Abstract
Background
We observed an increase in the number of rotavirus cases in the St. Louis area during the 2012–2013 rotavirus season compared with recent seasons. We aimed to determine whether the rotavirus cases during the 2012–2013 rotavirus season were of types not included in licensed vaccines.
Methods
Microbiology laboratories of 3 children's hospitals in St. Louis provided samples that were positive using rapid antigen tests from 2010 to 2013. The majority of samples were from St. Louis Children's Hospital. We determined rotavirus genotypes by polymerase chain reaction tests and further characterized a subset of viruses by genome sequencing and comparative sequence analysis.
Results
Eighty-six percent (24 of 28) of typed viruses analyzed from the 2012–2013 rotavirus season were G12. We performed whole genome sequencing on 8 G12 viruses, all of which were VP4 type P[8]. The sequenced viruses showed differences from vaccine strains in major antigenic epitopes on the VP7 protein, but most epitopes on VP4 were conserved. Rotavirus vaccine histories were available for 11 G12 cases, of whom 10 had not been vaccinated.
Conclusions
G12 was a dominant community-wide genotype in 2013. Most of the G12 cases for whom vaccine histories were available had not received rotavirus vaccine. The experience demonstrates the potential for rapid shifts in rotavirus genotype distribution and underscores the need for vigilant surveillance to detect unusual genotypes that might escape from vaccine protection.
Keywords: G12, genomic, rotavirus
(See the Editorial Commentary by Leshem and Parashar on pages e90–e92.)
Before the widespread utilization of rotavirus vaccine, rotavirus infection was a common cause of severe diarrhea in children under 5 years old, estimated to account for 200,000 emergency room visits and 55,000–70,000 hospitalizations each year in the United States [1]. Highly efficacious rotavirus vaccines have led to a dramatic decline in rotavirus disease in the United States [2–5]. However, rotavirus comprises an antigenically diverse group of viruses, causing concern for the emergence of rotavirus types for which infection may not be prevented by rotavirus vaccines that were directed at the types most common in the United States in the prevaccine era [6, 7].
Rotavirus typing is based on the 2 outer capsid proteins—the attachment protein VP4 (the P type) and the glycoprotein VP7 (the G type)—which contain epitopes that are important for eliciting immunity [8]. Ten G genotypes and 8 P genotypes have been detected in humans. Two rotavirus vaccines are currently licensed for use in the United States. RotaTeq® (RV5) (Merck & Co., Inc, Whitehouse Station, NJ) consists of a mixture of 5 bovine reassortant viruses that contain VP7 and VP4 genes from human G1, G2, G3, and G4 and P[8] viruses. Rotarix® (RV1) (GlaxoSmithKline Biologicals, King of Prussia, PA) consists of an attenuated virus derived from a human G1P[8] strain. The two vaccines offer comparable protection against commonly circulating rotavirus serotypes G1-4 [2–6, 9]. Types G9 and G12 rotaviruses, neither of which is represented in currently licensed vaccines, are less common types that have emerged and spread worldwide, beginning prior to vaccination [10, 11]. G12 has recently become prevalent in a number of countries in the developing world [12–15]. It has been unusual in the United States [16] with the exception of an outbreak in Rochester, New York in 2006 [16, 17] and a small set of adult samples from Chicago in 2010 [18]. The New Vaccine Surveillance Network surveyed 7 sites in the United States during the 2009–2010 and 2010–2011 seasons, and G12 viruses accounted for approximately 10% of the typed rotaviruses [5].
The Microbiology Laboratory at St. Louis Children's Hospital (SLCH) tests specimens from a large area around St. Louis. After the reintroduction of a rotavirus vaccine in 2006, the number of laboratory-confirmed rotavirus cases decreased through the 2009–2010 rotavirus season (December through May in St. Louis), but increased in the 2010–2011 and 2012–2013 seasons (Figure 1). Because of concern that the rotavirus resurgence might represent emergence of a strain not included in the vaccine, we genotyped available rotaviruses in samples from 2010 to 2013 and document the emergence of G12 rotavirus in St. Louis.
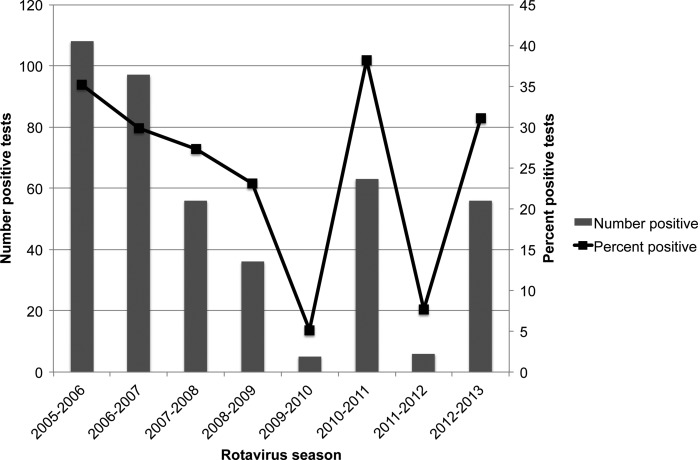
Figure 1.
Rotavirus-positive stool samples tested at St. Louis Children's Hospital, 2005–2013. The numbers of samples that tested positive for rotavirus during each rotavirus season are shown in the bar graphs (primary x-axis). The percentages of the total rotavirus tests that were positive each season are represented as points on the line (secondary x-axis). Note that the 2005–2006 season counts represent totals from January through May, and all other seasons represent December through May.
METHODS
Rotavirus testing was performed using the X/pect® Rotavirus rapid antigen test (Remel, Lenexa, KS) on stool samples submitted for rotavirus testing to the Microbiology Laboratory at SLCH. Some samples from the other 2 children's hospitals in the St. Louis area (Cardinal Glennon Children's Medical Center and Mercy Children's Hospital) were also tested. The samples that were typed were a convenience sample based on availability of residual sample material after clinical testing. No selection criteria were applied, and all of the samples available were typed. VP7 genotyping was carried out using reverse transcriptase–polymerase chain reaction (PCR) and sequencing [19]. A limited number of samples from 2012 to 2013 that contained high levels of virus as determined by PCR were selected for whole genome sequencing. Methods are fully described in Supplemental Material S1 (see online for Supplementary Material). This work was carried out under protocols approved by the Washington University Human Research Protection Office.
RESULTS
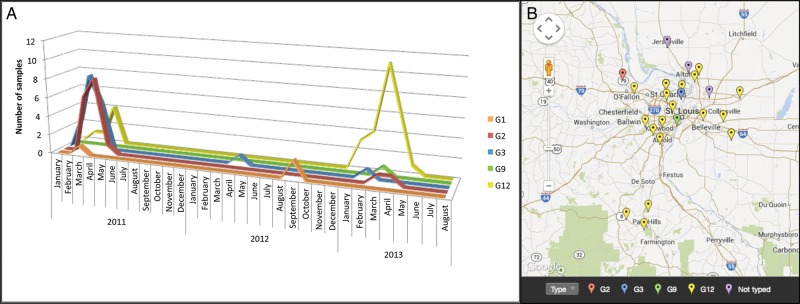
The numbers and the percentages of samples positive for rotavirus in the laboratory at St. Louis Children's Hospital are shown in Figure 1 for the rotavirus seasons from 2005 through 2013 (also see Supplementary Materials Table 1). We attempted VP7 genotyping on 77 available rotavirus-positive samples from 2010 to 2013, of which 68 were from St. Louis Children's Hospital (SLCH), 5 were from Cardinal Glennon Children's Medical Center (CGCMC), and 4 were from Mercy Children's Hospital (MCH). All of the samples from CGCMC and MCH were from the 2012 to 2013 rotavirus season. Eight of the SLCH samples could not be typed due to failure of PCR amplification, resulting in availability of typing results for 69 of the 77 available samples (Figure 2A and Supplementary Materials Table 2). G12 accounted for 24 (86%) of 28 typed samples from the 2012–2013 rotavirus season (95% confidence intervals 68%–95%), including 8 of the 9 samples from CGCMC and MCH. In contrast, genotypes detected in the 38 samples from the 2010–2011 winter–spring period were more varied: 15 (39%) were G2, 16 (42%) were G3, 6 (15%) were G12, and 1 was G1. The only sample available from the 2011–2012 rotavirus season was G3, and 2 samples collected in 2011 outside the rotavirus season were G1. The samples that were typed included 38 (60%) of the 63 rotavirus-positive samples tested by the SLCH laboratory during the 2010–2011 season, 16% (1 of 6) from the 2011–2012 season, 34% (19 of 56) from the 2012–2013 season, and 2 of 2 out-of-season samples (Figure 1 and Supplementary Materials Table 2).
G12-positive stool samples from the 2012–2013 season were submitted for testing between January 12 and May 1, 2013. The median patient age was 3.5 years (range 3 weeks–10 years). Patients with G12 resided throughout the hospital catchment area (Figure 2B). There were no recognized clusters of cases in households or geographic regions to suggest common exposures. Vaccine histories were obtained from 13 (57%) of the 23 subjects whose samples were tested at SLCH during the 2012-13 rotavirus season, 19 of which were successfully genotyped (Supplementary Materials Table 3). Of the 16 with G12, vaccine histories were available from 11, of whom 10 had not received rotavirus vaccine. The single vaccinated child with G12 was 21 months old at the time of illness. The unvaccinated children with G12 had a range of ages at the time of illness: 2 were ≤1 month of age and 4 were born in 2006 or before; the other ages were 18, 33, 49, and 73 months. Vaccine histories were not available from the 3 cases who had genotypes other than G12. Of the 4 subjects with undetermined rotavirus genotype, vaccine histories were available from 2, 1 of whom had not received rotavirus vaccine and 1 of whom had received 1 dose. Rotavirus vaccine uptake for Missouri children 19–35 months of age during 2012 was estimated at 69.3% by the National Immunization Survey [20]. RV5 was estimated to account for 57% and RV1 for 43% of rotavirus vaccine use in Missouri during 2012. In the Missouri counties included in the St. Louis metropolitan area, RV5 was estimated to account for 75% and RV1 for 25% of vaccine usage (data provided by the Missouri Department of Health and Senior Services) (Supplementary Materials Table 4).
Figure 2.
Rotavirus VP7 serotypes (G types) 2011–2013. (A) Samples determined to be positive for rotavirus (Fig. 1) were typed using a targeted polymerase chain reaction (PCR) and amplicon sequencing approach. Different colored lines represent each subtype detected. The number of samples of each subtype (y-axis) is plotted over time (x-axis). (B) The G12 serotype was distributed throughout the St. Louis metropolitan area in the 2012–2013 rotavirus season. Zip codes were used to map the cities of residence of the patients from the 2012–2013 rotavirus season. This includes patients from St. Louis Children's Hospital, Mercy Hospital, and Cardinal Glennon Children's Medical Center. Not shown are 3 patients from more remote locations in Missouri and Illinois (2 G12, 1 G2) and 1 patient from Arkansas (G12). Note that “not typed” indicates that the reverse transcriptase–PCR/sequencing reactions failed.
We performed whole genome shotgun sequencing on 8 G12 rotavirus-positive samples from 2012 to 2013. The sequences were used to type each gene, and each virus was determined to be P[8] and had a genotype constellation of I1-R1-C1-M1-A1-N1-T1-E1-H1, consistent with the Wa-like genogroup 1 [21]. Of the 6 G12P[8] viruses with nucleotide sequence coverage of at least 900 bases of the VP7 gene (Supplementary Materials Table 5), sequences from 3 samples (RVAA-3, -8, and -9) were 100% identical in the VP7 open reading frame. RVAA-6 was the most divergent from other St. Louis strains, with 96% nucleotide sequence identity. Phylogenetic trees based on VP7 showed that all of the G12 sequences were closely related to sequences from viruses detected in Rochester, New York in 2006–2007 [17] and Memphis, Tennessee in 2008 [22] or Sri Lanka in 2005 [23] (Supplementary Materials Figure 1). VP4 genes from the St. Louis strains were 99% identical to each other, except for RVAA-10, which was 97% identical.
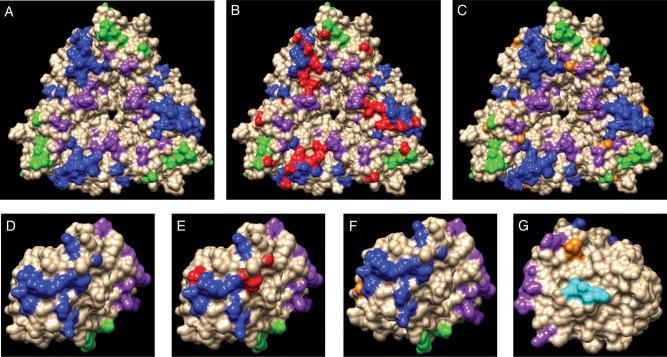
Comparison of G12 sequences corresponding to epitopes that are important in the immune response from St. Louis strains to sequences in RV5 and RV1 vaccine strains revealed multiple predicted amino acid differences within 3 well-characterized VP7 epitopes [24, 25]: 7-1a, 7-1b, and 7-2 (Figure 3A and B, Supplementary Material Table 6). The St. Louis G12 viruses also differed in 7 VP7 amino acids that were not within the critical epitopes but might be important to the immune response because of the proximity of 6 of them to epitopes (Figure 3C, Supplementary Material Table 6).
Examination of the VP4 genes from the St. Louis viruses showed 90–93% identity to the P[8] VP4 genes from the vaccine strains. Phylogenetic trees showed that most of the VP4 genes from St. Louis were closely related to the Rochester, New York strains from 2006–200717 and Memphis, Tennessee from 2008 (Supplementary Material Figure 2). We also examined changes in VP4 neutralization epitopes [22, 26–28]. The VP4 protein is cleaved by cellular proteases into two proteins, VP5* and VP8*. The St. Louis viruses matched the vaccine strains in epitopes 8-2, 8-4, 5-2, 5-3, 5-4, and 5-5 (Supplementary Material Table 6). However, the St. Louis strains differed from both vaccines in VP5* epitope 5-1 (Supplementary Material Table 6) and in VP8* epitopes 8-1 and 8-3 (in 1 virus) (Figure 3D and E and Supplementary Material Table 6). Furthermore, there were 11 additional differences shared among St. Louis strains at amino acid positions not associated directly with characterized epitopes (Figure 3F and G and Supplementary Material Table 6).
DISCUSSION
Rotavirus group A type G12, a globally emerging type [29] that has been unusual in the United States, accounted for most of the cases of rotavirus detected in children in the St. Louis metropolitan area during the 2012–2013 rotavirus season. To our knowledge, previous rotavirus serotype data from St. Louis is lacking, except for our finding that G12 accounted for 15% (6 of 38) of rotavirus-positive samples that were typed from the 2010–2011 rotavirus season. There were very few rotavirus-positive samples during the 2011–2012 season (Figure 1).
G12 rotaviruses were first reported in the United States in 1999 [30]. A survey of rotavirus serotypes in the United States from the years 1996–2005, prior to the introduction of current rotavirus vaccines, revealed only 1 G12 virus [31]. Surveys of samples collected after 2005 show G12 accounting for 2–11% of circulating rotaviruses [22, 32, 33], with the following 2 exceptions that we are aware of. First, G12 was predominant in Rochester, New York during the 2006–2007 rotavirus season, accounting for 69% of rotavirus-positive stool samples that were typed during that season, which was the rotavirus season during which RV5 was licensed [16, 17]. No G12-positive samples were detected in the preceding or the following rotavirus season. Second, G12 was also found to account for 53% of a small group of rotavirus-positive stool samples from adults in Chicago in 2010 [18].
The emergence or reemergence of G12P[8] rotavirus could be the result of vaccine pressure exerted on rotavirus types that are more similar to those in currently licensed vaccines or could represent genotype fluctuation occurring independently of the vaccine. Our study does not address which of these mechanisms was responsible. However, a recent study estimated that RV5 provides 83% protection against G12 [8]. Data were insufficient to allow a similar calculation for RV1 [5]. Antigenic overlap involving conserved epitopes of VP4 (P[8]) as well as epitopes on other proteins most likely accounts for the heterotypic protection against type G12.
This study has several limitations. The study was not a prospective study. Rather, we initiated the study when we observed an increase in rotavirus-positive stool samples during the 2012–2013 rotavirus season. Thus, genotyping of samples from before the early part of 2013 and all previous years was limited to the samples that were available in the clinical laboratory freezers. Because this was a convenience sample, biases may have been inadvertently introduced. Although we have no reason to suspect changes in the population being tested or the criteria used to send a specimen for rotavirus testing, we cannot exclude these possibilities. Additionally, because of the retrospective nature of the study, we were only able to obtain vaccine histories from a subset of patients. Nevertheless, the findings that (1) G12 accounted for 86% of the samples from the 2012–2013 rotavirus season that were typed; (2) 57% of the subjects from that season had vaccine histories available, with no obvious biases in the subset; and (3) 90% of the individuals with G12 and known vaccine status were unvaccinated, lead us to suspect that G12 was truly the dominant rotavirus genotype in the St. Louis area during the 2012–2013 winter–spring period and that most of the individuals with G12 had not been vaccinated.
Although there is no evidence that the G12-dominant season we report resulted from vaccine escape, the experience may have important long-term implications for rotavirus vaccine. The G12 viruses we sequenced and other G12 viruses differ from the licensed vaccine strains at multiple amino acids within important VP7 epitopes (Figure 3 and [6, 34–36]). Thus, G12 could provide a genomic platform from which recombination and/or mutation could give rise to a true escape virus. Vigilant virologic surveillance at many sites throughout the country is essential for maintaining the dramatic public health benefits that have been brought about by rotavirus vaccination.
Figure 3.
Amino acid changes in key epitopes of rotavirus VP7 and VP4. Amino acid changes mapped to the surface of VP7 (A–C) and VP4 (VP8*) (D–G). Panel A shows the epitopes 7-1a (blue), 7-1b (green), and 7-2 (purple) on a VP7 trimer (protein model PDB 3FMG). In panel B, amino acids within the epitopes that are different in serotype G12 viruses compared with vaccine strains are indicated in red. In panel C, additional amino acids that differed between the serotype G12 and vaccine strains but are not part of the epitopes are indicated in orange. Panel D shows the epitopes 8-1 (blue), 8-2 (green), 8-3 (purple) associated with VP8* (protein model PDB 1KQR). In Panel E, amino acids within the epitopes that are different in G12 viruses compared with vaccine strains are indicated in red. In Panel F, additional amino acids that differed between the G12 and vaccine strains but are not part of the epitopes are indicated in orange. In panel G, the VP8* protein is rotated to show epitope 8.4 (cyan) on the other side of the molecule. An amino acid in serotype G12 that differs from the vaccine strain is indicated in orange.
Supplementary Data
Supplementary materials are available at the Journal of The Pediatric Infectious Diseases Society online (http://jpids.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Acknowledgments
The authors would like to thank Brandi Herter and Maria Cannella for their contributions to the sequencing and virus subtyping work; Dr. Richard Buller and Stephanie Bledsoe from the Clinical Virology Laboratory at St. Louis Children's Hospital, Patricia Sellenriek and David Winkler from the Microbiology Laboratory at St. Louis Children's Hospital, Dr. Ella Swierkosz from Cardinal Glennon Children's Medical Center, and Rhonda Ferrett from Mercy Children's Hospital for supplying the rotavirus-positive stool samples; Kusal Mihindukulasuriya for obtaining vaccine histories from patients' families, and Damon Ferlazzo from the Missouri Department of Health and Senior Services for providing information about rotavirus vaccine utilization in Missouri.
The Virus Pathogen Database and Analysis Resource (ViPR) has been wholly funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200900041C. Molecular graphics images were produced using the UCSF Chimera package from the Computer Graphics Laboratory, University of California, San Francisco, supported by NIH P41 RR-01081.
Disclaimer. The National Institutes of Health had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Financial support. This work was supported by the National Institutes of Health [U54 HG003079 and U54 HG004968].
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Fischer TK, Viboud C, Parashar U, et al. Hospitalizations and deaths from diarrhea and rotavirus among children. J Infect Dis 2007; 195:1117–25. [DOI] [PubMed] [Google Scholar]
- 2.Vesikari T, Karvonen A, Prymula R, et al. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet 2007; 370:1757–63. [DOI] [PubMed] [Google Scholar]
- 3.Vesikari T, Matson DO, Dennehy P, et al. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med 2006; 354:23–33. [DOI] [PubMed] [Google Scholar]
- 4.Block SL, Vesikari T, Goveia MG, et al. Efficacy, immunogenicity, and safety of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine at the end of shelf life. Pediatrics 2007; 119:11–8. [DOI] [PubMed] [Google Scholar]
- 5.Payne DC, Boom JA, Staat MA, et al. Effectiveness of pentavalent and monovalent rotavirus vaccines in concurrent use among US children. Clin Infect Dis 2013; 57:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennehy PH. Rotavirus vaccines: an overview. Clin Microbiol Rev 2008; 21:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Ryan M. The ever-changing landscape of rotavirus serotypes. Pediatr Infect Dis J 2009; 28(3 Suppl):S60–2. [DOI] [PubMed] [Google Scholar]
- 8.Estes MK, Cohen J. Rotavirus gene structure and function. Microbiol Rev 1989; 53:410–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rha B, Tate JE, Payne DC, et al. Effectiveness and impact of rotavirus vaccines in the United States—2006-2012. Expert Rev Vaccines 2014; 13:365–76. [DOI] [PubMed] [Google Scholar]
- 10.Matthijnssens J, Bilcke J, Ciarlet M, et al. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol 2009; 4:1303–16. [DOI] [PubMed] [Google Scholar]
- 11.Banyai K, László B, Duque J, et al. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine 2012; 30(Suppl 1):A122–30. [DOI] [PubMed] [Google Scholar]
- 12.Kang G, Desai R, Arora R, et al. Diversity of circulating rotavirus strains in children hospitalized with diarrhea in India, 2005-2009. Vaccine 2013; 31:2879–2883. [DOI] [PubMed] [Google Scholar]
- 13.Wulan WN, Listiyaningsih E, Samsi KMK, Agtini MD, Kasper MR, Putnam SD. Identification of a rotavirus G12 strain, Indonesia. Emerging Infect Dis 2010; 16:159–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tra My PV, Rabaa MA, Vinh H, et al. The emergence of rotavirus G12 and the prevalence of enteric viruses in hospitalized pediatric diarrheal patients in southern Vietnam. Am J Trop Med Hyg 2011; 85:768–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ndze VN, Papp H, Achidi EA, et al. One year survey of human rotavirus strains suggests the emergence of genotype G12 in Cameroon. J Med Virol 2013; 85:1485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payne DC, Szilagyi PG, Staat MA, et al. Secular variation in United States rotavirus disease rates and serotypes: implications for assessing the rotavirus vaccination program. Pediatr Infect Dis J 2009; 28:948–53. [DOI] [PubMed] [Google Scholar]
- 17.Mijatovic-Rustempasic S, Teel EN, Kerin TK, et al. Genetic analysis of G12P [8] rotaviruses detected in the largest US G12 genotype outbreak on record. Infect Genet Evol 2013; 21c:214–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson EJ, Shippee DB, Weinrobe MH, et al. Indirect protection of adults from rotavirus by pediatric rotavirus vaccination. Clin Infect Dis 2013; 56:755–60. [DOI] [PubMed] [Google Scholar]
- 19.DiStefano DJ, Kraiouchkine N, Mallette L, et al. Novel rotavirus VP7 typing assay using a one-step reverse transcriptase PCR protocol and product sequencing and utility of the assay for epidemiological studies and strain characterization, including serotype subgroup analysis. J Clin Microbiol 2005; 43:5876–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Centers for Disease Control, ed. Centers for Disease Control. Available at: http://www.cdc.gov/vaccines/stats-surv/nis/tables/12/tab32_Rotavirus_race_iap_2012.pdf. Accessed August 1, 2014.
- 21.Matthijnssens J, Ciarlet M, Heiman E, et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 2008; 82:3204–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald SM, McKell AO, Rippinger CM, et al. Diversity and relationships of cocirculating modern human rotaviruses revealed using large-scale comparative genomics. J Virol 2012; 86:9148–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed K, Batuwanthudawe R, Chandrasena TGAN, et al. Rotavirus infections with multiple emerging genotypes in Sri Lanka. Arch Virol 2010; 155:71–5. [DOI] [PubMed] [Google Scholar]
- 24.Aoki ST, Settembre EC, Trask SD, Greenberg HB, Harrison SC, Dormitzer PR. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 2009; 324:1444–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald SM, Matthijnssens J, McAllen JK, et al. Evolutionary dynamics of human rotaviruses: balancing reassortment with preferred genome constellations. PLoS Pathog 2009; 5:e1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monnier N, Higo-Moriguchi K, Sun Z-YJ, Prasad BVV, Taniguchi K, Dormitzer PR. High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain. J Virol 2006; 80:1513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dormitzer PR, Nason EB, Prasad BVV, Harrison SC. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 2004; 430:1053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dormitzer PR, Sun Z-YJ, Wagner G, Harrison SC. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J 2002; 21:885–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthijnssens J, Heylen E, Zeller M, Rahman M, Lemey P, Van Ranst M. Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Mol Biol Evol 2010; 27:2431–2436. [DOI] [PubMed] [Google Scholar]
- 30.Griffin DD, Nakagomi T, Hoshino Y, et al. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6],G12) and rare P3[9] strains related to bovine rotaviruses. Virology 2002; 294:256–69. [DOI] [PubMed] [Google Scholar]
- 31.Gentsch JR, Hull JJ, Teel EN, et al. G and P types of circulating rotavirus strains in the United States during 1996-2005: nine years of prevaccine data. J Infect Dis 2009; 200(Suppl 1):S99–105. [DOI] [PubMed] [Google Scholar]
- 32.Hull JJ, Teel EN, Kerin TK, et al. United States rotavirus strain surveillance from 2005 to 2008: genotype prevalence before and after vaccine introduction. Pediatr Infect Dis J 2011; 30(1 Suppl):S42–7. [DOI] [PubMed] [Google Scholar]
- 33.Payne DC, Staat MA, Edwards KM, et al. Direct and indirect effects of rotavirus vaccination upon childhood hospitalizations in 3 US Counties, 2006-2009. Clin Infect Dis 2011; 53:245–53. [DOI] [PubMed] [Google Scholar]
- 34.Zeller M, Patton JT, Heylen E, et al. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq. J Clin Microbiol 2012; 50:966–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz-Palacios GM, Pérez-Schael I, Velázquez FR, et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med 2006; 354:11–22. [DOI] [PubMed] [Google Scholar]
- 36.Soares-Weiser K, Maclehose H, Bergman H, et al. Vaccines for preventing rotavirus diarrhoea: vaccines in use. Cochrane Database Syst Rev 2012; 11:CD008521. [DOI] [PubMed] [Google Scholar]