Abstract
Background
Beginning at birth, the microbes in the gut perform essential duties related to the digestion and metabolism of food, the development and activation of the immune system, and the production of neurotransmitters that affect behavior and cognitive function.
Objectives
The objectives of this review are to: (a) provide a brief overview of the microbiome and the “microbiome-gut-brain axis”; (b) discuss factors known to affect the composition of the infant microbiome: mode of delivery, antibiotic exposure, and infant feeding patterns; and (c) present research priorities for nursing science, and clinical implications for infant health and neurocognitive development.
Discussion
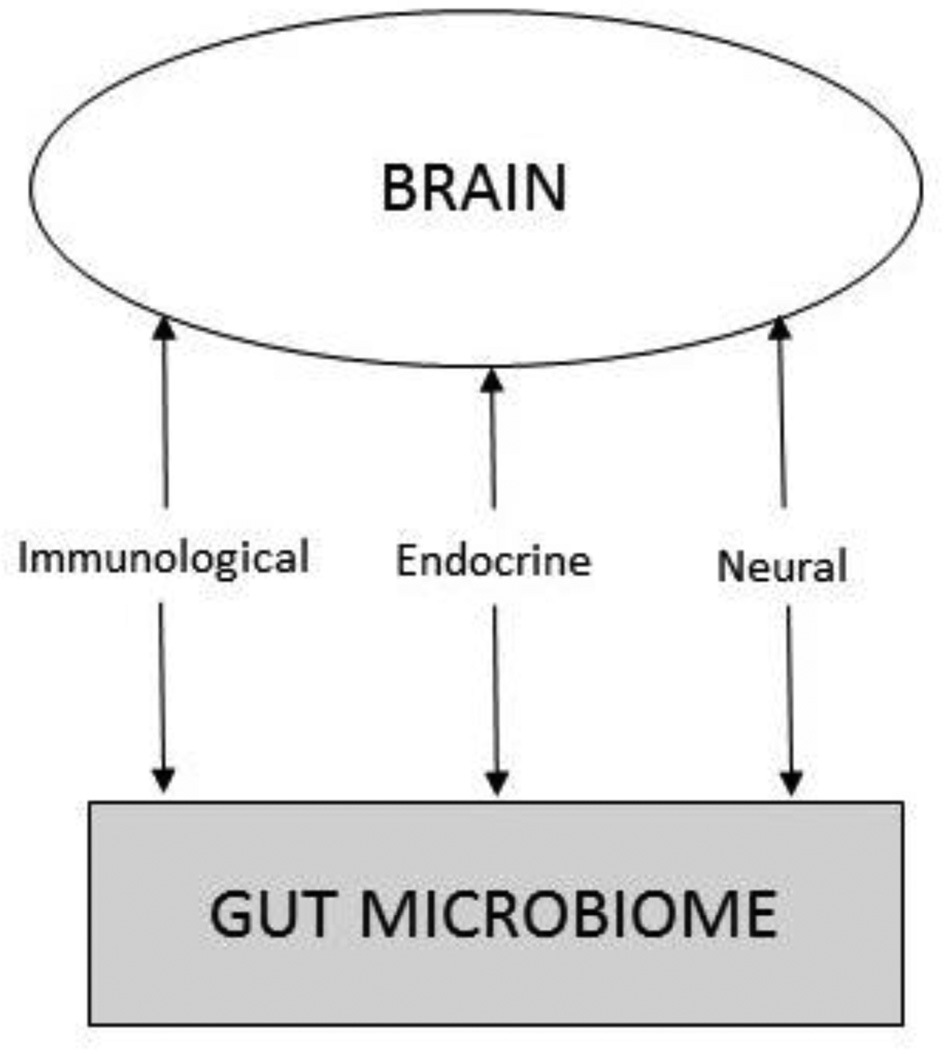
The gut microbiome influences immunological, endocrine, and neural pathways and plays an important role in infant development. Several factors influence colonization of the infant gut microbiome. Different microbial colonization patterns are associated with vaginal versus surgical birth, exposure to antibiotics, and infant feeding patterns. Because of extensive physiological influence, infant microbial colonization patterns have the potential to impact physical and neurocognitive development and life course disease risk. Understanding these influences will inform newborn care and parental education.
Keywords: biology, birth, microbiome, newborn
This article provides a brief overview of the microbiome and the gut-brain axis, followed by a focus on factors known to affect the composition of the infant microbiome during the birth process and the first 1,000 days of life. These include: mode of delivery (vaginal or surgical), antibiotic exposure, and infant feeding patterns. New evidence suggesting the influence of maternal stress on the composition of the infant microbiome, the state of the science, implications for nursing science, and long-term implications for clinical practice and infant health are identified as well.
The Microbiome
The human body is host to millions of microorganisms that live on us and in us, and function synergistically with our own cells to influence health outcomes across the lifespan—and potentially across generations. These microorganisms, referred to as “microbiota” (the organisms) or the “microbiome” (the organisms and their collective genetic makeup) carry out their actions by influencing immunologic, endocrine, and neural pathways. Although a wide variety of microorganisms flourish on the skin, in the oral cavity, and the urogenital tract, those in the gut are the most diverse and abundant and their functions are the best understood.
Starting from birth, the gut microbiota has three essential roles: protective, metabolic, and trophic (Guarner & Malagelada, 2003). First, gut microorganisms serve as a barrier against the proliferation of pathogenic organisms. Second, they play an important role in: the digestion and metabolism of colostrum, breast milk, formula, and weaning foods in infants, and a wide variety of food in adults; the breakdown of toxins and drugs; vitamin synthesis; and ion absorption. Trophic functions include the growth and differentiation of the epithelial cells lining the intestinal lumen, and the homeostatic maintenance of the immune system including tolerance to food antigens (Guarner & Malagelada, 2003). With complete colonization of the gut, which occurs within approximately three years of life (Koenig et al., 2011; Weng & Walker, 2013), immune homeostasis is achieved. In a healthy individual, the gut is in a state of eubiosis, populated by a diverse array of microorganisms and marked by oral tolerance to commensal bacteria and benign antigens (Walker, 2013). Inadequate colonization during this early period, however, may lead to dysbiosis (or an imbalance between commensal and pathogenic organisms) which may increase susceptibility to a variety of immune-related pathogenic states (Renz, Brandtzaeg, & Hornef, 2012; Walker, 2013) and other adverse metabolic or immune outcomes. In this way, the gut microbiome may be viewed as a key mediator between exposures to internal and external environmental factors, including diet and stress (Sudo, 2014), and health and developmental outcomes (Grenham, Clarke, Cryan, & Dinan, 2011).
In an attempt to further understand the role of gut microbial communities on human health, researchers have attempted to characterize its resident composition and diversity. In the past, studies of gut microflora utilized in vitro culture techniques—which identified a limited range of organisms due to the challenging requirements needed for their growth—as well as the lack of technology to detect them. Specimen transportation issues and the distribution of specific organisms within a given specimen also limited the meaningfulness of culture results (Finegold, Sutter, & Mathisen, 1983). More recently, however, researchers have begun using culture-independent analysis such as fluorescent in situ hybridization, gel electrophoresis, 16S ribosomal RNA cloning, and sequencing and quantitative polymerase chain reaction (PCR) (Brooks, 2013). (See Table 2 for definitions of terms.) Overall, culture and DNA methodology show some agreement; however, nuances in taxonomy have been identified with several gut microorganisms not grown in culture shown to be highly prominent when using newer technologies (Vael & Desager, 2009). From these more advanced studies, the organisms identified as the most prominent in the gut are Firmicutes (such as Clostridium, Enterococcus, Lactobacillus, and Ruminococcus), Bacteroidetes (such as Bacteroides and Prevotella) and, to a lesser extent, Proteobacteria and Actinobacteria (Brown et al., 2013; Power, O’Toole, Stanton, Ross, & Fitzgerald, 2014). In most circumstances, a highly diverse microbiome is advantageous for optimal health.
TABLE 2.
Glossary of Terms
| Term | Definition |
|---|---|
| 16S rRNA | 16S ribosomal RNA is a subunit of the ribosomal RNA containing specific signature sequences useful for bacterial identification. |
| Diversity | The range of different types of organisms and their relative abundance in a particular environment. |
| Dysbiosis | An unhealthy alteration or imbalance in microbial composition of a part of the body. |
| Eubiosis | An optimum balance of microflora in the gastrointestinal tract. |
| FISH | A technique using fluorescent probes to bind to specific portions of DNA, allowing for detection of specific DNA sequences. |
| Metagenomics | Field of research that transcends the individual organism to look at the collection of genomes within complex microbial communities. |
| Microbial profile | General term to describe the diversity of a given environment; i.e., identified species and their relative abundance. |
| Microbiome | Combined genetic material of microorganisms in a particular environment. May be used interchangeably with “microbiota.” |
| Microbiota | Microbial community in a particular environment. May be used interchangeably with “microbiome.” |
| PCR | A technique used to make multiple copies of DNA segments. |
| Taxonomy | A classification of organisms. |
Note. DNA = deoxyribonucleic acid; FISH = fluorescent in situ hybridization; PCR = polymerase chain reaction; RNA = ribonucleic acid; rRNA = ribosomal ribonucleic acid.
Gut-Brain Communication
While the brain’s regulation of gut function has long been recognized, only in the last decade has the bidirectional nature of this relationship been elucidated (Cryan & Dinan, 2012). As a result of the Human Microbiome Project (Peterson et al., 2009), evidence has mounted that gut-brain communication primarily occurs via interactions between the gut microbes and established psychoneuroimmunologic (PNI) pathways, including immunological (cytokines), endocrine (hypothalamic-pituitary-adrenal [HPA]), and neural (vagus) pathways (Grenham et al., 2011). These bidirectional relationships are illustrated in Figure 1.
FIGURE 1.
The gut-brain axis.
The Immune System
While a breach of gut integrity and exposure of gut microorganisms to host sites outside the gut clearly stimulates a robust inflammatory-immune response—even without an actual breach—the gut microbiome contributes to the systemic inflammatory milieu of the host. This is due to low-level leakage of bacteria and bacterial cell wall components like the powerful inflammatory stimulus, lipopolysaccharide (LPS). These cross the luminal intestinal host-microbial interface and gain access to the peritoneal cavity stimulating a systemic inflammatory response (Duerkop, Vaishnava, & Hooper, 2009). Moreover, certain microbes, including Gram-negative bacteria such as Enterobacteriaceae and Pseudomonadaceae, are inherently more likely when leaked, to stimulate a robust systemic inflammatory response (Bengmark, 2013); as such, when these microbes are the dominant species in the gut, a chronic, low-grade systemic pro-inflammatory state may result. In contrast, other bacteria, when present, maintain a more anti-inflammatory milieu in the gut, often by secreting chemicals that are inhospitable to colonization by more inflammatory microorganisms. An example of protective bacteria is Lactobacillus (which secretes lactic acid), lowering gut pH to a range prohibitive to the colonization of more inflammatory microbes (Adams & Hall, 1988; Haarman & Knol, 2006). In human infants, an inverse relationship between gut colonization with Lactobacillus and levels of interferon-gamma, interleukin-10, and interleukin-4 has been reported, while colonization with Staphylococcus aureus has been associated with elevated cytokine levels (Johansson et al., 2012), suggesting that early life dysbiosis of the microbiome may impact the developing immune system—perhaps increasing an infant’s tendency toward developing an inflammatory disease. Once stimulated, systemic inflammatory cytokines then exert effects on the central nervous system (CNS), shaping mood, stress response, and behavior via the initiation of what is often referred to as “sickness behaviors” (e.g., fatigue, insomnia, lack of appetite and depression) (Dantzer & Kelley, 2007; Elenkov & Chrousos, 2002; Raison, Capuron, & Miller, 2006).
The Endocrine Pathway
The composition of the microbiome influences the hypothalamic-pituitary-adrenal (HPA) axis by influencing cortisol secretion and the normal development of the stress response (Sudo, 2014; Sudo et al., 2004). Whether or not the microbiome directly affects the HPA axis remains under investigation; however, given the ability of pro-inflammatory cytokines to activate all levels of the HPA axis, (Elenkov & Chrousos, 2002), indirect effects via the microbiome-induced stimulation of pro-inflammatory cytokines, as described above, do occur. Furthermore, animal experiments suggest that the increased cortisol levels that accompany acute or chronic stress themselves increase gut permeability and bacterial LPS leakage across the gut wall (Santos, Yang, Söderholm, Benjamin, & Perdue, 2001), setting up a dangerous positive feedback cycle between chronic stress, the microbiome, and systemic inflammation. Animal studies have also shown that exposure to chronic stress reduces the diversity of the gut microbiome and affects the relative abundance of various types of resident bacteria in a manner that correlates with increases in pro-inflammatory cytokines, including interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-a) (Bailey et al., 2011). Stress-induced changes in the gut microbiome, including a decrease in the protective gut genus Lactobacillus, were also significantly related to an increased production of IL-6 and increased anxiety-like behavior (Bailey et al., 2011).
Neural Pathway
The third pathway by which the gut microbiome and the brain communicate is through the afferent and efferent fibers of the vagus nerve. The vagus nerve (the parasympathetic nerve of the autonomic nervous system) innervates and regulates the gut, maintains systemic homeostasis, promotes anti-inflammatory activity, and influences the CNS and behavior directly, and via interaction with the HPA axis and inflammatory mediators (Bailey et al., 2011).
Bacteria in the gut interact with cells in the gut wall to stimulate production of peptides that activate afferent endings of the vagus nerve. The resultant signals are transmitted to the CNS, affecting behavior and efferent neural activity. Similarly, pro-inflammatory cytokines appear to activate vagal afferent fibers, with vagal transmission of inflammatory signals believed to be a key mechanism by which the brain receives information regarding systemic inflammation, contributing to affective symptoms and initiating behavioral responses including depression and other sickness behaviors (Dantzer & Kelley, 2007; Miller, Maletic, & Raison, 2009; Raison et al., 2006)—giving rise to the notions of “gut instinct” or that “sinking feeling in the pit of your stomach” (Sonnenburg & Sonnenburg, 2015). Vagal afferents traveling from the gut are also thought to be responsible for “gut feelings” which act as signals to the brain that an environment may be threatening or anxiety provoking (Forsythe, Sudo, Dinan, Taylor, & Bienenstock, 2010; Mayer, 2011). Activation of the efferent fibers of the vagus, in turn, carry anti-inflammatory signals to the periphery, via what is termed the “cholinergic anti-inflammatory pathway,” ultimately reducing the production of pro-inflammatory cytokines, including tumor necrosis factor-alpha (TNF-α) (Pavlov, Wang, Czura, Friedman, & Tracey, 2003; Tracey, 2009; Vijayaraghavan et al., 2013).
Factors Influencing the Infant Microbiome
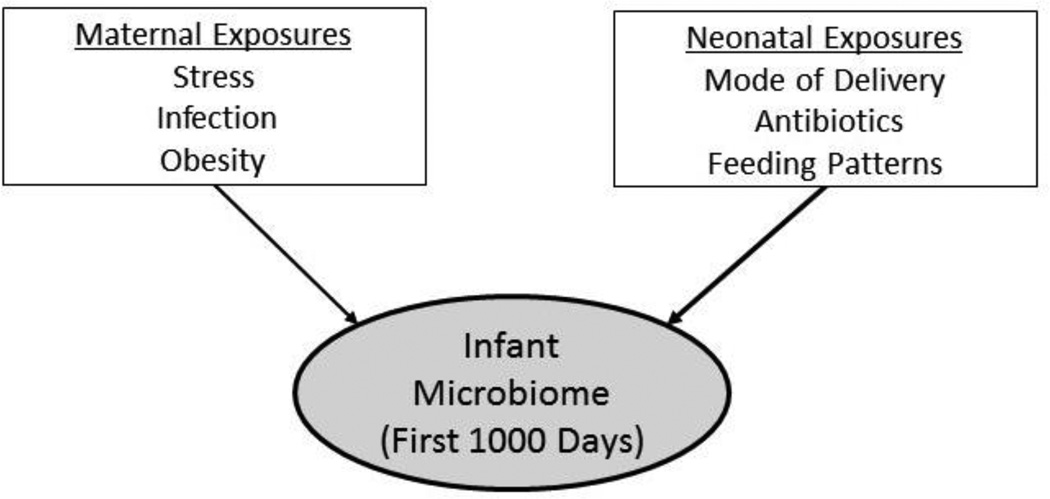
The newborn gut microbiome is less diverse than that of the adult. During the first three years of life, the development of the gut microbiome is influenced by the gut-brain axis (as described previously), and by maternal and neonatal exposures, including mode of delivery, antibiotic exposure, and feeding patterns (Figure 2). By the end of this period, the infant gut microbiome has assumed the diversity and composition of the adult gut and is generally characterized by species from four main phyla: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria (see Table 1).
FIGURE 2.
Maternal and neonatal factors influencing the development of the infant microbiome.
TABLE 1.
Phyla Identified in Infant Gut
| Phylum | Class | Genus examples | Species examples | Characteristics |
|---|---|---|---|---|
| Actinobacteria | Actinobacteria | Bifidobacterium |
|
|
| Atopobium |
|
|
||
| Corynebacterium |
|
|
||
| Propionibacterium |
|
|
||
| Bacteroidetes | Bacteroidetes | Bacteroides |
|
|
| Prevotella |
|
|
||
| Firmicutes | Bacilli | Lactobacillus |
|
|
| Enterococcus |
|
|
||
| Clostridium |
|
|
||
| Ruminococcus |
|
|
||
| Streptococcus |
|
|
||
| Staphylococcus |
|
|
||
| Clostridia | Escherichia |
|
|
|
| Klebsiella |
|
|
||
| Proteobacteria | Gammaproteobacteria | Enterobacter |
|
|
Mode of Delivery
A major factor contributing to the variation in the infant microbiome is the mode of delivery at birth. Infants born vaginally have a gut microbiome very similar to that of their mother’s vaginal and fecal flora. This occurs through vertical transfer of the vaginal-perianal microbes of the mother as the infant passes through the birth canal (Dominguez-Bello et al., 2010). For vaginally born infants, Lactobacillus, Prevotella, or Sneathia spp. dominates the gut, but within months there is a greater distribution of Bifidobacterium and Bacteroides (Azad et al., 2013; Gregory, 2011; Karlsson, Molin, Cilio, & Ahrné, 2011; Mitsou, Kirtzalidou, Oikonomou, Liosis, & Kyriacou, 2008; Penders et al., 2006). In addition to these genera of microbes, other studies have also found species of Atopobium, Streptococcus, Enterococcus, and Enterobacteriaceae within the first six weeks of development in vaginally delivered infants (Bezirtzoglou, 1997; Dominguez-Bello et al., 2010; Fallani et al., 2010; Rotimi & Duerden, 1981).
In contrast, the gut microbiome of an infant born by cesarean section comprised bacteria transferred horizontally from the mother’s and others’ skin surfaces and, to a lesser extent, the place of birth (Dominguez-Bello et al., 2010; Grölund, Lehtonen, Eerola, & Kero, 1999; Penders et al., 2006). This tends to result in an infant microbiome dominated by Staphylococcus, Corynebacteria, and Propionibacterium spp. with lower proportions of Bifidobacteria and Bacteroides spp. (Azad et al., 2013; Chen, Cai, & Feng, 2007; Dominguez-Bello et al., 2010; Fallani et al., 2010; Grölund et al., 1999; Huurre et al., 2008; Penders et al., 2006). These differences in initial newborn communities may have important health consequences since the genera Bifidobacterium and Lactobacillus are considered to be health protective (Rastall, 2004), whereas some Staphylococcus spp. and Clostridium spp. have pathogenic potential (Adlerberth & Wold, 2009).
In addition to differences in bacterial genera, the intestinal microbiome of infants born surgically shows less diversity compared to vaginally delivered infants (Adlerberth & Wold, 2009; Azad et al., 2013; Lif Holgerson, Harnevik, Hernell, Tanner, & Johansson, 2011). This is potentially important since, in general, increased diversity of microbes within the gut is considered protective, while low diversity has been linked to a variety of human diseases, including inflammatory bowel disease and obesity (Qin et al., 2010; Turnbaugh et al., 2009). Variances in bacterial composition among infants born surgically compared to vaginally appear to continue in a child up to seven years (Azad et al., 2013; Fanaro, Chierici, Guerrini, & Vigi, 2003; Grölund et al., 1999; Huurre et al., 2008; Salminen, Gibson, McCartney, & Isolauri, 2004). Additionally, it can take months to establish a stable gut microbiome in a surgically delivered neonate (Fanaro et al., 2003; Grölund et al., 1999; Huurre et al., 2008; Salminen et al., 2004) in light of the multiple-associated sequelae related to surgical birth. Thus, delivery mode appears to be an important factor in the development of the infant gut microbiome, since surgically delivered infants miss the unique opportunity to be inculcated with their mother’s vaginal microbiome via the birth canal (Gregory, 2011; Salminen et al., 2004). Additionally, in those cases where they are separated from their mothers after birth for an extended period of time, they may also miss the opportunity for immediate colonization with the maternal skin microbiome.
Delivery mode data, however, must be interpreted with caution due to several confounding factors that may impact the course of development of an infant’s gut microbiome. For example, the type of cesarean section, elective versus indicated after trial of labor, may affect an infant’s maternal microbial exposure. An infant who has come in contact with microbes in the vaginal canal during a trial of labor has a degree of exposure that is very different from the infant of an elective cesarean section—born without labor or prolonged rupture of membranes (Neu & Rushing, 2011). Some indications for cesarean section, such as infant being large for gestational age, maternal obesity, or fetal distress, may also suggest a dissemblance between the infant and mother, accounting for variability in the microbiome (Barau et al., 2006; Karlsson et al., 2011). Additionally, infants of mothers who undergo cesarean section are more likely to have been exposed to antibiotics in utero (Cho & Norman, 2013), which may significantly alter their enteric microflora (Jakobsson et al., 2010; Jernberg, Löfmark, Edlund, & Jansson, 2010).
Pre- and Postnatal Antibiotic Exposure
Exposure to antibiotic therapy and its modulatory effects on the human microbiome can begin in utero and continue throughout critical growth and developmental stages. Factors under consideration and expounded in the discussion include the effects of antibiotic exposure on the trajectory of microbial colonization pattern and diversity, as well as their possible contribution to early-onset obesity and neurodevelopmental delays. Ultimately, an avenue of future inquiry includes the long-term implications of altering a fragile microbial ecosystem through potentially unnecessary antibiotic exposure.
Healthy infants born to mothers receiving ampicillin prior to delivery to treat group B Streptococcus display a significant decrease in Bifidobacterium by the seventh day of life, highlighting and confirming the modulatory effects of intrapartum antibiotic interventions (Aloisio et al., 2014). Similarly, prophylactic antibiotic treatment in preterm infants is commonly practiced as a safeguard against colonization by pathogenic microorganisms in this vulnerable group. Consequently, this intervention reduces the diversity of gut flora (Fricke, 2014) and delays the colonization of commensal flora (La Rosa et al., 2014). La Rosa et al. (2014) demonstrated, however, that the delay in commensal gut flora was recovered in the sampled population by 36 weeks postconceptional age, suggesting a short-term decrease and sequential rebound of microbial diversity. This rebound property has not been shown to mitigate the risk for infections such as necrotizing enterocolitis (NEC), however, which remains one of the leading causes of death in preterm infants (Panigrahi, 2006). In fact, NEC occurs most frequently during periods of decreased microbial diversity (Wang et al., 2009). Associations between antibiotic use and common intestinal infections plaguing preterm infants are important considerations in the early treatment of preterm infants. Recurrent perturbations of the gut flora could potentiate windows of opportunity for pathogenic and antibiotic-resistant bacterial challenge and possible chronic disruption of microbial diversity, leading to a diseased state.
For example, intestinal dysbiosis related to ampicillin use has been shown to displace beneficial enterobacteria with that of ampicillin-resistant enterobacteria (Yu et al., 2014). Animal studies have shown that even after termination of low-dose penicillin, mice displayed altered microbial phenotypes and higher ratios of fat mass, with a decrease in fat mass attributed to later exposure to the antibiotic (Cox et al., 2014). Further, both pathogenic and normal microbial profiles have been linked to altered physical, behavioral, and memory functioning in recent animal models (Bilbo et al., 2005; Heijtz et al., 2011). The negative impact of antibiotic exposure on newborn intestinal flora should be recognized and considered in clinical decision making for laboring women. These considerations should inform the development of guidelines for risk management in laboring women requiring antibiotics.
Infant-Feeding Patterns
The most dramatic changes in the composition of the intestinal microbiome begin during the first year of life with the rapid microbial colonization of the newborn intestinal tract (Palmer, Bik, DiGiulio, Relman, & Brown, 2007). Research suggests that the microbial population that develops during the early months of a newborn’s life varies highly from infant to infant (Palmer et al., 2007; Stark & Lee, 1982), and that the newborn diet represents an essential extrinsic factor related to the establishment of the gut microbiota (Fanaro et al., 2003).
Diet Composition
Many studies suggest that the gut microbial profile of breastfed infants is dominated by Bifidobacterium (Bezirtzoglou, Tsiotsias, & Welling, 2011; Harmsen et al., 2000; Kleessen, Bunke, Tovar, Noack, & Sawatzki, 1995; Knol et al., 2005; Stark & Lee, 1982), with the addition of a few other anaerobes and small numbers of facultative anaerobic bacteria (Stark & Lee, 1982). It is thought that the colonization of Bifidobacterium and Bacteroides (another commonly found organism in the breastfed infant gut) is stimulated by the presence of human milk oligosaccharides (HMOs), the most abundant carbohydrate component in breast milk. Infants lack the enzymes necessary to digest HMOs causing them to pass into the lower intestinal tract where they are thought to function as a prebiotic, stimulating the growth of Bifidobacterium and Bacteroides (Marcobal & Sonnenburg, 2012). In addition to Bifidobacterium and Bacteroides, Streptococcus, and Lactobacillus have been found in breastfed infants (Harmsen et al., 2000).
Formula-fed infants exhibit a more diverse flora with the presence of species of Staphylococcus, anaerobic Streptococcus, and Clostridium in addition to Bifidobacterium (Harmsen et al., 2000; Stark & Lee, 1982). Some studies have shown that exclusively formula-fed infants are more often colonized with E. coli, C. difficile, B. fragilis group, and Lactobacilli than those that are exclusively breastfed (Penders et al., 2006; Penders et al., 2005).
The expectation of a dominance of Bifidobacterium spp. in breastfed infants, however, has been challenged by the results of more recent studies that show that both breastfed and formula-fed infants have similar counts of this genus (Adlerberth & Wold, 2009; Penders et al., 2005). Adlerberth and Wold (2009) reviewed 34 studies from the last 30 years and found that Bifidobacterium are found equally often in breast and formula-fed infants in most studies after 1980. Penders et al. (2005) suggest that this discrepancy may be due to changes in formula content and/or microbial identification techniques. Most studies over the last several decades are in agreement, however, that numbers of Clostridium are lower in breastfed infants. Additional trends observed included an increased prevalence of Bacteroides, Enterococci, and Enterobacteriaceae in formula-fed infants and increased Staphylococci in breastfed infants (Adlerberth & Wold, 2009).
The greater abundance of Clostridium spp., and in particular, C. difficile in the gut microbiota of formula-fed infants has implications for subsequent development of atopy. Penders, Stobberingh, van den Brandt, and Thijs (2007) demonstrated that infants colonized with C. difficile were at higher risk of developing several atopic symptoms, including eczema, recurrent wheeze, allergic sensitization, and diagnosis of atopic dermatitis.
Diet continues to play a primary role in generating compositional change and diversity in the microbiome as dietary patterns progress over the first three years. Studies have shown that major shifts in the taxonomic groups of the microbiome have been observed with changes in diet such as weaning to solid foods (Koenig et al., 2011). The introduction of table food to the breastfed infant causes a rapid rise in the number of enterobacteria and enterococci, followed by progressive colonization by Bacteroides spp., Clostridium, and anaerobic Streptococcus. In formula-fed infants, however, the transition to solid food does not have as great an impact on gastrointestinal flora (Stark & Lee, 1982). As the amount of solid food in the diet increases, the bacterial flora of both breast and bottle-fed babies approach that of adults (Stark & Lee, 1982) with a sustained increase in the abundance of Bacteroidetes, elevated fecal short chain fatty acid levels, enrichment of genes associated with carbohydrate utilization, vitamin biosynthesis, xenobiotic degradation, and a more stable community composition characteristic of the adult microbiota (Koenig et al., 2011).
Beyond the transition to table foods, we know that types of food and dietary habits influence the gut microbiome (Kau, Ahern, Griffin, Goodman, & Gordon, 2011). In a comparison of rural African and European children, rural African children who consumed a low-fat, predominantly vegetarian diet had more of the Bacteroides enterotype and less Firmicutes. The European cohort, however, demonstrated the opposite with a significantly increased Firmicutes to Bacteroidetes ratio (De Filippo et al., 2010). The ratio of Firmicutes to Bacteroidetes (F/B) differs between obese and lean humans, with obese people having fewer Bacteroidetes and more Firmicutes than their lean counterparts (Ley, Turnbaugh, Klein, & Gordon, 2006). It has been theorized, therefore, that the increase in the F/B ratio in children in the European Union, probably driven by their high-calorie diet, might predispose them to future obesity (De Filippo et al., 2010). In addition, the gut microbiome in the rural African children contained an abundance of two bacterial species (Prevotella and Xylanibacter) absent from the Western cohort’s microbiome. These two species have enzymes necessary for the hydrolysis of starchy fibers (De Filippo et al., 2010). The fermentation of these starchy fibers produces large amounts of short-chain fatty acids which have been shown in preclinical studies to be critically important for immune-regulation (Scheppach & Weiler, 2004).
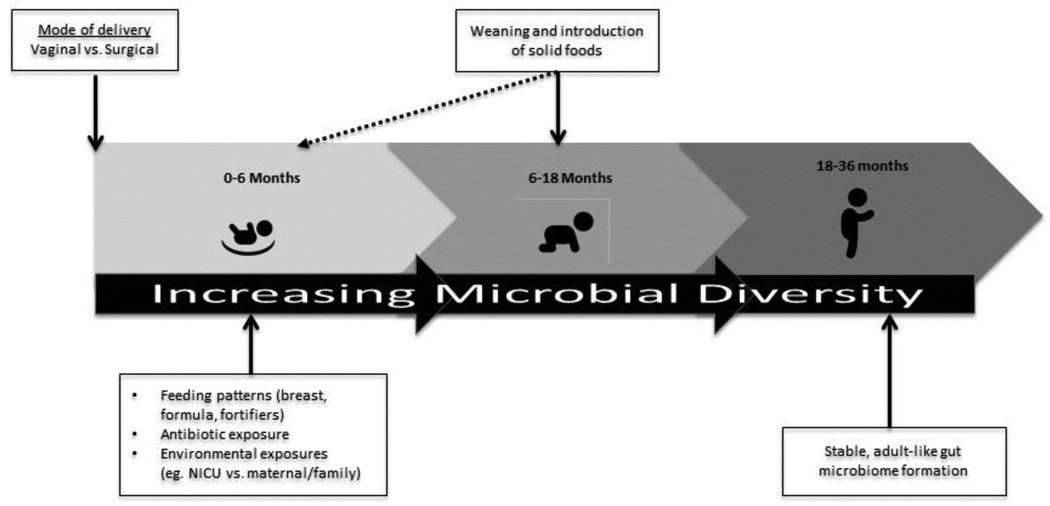
In summary, diet composition and patterns during the first three years of life may impact the diversity and functional capacity of the gut microbiome with potential downstream effects (as detailed below) on infant development and disease risk (Johnson & Versalovic, 2012). Understanding the colonization patterns of the gut microbiota during infancy and early childhood, the factors that influence colonization (see Figure 3), and the mechanisms through which the gut microbiota interact with immune regulation, the endocrine system, and metabolism may help in the development of strategies to guide the formation of health-promoting microbiotas that could then be maintained throughout the lifespan (Koenig et al., 2011).
FIGURE 3.
Timeline of factors influencing the development of the infant gut from birth to three years.
Preterm Birth
Because of the relationship between each of the above-described factors and preterm birth, premature infants and those at a very low birth weight are at risk for gut microbial dysbiosis. Premature infants most likely have come into the world by rapid vaginal birth or surgical birth. Both of these delivery modes reduce their exposure to maternal enteric and vaginal flora. They are more likely to have both in utero and neonatal antibiotic exposure, and are more likely to be formula fed and given human nutrition fortifiers. All of these prenatal and postnatal factors may affect the development of the premature infant’s gut microbiome. In addition, preterm birth is associated with several inflammatory factors: prenatal maternal illness, infections, smoking, and stress. After birth, these infants are more likely to have increased exposure to invasive procedures, flora of the environment of the newborn intensive care unit, medications that alter the pH of the intestinal environment, and decreased exposure to maternal and home microbial environments (Groer et al., 2014).
The Microbiome-Gut-Brain Axis and Implications for Infant Development
The first years of life are a time of rapid change in both the gut microbiome and the developing infant CNS. As a result, the microbiome has increasingly become a focus of clinical and preclinical studies of neurocognitive and emotional development. While studies in humans are just beginning, research on rodents indicate that through perturbation of the three psychoneuroimmune pathways described previously (immune, HPA axis, and vagus nerve), the gut microbiome modulates brain development, neurotransmitter systems, canonical signaling pathways, synaptic related proteins, and behavior (Clarke, O’Mahony, Dinan, & Cryan, 2014). As a result, growing evidence suggests the gut microbiome exerts influence over a range of developmental indices, from cognition to anxiety, mood, and sociability (Borre et al., 2014; Moloney, Desbonnet, Clarke, Dinan, & Cryan, 2014). An enhanced understanding of how the microbiome-gut-brain axis operates during infancy may thus provide novel insights into early-life neurocognitive and emotional development.
Rodent studies have identified deficits in social functioning (Desbonnet, Clarke, Shanahan, Dinan, & Cryan, 2014) and working memory (Gareau et al., 2011) in animals born microbiota-free. Notably, in these animals, postweaning microbial colonization of the gut resulted in a reversal of the previous social deficits (Desbonnet et al., 2014), suggesting that microbiome-associated developmental delays might be modifiable through treatment. In humans, correlational studies have linked the neurodevelopmental disorder autism to an abnormal gut microbiome composition (Mulle, Sharp, & Cubells, 2013). However, these studies have been completed on previously diagnosed individuals making it difficult to control for confounders such as antibiotic use and diet that may have impacted the microbiome.
Both animal and human studies also have demonstrated that high levels of glucocorticoids negatively impact brain structure and function in areas relevant to cognitive and emotional development (Carrion & Wong, 2012; Gunnar, 1998), and, as described previously, studies of germ-free animals have demonstrated the importance of the microbiome in regulating the HPA axis (Dinan & Cryan, 2012). One largely unexplored, but potentially important influence on the HPA axis and the infant microbiome, is maternal prenatal stress. Animal (Bailey et al., 2011; Wenzl, Schimpl, Feierl, & Steinwender, 2003) and more recent human studies (Zijlmans, Korpela, Riksen-Walraven, de Vos, & de Weerth, 2015) have shown a positive association between maternal stress levels and levels of pathogenic strains of Escherichia and Enterobacter, and an inverse association with levels of beneficial bacteria such as Lactobacillus or Bifidobacteria in the infant gut microbiome. These stress-induced changes in gut microbial composition are significantly related to increased production of pro-inflammatory cytokines like IL-6 (Bailey et al., 2011; Bailey, Lubach, & Coe, 2004; Galley et al., 2014), and exposure to these pro-inflammatory cytokines has been associated with damage to the developing brain, interfering with white matter and brain plasticity (Dammann & O’Shea, 2008).
In summary, the gut microbiome plays a major role in key systems that have the capacity to influence CNS development. Although the field of microbiome-gut-brain axis is still new, growing evidence suggests the gut microbiome plays a key role in brain functioning, ranging from cognition to anxiety, mood, and sociability (Borre et al., 2014; Moloney et al., 2014) and, therefore, an enhanced understanding of how the microbiome-gut-brain axis operates during infancy may provide novel insights into early-life neurocognitive and social-emotional development. In addition, a better understanding and appreciation of several potentially modifiable factors that influence the newborn and infant microbiome is essential, particularly for midwives and neonatal nurses who care for women and their infants during the crucial first 1,000 days of life.
State of the Science
The composition of the adult gut microbiota has been intensely studied using both classical culture techniques and more recently developed ribosomal DNA sequence-based methods (Palmer et al., 2007). Findings suggest that the adult microbial gut is made up of over 400 bacterial species belonging to a small number of broad taxonomic divisions (Eckburg et al., 2005; Palmer et al., 2007). There is significant interindividual variation in the adult gut microbial profile. However, the intra-individual profile appears to remain stable for months at a time (Eckburg et al., 2005; Palmer et al., 2007), although acute changes in response to new dietary practices, antibiotics, or infection can be seen and, in some instances, persist. By contrast, we know that the infant intestinal microbiota differs significantly from the adult microbiome. Infant gut microbiome studies have begun to elucidate the progression of diversity from birth to a relatively stable gut environment by the age of three. Major external influences on the infant gut microbiome are mode of delivery, antibiotic exposure, and infant feeding patterns. Animal research suggests infant stress exposure also exerts significant influence. Studies also suggest that, during this period, the gradual growth in diversity is interspersed with periods of large shifts in the relative abundance of taxonomic groups. These shifts are theorized to be related to life events such as drastic diet changes or medication intake (Koenig et al., 2011; Palmer et al., 2007). Emerging evidence also suggests that inadequate colonization or dysbiosis in this early period may be associated with several disease processes like necrotizing enterocolitis, inflammatory bowel disease, obesity, and atopy and asthma (Arrieta, Stiemsma, Amenyogbe, Brown, & Finlay, 2014).
The bulk of knowledge on the infant gut microbiome relies on research utilizing traditional culture methods. Only few, recent studies have used high throughput metagenomic analysis (Clarke et al., 2014). Advances in DNA methodologies now allow for more accurate analysis of the gut microbiota in a culture-independent way using genetic analysis. With techniques like fluorescence in situ hybridization (FISH) or real-time polymerase chain reaction (PCR), bacterial species that were previously undetectable by culture, are now able to be identified, giving us a better picture of the infant gut microbial profile (Adlerberth & Wold, 2009; Bezirtzoglou et al., 2011).
Research Priorities for Nursing Science
Given the “critical window” of development that occurs for the infant gut microbiota, there is a need for large cohort studies that survey the infant microbiome from birth and throughout the first few years of life. Understanding the microbial and metabolic changes that occur during this period may explain the development of later pathologies that emerge (Arrieta et al., 2014). Priorities for research include investigating basic questions on what constitutes a “healthy” gut microbial profile. Do desirable taxonomic combinations exist? Or is diversity of fundamental importance (Azad et al., 2013)? Other important questions concern variables that influence the development of the infant gut microbiome. For example, we know that mode of delivery, antibiotic exposure, and infant-feeding patterns impact gut microbial composition, but what other variables impacting colonization patterns are we missing? How important is the early infant environment and/or life events (family, pets, daycare, early childhood illness, medications, secondhand smoke exposure) (Azad et al., 2013)? Do these variables impact the gut microbiome of term versus preterm infants differently? Do they interact and if so, how? Finally, the potential to longitudinally track relationships between early gut colonization patterns, diversity, or dysbiosis and neurocognitive and social-emotional development will provide critical insights for future interventions.
Implications for Clinical Practice and Health
The literature to date suggests important information for perinatal and pediatric healthcare providers to consider in order to support development of a healthy and diverse infant gut microbiome. This review suggests that the vertical transfer of beneficial bacteria that occurs with vaginal birth is preferable to the horizontal transfer of potentially pathogenic organisms that can occur with surgical births. The gut microbiome of vaginally delivered infants also displays greater diversity, which is considered health protective. Of course, surgical delivery is sometimes warranted. In those cases, keeping mother and baby in close physical contact when possible, and supporting breast rather than formula feeding, should be a priority.
Antibiotics should be used judiciously across the lifespan, but perhaps especially so during the first 1,000 days. In cases where antibiotic treatment is necessary, practitioners might consider probiotic supplements as early evidence suggests this may lessen the deleterious impact of antibiotics on the infant gut microbiome (Johnston, Goldenberg, Vandvik, Sun, & Guyatt, 2011).
Finally, perinatal and pediatric healthcare providers should support the early initiation of breastfeeding for all newborns since breastfed infants are less likely to be colonized by potentially pathogenic organisms like C. difficile. The promotion of breastfeeding may support the proliferation of beneficial microbes, thus providing protection from those linked with atopy or NEC. Emerging research on the influence of diet on the newborn gut microbiome lends support to the widely held belief that breastfeeding is beneficial for infants. Breastfed infants have reduced colonization by C. difficile and E. Coli bacteria (Adlerberth & Wold, 2009; Penders, Thijs, et al., 2007), both of which have been associated with atopic manifestations (Penders, Thijs, et al., 2007). Many of the studies linking the infant gut microbiota with atopic disease emphasize the first six months as a “critical window period” (Penders, Stobberingh, et al., 2007), suggesting that colonization of the gut microbiota during this period functions as an important determinant of future health status (Penders, Stobberingh, et al., 2007). Beyond the early months of infancy, a diet high in fiber is associated with a more diverse gut microbiome with potentially beneficial bacterial genomes. The introduction of a diet rich in complex carbohydrates may continue the diversification of gut microbiota with corollary benefits of protecting the young child from pathogenic gastrointestinal organisms (De Filippo et al., 2010).
Conclusion
Although studies examining the human microbiome-gut-brain axis are still relatively few, growing evidence suggests the influence of the gut microbiome on infant development and health outcomes across the lifespan. One key area where the microbiome may be particularly important is in the development of the newborn brain, with potential outcomes ranging from effects on cognition, anxiety, mood, and sociability. A better understanding and appreciation of several potentially modifiable factors that influence the newborn and infant microbiome is essential, particularly for midwives and neonatal nurses who care for women and their infants during the crucial first 1,000 days of life.
Acknowledgments
The authors acknowledge this work was supported by a grant from the National Institutes of Health, National Institute of Nursing Research (R01NR014800).
Footnotes
The authors have no conflicts of interest to report.
Contributor Information
Irene Yang, Nell Hodgson Woodruff School of Nursing, Emory University, Atlanta, GA..
Elizabeth J. Corwin, Nell Hodgson Woodruff School of Nursing, Atlanta, GA..
Patricia A. Brennan, Department of Psychology, Emory University, Atlanta, GA..
Sheila Jordan, Nell Hodgson Woodruff School of Nursing, Emory University, Atlanta, GA..
Jordan R. Murphy, Nell Hodgson Woodruff School of Nursing, Emory University, Atlanta, GA..
Anne Dunlop, Nell Hodgson Woodruff School of Nursing, Emory University, Atlanta, GA..
References
- Adams MR, Hall CJ. Growth inhibition of food-borne pathogens by lactic and acetic acids and their mixtures. International Journal of Food Science & Technology. 1988;23:287–292. [Google Scholar]
- Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Pædiatrica. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- Aloisio I, Mazzola G, Corvaglia LT, Tonti G, Faldella G, Biavati B, Di Gioia D. Influence of intrapartum antibiotic prophylaxis against group B Streptococcus on the early newborn gut composition and evaluation of the anti-Streptococcus activity of Bifidobacterium strains. Applied Microbiology and Biotechnology. 2014;98:6051–6060. doi: 10.1007/s00253-014-5712-9. [DOI] [PubMed] [Google Scholar]
- Arrieta M-C, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: Health and disease. Frontiers in Immunology. 2014;5 doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Kozyrskyj AL. Gut microbiota of healthy Canadian infants: Profiles by mode of delivery and infant diet at 4 months. Canadian Medical Association Journal. 2013;185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain, Behavior, and Immunity. 2011;25:397–407. doi: 10.1016/j.bbi.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MT, Lubach GR, Coe CL. Prenatal stress alters bacterial colonization of the gut in infant monkeys. Journal of Pediatric Gastroenterology and Nutrition. 2004;38:414–421. doi: 10.1097/00005176-200404000-00009. [DOI] [PubMed] [Google Scholar]
- Barau G, Robillard P-Y, Hulsey TC, Dedecker F, Laffite A, Gérardin P, Kauffmann E. Linear association between maternal pre-pregnancy body mass index and risk of caesarean section in term deliveries. BJOG: An International Journal of Obstetrics & Gynaecology. 2006;113:1173–1177. doi: 10.1111/j.1471-0528.2006.01038.x. [DOI] [PubMed] [Google Scholar]
- Bengmark S. Gut microbiota, immune development and function. Pharmacological Research. 2013;69:87–113. doi: 10.1016/j.phrs.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Bezirtzoglou E. The intestinal microflora during the first weeks of life. Anaerobe. 1997;3:173–177. doi: 10.1006/anae.1997.0102. [DOI] [PubMed] [Google Scholar]
- Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast-and formula-fed newborns by using fluorescence in situ hybridization (FISH) Anaerobe. 2011;17:478–482. doi: 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Bilbo SD, Levkoff LH, Mahoney JH, Watkins LR, Rudy JW, Maier SF. Neonatal infection induces memory impairments following an immune challenge in adulthood. Behavioral Neuroscience. 2005;119:293–301. doi: 10.1037/0735-7044.119.1.293. [DOI] [PubMed] [Google Scholar]
- Borre YE, O’Keeffe GW, Clarke G, Stanton C, Dinan TG, Cryan JF. Microbiota and neurodevelopmental windows: Implications for brain disorders. Trends in Molecular Medicine. 2014;20:509–518. doi: 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Brooks HJL. Modern microbiology—A quiet revolution with many benefits. Australasian Medical Journal. 2013;6:378–381. doi: 10.4066/AMJ.2013.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, de Vos WM, DiStefano PS, Doré J, Huttenhower C, Knight R, Turnbaugh P. Translating the human microbiome. Nature Biotechnology. 2013;31:304–308. doi: 10.1038/nbt.2543. [DOI] [PubMed] [Google Scholar]
- Carrion VG, Wong SS. Can traumatic stress alter the brain? Understanding the implications of early trauma on brain development and learning. Journal of Adolescent Health. 2012;51:S23–S28. doi: 10.1016/j.jadohealth.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Chen J, Cai W, Feng Y. Development of intestinal bifidobacteria and lactobacilli in breast-fed neonates. Clinical Nutrition. 2007;26:559–566. doi: 10.1016/j.clnu.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Cho CE, Norman M. Cesarean section and development of the immune system in the offspring. American Journal of Obstetrics and Gynecology. 2013;208:249–254. doi: 10.1016/j.ajog.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Clarke G, O’Mahony SM, Dinan TG, Cryan JF. Priming for health: Gut microbiota acquired in early life regulates physiology, brain and behaviour. Acta Paediatrica. 2014;103:812–819. doi: 10.1111/apa.12674. [DOI] [PubMed] [Google Scholar]
- Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Blaser MJ. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nature Reviews Neuroscience. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- Dammann O, O’Shea TM. Cytokines and perinatal brain damage. Clinics in Perinatology. 2008;35:643–663. doi: 10.1016/j.clp.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R, Kelley KW. Twenty years of research on cytokine-induced sickness behavior. Brain, Behavior, and Immunity. 2007;21:153–160. doi: 10.1016/j.bbi.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Molecular Psychiatry. 2014;19:146–148. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan TG, Cryan JF. Regulation of the stress response by the gut microbiota: Implications for psychoneuroendocrinology. Psychoneuroendocrinology. 2012;37:1369–1378. doi: 10.1016/j.psyneuen.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenkov IJ, Chrousos GP. Stress hormones, proinflammatory and antiinflammatory cytokines, and autoimmunity. Annals of the New York Academy of Sciences. 2002;966:290–303. doi: 10.1111/j.1749-6632.2002.tb04229.x. [DOI] [PubMed] [Google Scholar]
- Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R INFABIO team. Intestinal microbiota of 6-week-old infants across Europe: Geographic influence beyond delivery mode, breast-feeding, and antibiotics. Journal of Pediatric Gastroenterology and Nutrition. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: Composition and development. Acta Paediatrica. 2003;92:48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x. [DOI] [PubMed] [Google Scholar]
- Finegold SM, Sutter VL, Mathisen GE. Normal indigenous intestinal flora. New York, NY: Academic Press; 1983. [Google Scholar]
- Forsythe P, Sudo N, Dinan T, Taylor VH, Bienenstock J. Mood and gut feelings. Brain, Behavior, and Immunity. 2010;24:9–16. doi: 10.1016/j.bbi.2009.05.058. [DOI] [PubMed] [Google Scholar]
- Fricke WF. The more the merrier? Reduced fecal microbiota diversity in preterm infants treated with antibiotics. Journal of Pediatrics. 2014;165:8–10. doi: 10.1016/j.jpeds.2014.03.022. [DOI] [PubMed] [Google Scholar]
- Galley JD, Nelson MC, Yu Z, Dowd SE, Walter J, Kumar PS, Bailey MT. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiology. 2014;14:189. doi: 10.1186/1471-2180-14-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, Sherman PM. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60:307–317. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- Gregory KE. Microbiome aspects of perinatal and neonatal health. Journal of Perinatal & Neonatal Nursing. 2011;25:158–164. doi: 10.1097/JPN.0b013e3182169346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenham S, Clarke G, Cryan JF, Dinan TG. Brain-gut-microbe communication in health and disease. Frontiers in Physiology. 2011;2:94. doi: 10.3389/fphys.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groer MW, Luciano AA, Dishaw LJ, Ashmeade TL, Miller E, Gilbert JA. Development of the preterm infant gut microbiome: A research priority. Microbiome. 2014;2:38. doi: 10.1186/2049-2618-2-38. http://www.biomedcentral.com/content/pdf/2049-2618-2-38.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grölund M-M, Lehtonen O-P, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: Permanent changes in intestinal flora after cesarean delivery. Journal of Pediatric Gastroenterology and Nutrition. 1999;28:19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- Gunnar MR. Quality of early care and buffering of neuroendocrine stress reactions: Potential effects on the developing human brain. Preventive Medicine. 1998;27:208–211. doi: 10.1006/pmed.1998.0276. [DOI] [PubMed] [Google Scholar]
- Haarman M, Knol J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Applied and Environmental Microbiology. 2006;72:2359–2365. doi: 10.1128/AEM.72.4.2359-2365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen HJM, Wildeboer-Veloo ACM, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. Journal of Pediatric Gastroenterology and Nutrition. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- Heijtz RD, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huurre A, Kalliomäki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of delivery—Effects on gut microbiota and humoral immunity. Neonatology. 2008;93:236–240. doi: 10.1159/000111102. [DOI] [PubMed] [Google Scholar]
- Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLOS ONE. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- Johansson MA, Saghafian-Hedengren S, Haileselassie Y, Roos S, Troye-Blomberg M, Nilsson C, Sverremark-Ekstrom E. Early-life gut bacteria associate with IL-4-, IL-10- and IFN-γ production at two years of age. PLOS ONE. 2012;7:e49315. doi: 10.1371/journal.pone.0049315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Versalovic J. The human microbiome and its potential importance to pediatrics. Pediatrics. 2012;129:950–960. doi: 10.1542/peds.2011-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston BC, Goldenberg JZ, Vandvik PO, Sun X, Guyatt GH. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Library. 2011 doi: 10.1002/14651858.CD004827.pub3. [DOI] [PubMed] [Google Scholar]
- Karlsson CLJ, Molin G, Cilio CM, Ahrné S. The pioneer gut microbiota in human neonates vaginally born at term—A pilot study. Pediatric Research. 2011;70:282–286. doi: 10.1203/PDR.0b013e318225f765. [DOI] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleessen B, Bunke H, Tovar K, Noack J, Sawatzki G. Influence of two infant formulas and human milk on the development of the faecal flora in newborn infants. Acta Paediatrica. 1995;84:1347–1356. doi: 10.1111/j.1651-2227.1995.tb13567.x. [DOI] [PubMed] [Google Scholar]
- Knol J, Scholtens P, Kafka C, Steenbakkers J, Gro S, Helm K, Wells J. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: More like breast-fed infants. Journal of Pediatric Gastroenterology and Nutrition. 2005;40:36–42. doi: 10.1097/00005176-200501000-00007. [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, Tarr PI. Patterned progression of bacterial populations in the premature infant gut. Proceedings of the National Academy of Sciences. 2014;111:12522–12527. doi: 10.1073/pnas.1409497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Lif Holgerson P, Harnevik L, Hernell O, Tanner ACR, Johansson I. Mode of birth delivery affects oral microbiota in infants. Journal of Dental Research. 2011;90:1183–1188. doi: 10.1177/0022034511418973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clinical Microbiology and Infection. 2012;18:12–15. doi: 10.1111/j.1469-0691.2012.03863.x. http://onlinelibrary.wiley.com/doi/10.1111/j.1469-0691.2012.03863.x/pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer EA. Gut feelings: The emerging biology of gut-brain communication. Nature Reviews Neuroscience. 2011;12:453–466. doi: 10.1038/nrn3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AH, Maletic V, Raison CL. Inflammation and its discontents: The role of cytokines in the pathophysiology of major depression. Biological Psychiatry. 2009;65:732–741. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsou EK, Kirtzalidou E, Oikonomou I, Liosis G, Kyriacou A. Fecal microflora of Greek healthy neonates. Anaerobe. 2008;14:94–101. doi: 10.1016/j.anaerobe.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Moloney RD, Desbonnet L, Clarke G, Dinan TG, Cryan JF. The microbiome: Stress, health and disease. Mammalian Genome. 2014;25:49–74. doi: 10.1007/s00335-013-9488-5. [DOI] [PubMed] [Google Scholar]
- Mulle JG, Sharp WG, Cubells JF. The gut microbiome: A new frontier in autism research. Current Psychiatry Reports. 2013;15:337. doi: 10.1007/s11920-012-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu J, Rushing J. Cesarean versus vaginal delivery: Long-term infant outcomes and the hygiene hypothesis. Clinics in Perinatology. 2011;38:321–331. doi: 10.1016/j.clp.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLOS Biology. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi P. Necrotizing enterocolitis. Pediatric Drugs. 2006;8:151–165. doi: 10.2165/00148581-200608030-00002. [DOI] [PubMed] [Google Scholar]
- Pavlov VA, Wang H, Czura CJ, Friedman SG, Tracey KJ. The cholinergic anti-inflammatory pathway: A missing link in neuroimmunomodulation. Molecular Medicine. 2003;9:125–134. [PMC free article] [PubMed] [Google Scholar]
- Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, Stobberingh EE. Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut. 2007;56:661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiology Letters. 2005;243:141–147. doi: 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Guyer M. The NIH human microbiome project. Genome Research. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power SE, O’Toole PW, Stanton C, Ross RP, Fitzgerald GF. Intestinal microbiota, diet and health. British Journal of Nutrition. 2014;111:387–402. doi: 10.1017/S0007114513002560. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Capuron L, Miller AH. Cytokines sing the blues: Inflammation and the pathogenesis of depression. Trends in Immunology. 2006;27:24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastall RA. Bacteria in the gut: Friends and foes and how to alter the balance. Journal of Nutrition. 2004;134:2022S–2026S. doi: 10.1093/jn/134.8.2022S. [DOI] [PubMed] [Google Scholar]
- Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nature Reviews Immunology. 2012;12:9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- Rotimi VO, Duerden BI. The development of the bacterial flora in normal neonates. Journal of Medical Microbiology. 1981;14:51–62. doi: 10.1099/00222615-14-1-51. [DOI] [PubMed] [Google Scholar]
- Salminen S, Gibson G, McCartney AL, Isolauri E. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. 2004;53:1388–1389. doi: 10.1136/gut.2004.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J, Yang P-C, Söderholm JD, Benjamin M, Perdue MH. Role of mast cells in chronic stress induced colonic epithelial barrier dysfunction in the rat. Gut. 2001;48:630–636. doi: 10.1136/gut.48.5.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheppach W, Weiler F. The butyrate story: Old wine in new bottles? Current Opinion in Clinical Nutrition & Metabolic Care. 2004;7:563–567. doi: 10.1097/00075197-200409000-00009. [DOI] [PubMed] [Google Scholar]
- Sonnenburg J, Sonnenburg E. The good gut: Taking control of your weight, your mood, and your long term health. New York, NY: Penguin Press; 2015. [Google Scholar]
- Stark PL, Lee A. The microbial ecology of the large bowel of breastfed and formula-fed infants during the first year of life. Journal of Medical Microbiology. 1982;15:189–203. doi: 10.1099/00222615-15-2-189. [DOI] [PubMed] [Google Scholar]
- Sudo N. Microbiome, HPA axis and production of endocrine hormones in the gut. In: Lyte M, Cryan JF, editors. Microbial endocrinology: The microbiota-gut-brain axis in health and disease. New York, NY: Springer; 2014. pp. 177–194. [Google Scholar]
- Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. Journal of Physiology. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey KJ. Reflex control of immunity. Nature Reviews Immunology. 2009;9:418–428. doi: 10.1038/nri2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vael C, Desager K. The importance of the development of the intestinal microbiota in infancy. Current Opinion in Pediatrics. 2009;21:794–800. doi: 10.1097/MOP.0b013e328332351b. [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan S, Karami A, Aeinehband S, Behbahani H, Grandien A, Nilsson B, Darreh-Shori T. Regulated extracellular choline acetyltransferase activity—The plausible missing link of the distant action of acetylcholine in the cholinergic anti-inflammatory pathway. PLOS ONE. 2013;8:e65936. doi: 10.1371/journal.pone.0065936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker WA. Initial intestinal colonization in the human infant and immune homeostasis. Annals of Nutrition and Metabolism. 2013;63:8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME Journal. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M, Walker W. The role of gut microbiota in programming the immune phenotype. Journal of Developmental Origins of Health and Disease. 2013;4:203–214. doi: 10.1017/S2040174412000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzl HH, Schimpl G, Feierl G, Steinwender G. Effect of prenatal cortisone on spontaneous bacterial translocation from gastrointestinal tract in neonatal rat. Digestive Diseases and Sciences. 2003;48:1171–1176. doi: 10.1023/a:1023789301547. [DOI] [PubMed] [Google Scholar]
- Yu LC-H, Shih Y-A, Wu L-L, Lin Y-D, Kuo W-T, Peng W-H, Ni Y-H. Enteric dysbiosis promotes antibiotic-resistant bacterial infection: Systemic dissemination of resistant and commensal bacteria through epithelial transcytosis. American Journal of Physiology—Gastrointestinal and Liver Physiology. 2014;307:G824–G835. doi: 10.1152/ajpgi.00070.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlmans MA, Korpela K, Riksen-Walraven JM, de Vos WM, de Weerth C. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology. 2015;53:233–245. doi: 10.1016/j.psyneuen.2015.01.006. [DOI] [PubMed] [Google Scholar]