 180 to 220 kb and were associated with three blaIMP genes (blaIMP-4, blaIMP-26, blaIMP-38) within the typical Australian cassette array. The gene blaIMP-4 was found on replicon A/C in E. coli of various genotypes (ST58, ST167, ST189 and ST1144) as well as on multi-replicon A/C-Y in one E. coli ST1178 and one K. pneumoniae ST1734. The presence of the two A/C and Y replicons on a single plasmid did not occur as a result of plasmid fusion during the conjugative experiment. IncL/M plasmids of 75 kb carrying blaIMP-4 within the typical Australian array were found in one isolate of K. pneumoniae ST584, three E. aerogenes isolates and one E. cloacae isolate. Six strains, including E. coli ST542, K. pneumoniae ST1736, E. aerogenes, P. mirabilis and P. penneri, contained the gene on plasmids ranging between 140 and 400 kb that were not typeable by the PBRT scheme. Apart from two Proteus isolates that carried the gene in a class 1 integron with a different cassette array (blaIMP-4-qacG-aacA4), the typical Australian array was identified for these plasmids.
The most common replicon associated with blaIMP-4 was HI2, found in 41 transconjugants of donor strains, including E. coli (n = 33; ST48, ST58, ST216, ST746 and ST2178), E. fergusonii (7) and K. pneumoniae ST1737 (1). E. coli ST58 isolates with two blaIMP variants (blaIMP-4 and blaIMP-38) produced transconjugants that carried the gene blaIMP-4 on HI2-N replicons. The gene blaIMP-38 was not transferred by conjugation. IncHI2 plasmids ranged from 150 up to 320 kb and contained the gene in both the typical Australian array (47%) or as a single-gene cassette with unknown structure of the 3′ parts of the integron (53%). Twenty-two transconjugants contained plasmids of higher molecular weight than their donors, indicating frequent fusions during conjugation, likely occurring between blaIMP-carrying IncHI2 or IncHI2-N plasmids and other plasmids, co-resident within the donor strain. The majority (84%) of IncHI2 plasmids were transferred at 37°C and no relationship between the change in plasmid size and temperature used for conjugative transfer was observed (data not shown).
Discussion
Our study demonstrates a high level of colonization by Salmonella and carbapenemase-producing bacteria of gulls on Five Islands. This is the first known report of extensive transmission of carbapenemase-producing bacteria into wild animals. Feeding habits related to garbage and sewage have been largely assumed to increase the risk of transmission of bacteria originating from humans or farm animals to wildlife. The long-term observations of the colony on Five Islands demonstrated the role of human refuse from the local depot near the city of Wollongong as an important source of food for breeding gulls in the colony.12 We assume that the source of IMP-4-producing isolates for gulls could be clinical material contaminating this local waste depot, supported by the demonstrated significant similarities of gull isolates with those reported from humans in Australia and the fact that no carbapenemase-producing bacteria have been reported from livestock and meat in that continent.
The distribution of Salmonella serovars in Australia varies geographically; however, Salmonella Typhimurium is one of the most commonly reported serovars from human infections. Three phage types of Salmonella Typhimurium, DT135, DT170/108 and DT9, are predominantly isolated from human infections, but also from animals.26 Although Salmonella Typhimurium dominated in the gulls in our study, none of these phage types was identified. The most prevalent phage type found in gulls, DT2, is commonly isolated from pigeons worldwide.27 The second most prevalent phage type, DT12, used to be prevalent in human infections in Australia, but it has declined in recent years.26
Carbapenemase producers belonged to 10 different species of the Enterobacteriaceae family and showed resistance to several groups of antimicrobials. Most isolates were selected by cultivation on in-house MacConkey agar supplemented with meropenem (0.125 mg/L) and ZnSO4 (100 mg/L) compared with commercially available CHROMagar KPC, suggesting that media with a low concentration of carbapenems are more suitable for environmental samples, where carbapenemase-producing bacteria can be present at very low concentration. CPE are infrequent in Australia, with overall prevalence of 0.3% and <0.1% in hospitals and the community, respectively.28 Nevertheless, blaIMP-4, found in the majority of the isolates from gulls, is also the most prevalent among carbapenemase-producing clinical Enterobacteriaceae isolates in Australia. IMP-4-producing E. coli isolates from gulls showed significant clonal similarity with five predominant STs (ST58, ST167, ST216, ST224 and ST354), suggesting their common source in the environment. The majority of them belonged to widely disseminated pathogenic and commensal MDR clones contributing to the spread of ESBL.29–31 Of note is that E. coli ST354 is the most common clonal lineage found among colonized and infected dogs in Australia.32 ST216, the most prevalent ST in our study, is not frequently reported in association with emerging resistance mechanisms.
Various plasmids and class 1 integrons were described to play a role in the dissemination of blaIMP-4 between clinical isolates in Australia, including the blaIMP-4-qacG-aacA4-catB3 array carried by IncA/C and IncL/M plasmids in Sydney and Melbourne22 and the blaIMP-4-aacA4 array on IncHI2 or IncL/M plasmids in the state of Queensland.33 The same blaIMP-4-qacG-aacA4-catB3 array was present in the majority of the isolates from gulls in our study and it was carried by conjugative plasmids of various sizes and replicon types, including IncHI2, IncA/C, IncL/M, IncI1 and multi-replicon (HI2-N, A/C-Y) and non-typeable plasmids. We also identified this integron array in single-nucleotide variants of the blaIMP-4 gene, the genes blaIMP-26 and blaIMP-38, carried by IncA/C plasmids. In Australia and the Philippines, blaIMP-26 has been identified in various STs of K. pneumoniae with the blaIMP-26-qac-aacA4 cassette array on A/C or L/M replicons.6 A novel blaIMP-4-qacG-aacA4 cassette array, recently reported for blaIMP-26,6 was found in Proteus spp. isolates. In 14 E. coli isolates, blaIMP-4 was found as a single cassette of the integron missing the 3′-conserved region and carried by multi-replicon HI2-N plasmids. These observations point out the high plasticity of class 1 integrons carrying genes of the blaIMP-4 family and their ability to be transferred between various plasmid families, likely increasing their dissemination potential in bacterial communities.
The most prevalent plasmids associated with the blaIMP gene found in 57% of transconjugants belonged to IncHI2. Recently, a high incidence of IncHI2 plasmids carrying blaIMP-4 was identified in various human clinical isolates of Enterobacteriaceae in Queensland,33 suggesting a growing role of this plasmid family in dissemination of blaIMP-4 in Australia. In contrast with this report, most of our plasmids showed a second replicon specific for plasmids of the IncN family and contained the blaIMP-4-integron within a different cassette array. IncN plasmid pIMP-HZ1, carrying blaIMP-4 in a class 1 integron with a truncated 3′-conserved region, as observed in our IncHI2-N plasmids, has been recently described in China.34 Ability of IncHI2 plasmids to conjugate at low temperature and to fuse with highly conjugative epidemic plasmids with a broad host range, such as IncN, may facilitate their dissemination in the environment. We identified several plasmids, such as IncI1 or non-typeable, that have not been previously associated with the blaIMP-4 gene. Demonstrated variability of Enterobacteriaceae isolates and plasmids carrying blaIMP-4 as well as the ability of the plasmids to be efficiently transferred by conjugation even at low temperature may indicate the further movement of the gene between bacteria and replicons after transmission to the environment of the gull nesting colony.
Overall, the findings from the silver gull reflect the current epidemiological situation in Australia, indicating the human origin of IMP-4-producing Enterobacteriaceae. Differences from reported data on human isolates may reflect unexplored complexity of epidemiology of IMP-4-producing bacteria in that continent and the important role of the environment in dissemination of the gene. This study documents a significant level of environmental contamination by bacteria resistant to clinically important carbapenems, which may potentially pose a risk for public health. As migratory birds, gulls may be potential vectors of IMP-producing strains along the coast of Australia. The cloacal samples were taken only from gull chicks in the colony, and the level of colonization by CPE in adult birds as possible vectors is unknown and should be explored in the future studies to fully understand the importance of these findings and the real risks indicated by them. The source of these MDR bacterial strains among the gulls nesting on Five Islands should be explored and proper measures applied to stop the transmission of this pathogen from the human population into the environment.
Funding
This work was funded by the Czech Science Foundation (15-14683Y/P502) and CEITEC (CZ.1.05/1.1.00/02.0068) from the European Regional Development Fund. I. J. and H. D. were supported by the Internal Grant Agency of the University of Veterinary and Pharmaceutical Sciences (projects 12/2014/FVHE and 242/2015/FVHE). Characterization of Salmonella isolates was partially funded by project LO1218 of MEYS of the CR under the NPU I programme.
Transparency declarations
None to declare.
Supplementary data
Acknowledgements
We thank Jana Hofirkova, Katerina Kachlikova, Katerina Kralova, Magdalena Sevcikova, Jiri Sedmik, Veronika Hradilova, Monika Novakova, Nicol Janecko, Dana Halova, Eva Suchanova and Petra Myskova for excellent cooperation in the laboratory. Our thanks go to Alessandra Carattoli and Sally Partridge for providing control strains. We thank the team of the curators of the Institut Pasteur MLST system (Paris, France) for importing novel alleles, profiles and/or isolates at http://bigsdb.web.pasteur.fr. This publication made use of the E. coli/MLST database developed by Mark Achtman and colleagues at University College Cork, formally hosted at http://mlst.ucc.ie/mlst/dbs/Ecoli.
References
- 1.Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis 2011; 11: 381–93. [DOI] [PubMed] [Google Scholar]
- 2.Zhao WH, Hu ZQ. IMP-type metallo-β-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol 2011; 37: 214–26. [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011; 17: 1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu L, Zhong Q, Shang Y et al. The prevalence of carbapenemase genes and plasmid-mediated quinolone resistance determinants in carbapenem-resistant Enterobacteriaceae from five teaching hospitals in central China. Epidemiol Infect 2014; 142: 1972–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong JS, Kim JO, Lee HB. Characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa in Korea. Infect Chemother 2015; 47: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peirano G, Lascols C, Hackel M et al. Molecular epidemiology of Enterobacteriaceae that produce VIMs and IMPs from the SMART surveillance program. Diagn Microbiol Infect Dis 2014; 78: 277–81. [DOI] [PubMed] [Google Scholar]
- 7.Leung GH, Gray TJ, Cheong EY et al. Persistence of related blaIMP-4 metallo-β-lactamase producing Enterobacteriaceae from clinical and environmental specimens within a burns unit in Australia—a six-year retrospective study. Antimicrob Resist Infect Control 2013; 2: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerra B, Fischer J, Helmuth R. An emerging public health problem: acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet Microbiol 2014; 171: 290–7. [DOI] [PubMed] [Google Scholar]
- 9.Palmgren H, Aspan A, Broman T et al. Salmonella in black-headed gulls (Larus ridibundus); prevalence, genotypes and influence on Salmonella epidemiology. Epidemiol Infect 2006; 134: 635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iveson JB, Shellam GR, Bradshaw SD et al. Salmonella infections in Antarctic fauna and island populations of wildlife exposed to human activities in coastal areas of Australia. Epidemiol Infect 2009; 137: 858–70. [DOI] [PubMed] [Google Scholar]
- 11.Smith GC, Carlile N. Habitat use by silver gulls Larus novaehollandiae in the Sydney-Wollongong region, New South Wales. Wetlands (Australia) 1992; 11: 33–46. [Google Scholar]
- 12.Smith GC, Carlile N. Food and feeding ecology of breeding silver gulls (Larus novaehollandiae) in urban Australia. Colonial Waterbirds 1993; 16: 9–17. [Google Scholar]
- 13.Grimont P, Weill F. Antigenic Formulae of the Salmonella Serovars. 9th edn Paris, France: WHO Collaborating Centre for Reference and Research on Salmonella, 2007. [Google Scholar]
- 14.Anderson ES, Ward LR, Saxe MJ et al. Bacteriophage-typing designations of Salmonella typhimurium. J Hyg (Lond) 1977; 78: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward LR, de Sa JD, Rowe B. A phage-typing scheme for Salmonella enteritidis. Epidemiol Infect 1987; 99: 291–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fourth Informational Supplement M100-S24. CLSI, Wayne, PA, USA, 2014. [Google Scholar]
- 17.Dunne MW, Hardin DJ. Use of several inducer and substrate antibiotic combinations in a disk approximation assay format to screen for AmpC induction in patient isolates of Pseudomonas aeruginosa, Enterobacter spp., Citrobacter spp., and Serratia spp. J Clin Microbiol 2005; 43: 5945–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giske CG, Gezelius L, Samuelsen Ø et al. A sensitive and specific phenotypic assay for detection of metallo-β-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin Microbiol Infect 2011; 17: 552–6. [DOI] [PubMed] [Google Scholar]
- 19.Diancourt L, Passet V, Verhoef J et al. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 2005; 43: 4178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirth T, Falush D, Lan R et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 2006; 60: 1136–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 2000; 66: 4555–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espedido BA, Partridge SR, Iredell JR. blaIMP-4 in different genetic contexts in Enterobacteriaceae isolates from Australia. Antimicrob Agents Chemother 2008; 52: 2984–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maher D, Taylor DE. Host range and transfer efficiency of incompatibility group HI plasmid. Can J Microbiol 1993; 39: 581–7. [DOI] [PubMed] [Google Scholar]
- 24.Carattoli A, Bertini A, Villa L et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 2005; 63: 219–28. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Fernandez A, Fortini D, Veldman K et al. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 2009; 63: 274–81. [DOI] [PubMed] [Google Scholar]
- 26.Powling J. National Enteric Pathogens Surveillance Scheme, Human Annual Report 1996–2010. Victoria: Microbiological Diagnostic Unit, University of Melbourne, 2010. [Google Scholar]
- 27.Rabsch W, Andrews HL, Kingsley RA et al. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect Immun 2002; 70: 2249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Australian Group on Antimicrobial Resistance. The Evolution of Carbapenemases in Enterobacteriaceae in Australia. Updated 20 October 2014. http://www.agargroup.org/files/AGAR_Carbapenemase_evolution%20Final.pdf.
- 29.Dahmen S, Metayer V, Gay E et al. Characterization of extended-spectrum β-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Vet Microbiol 2013; 162: 793–9. [DOI] [PubMed] [Google Scholar]
- 30.Fischer J, Rodriguez I, Baumann B et al. blaCTX-M-15-carrying Escherichia coli and Salmonella isolates from livestock and food in Germany. J Antimicrob Chemother 2014; 69: 2951–8. [DOI] [PubMed] [Google Scholar]
- 31.Izdebski R, Baraniak A, Fiett J et al. Clonal structure, extended-spectrum β-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob Agents Chemother 2013; 57: 309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo S, Wakeham D, Brouwers HJ et al. Human-associated fluoroquinolone-resistant Escherichia coli clonal lineages, including ST354, isolated from canine feces and extraintestinal infections in Australia. Microbes Infect 2015; 17: 266–74. [DOI] [PubMed] [Google Scholar]
- 33.Sidjabat HE, Townell N, Nimmo GR et al. Dominance of IMP-4-producing Enterobacter cloacae amongst carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob Agents Chemother 2015; 59: 4059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo WU, Cheung YY, Lai E et al. Complete sequence of an IncN plasmid, pIMP-HZ1, carrying blaIMP-4 in a Klebsiella pneumoniae strain associated with medical travel to China. Antimicrob Agents Chemother 2013; 57: 1561–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.