Abstract
Although it is generally accepted that cellular differentiation requires changes to transcriptional networks, dynamic regulation of promoters and enhancers at specific sets of genes has not been previously studied en masse. Exploiting the fact that active promoters and enhancers are transcribed, we simultaneously measured their activity in 19 human and 14 mouse time courses covering a wide range of cell types and biological stimuli. Enhancer RNAs, then messenger RNAs encoding transcription factors, dominated the earliest responses. Binding sites for key lineage transcription factors were simultaneously overrepresented in enhancers and promoters active in each cellular system. Our data support a highly generalizable model in which enhancer transcription is the earliest event in successive waves of transcriptional change during cellular differentiation or activation.
Regulated transcription initiation underlies state changes in cell phenotype and is coordinated by transcription factors binding to gene-proximal promoters or distal regulatory regions such as enhancers. The interaction between enhancers and transcription induction during cellular differentiation has been cited as one of the outstanding mysteries of modern biology (1). Enhancer chromatin landscapes change drastically between developing tissues and differentiated cells (2–4). Active enhancers initiate production of RNAs (eRNAs) (5) and enhancer action during differentiation can be assessed by sequencing of steady-state (6, 7) or nascent RNA (8–10), demonstrating that eRNA and target gene expression are correlated. eRNA production is also correlated to physical proximity between enhancers and promoters (8, 9). However, the general temporal relationship between enhancer and promoter activation across biological system is unknown.
Genome-scale 5′ rapid amplification of cDNA ends (cap analysis of gene expression, or CAGE) detects transcription start sites (TSSs), including the bidirectional TSS characteristic of active enhancers (11). Based on a large set of reporter assays, CAGE-defined enhancers are two to three times as likely to validate (12) as untranscribed chromatin-defined enhancer candidates from the ENCODE (Encyclopedia of DNA Elements) consortium (13). Here, we used CAGE to dissect the relationship between dynamic changes in mRNA and eRNA in 33 time courses of differentiation and activation. The time courses included stem cells (embryonic, induced pluripotent, trophoblastic, and mesenchymal stem cells) and committed progenitors undergoing terminal differentiation toward mesodermal, endodermal, and ectodermal fates, as well as fully differentiated primary cells and cell lines responding to stimuli (growth factors and pathogens) (Fig. 1, A and B; tables S1 to S3; and supplementary methods). In total, 1189 CAGE libraries from 408 distinct time points in the 33 time courses were analyzed (Fig. 1B and auxiliary data tables S1 and S2). Differentiation or response to stimulus was assessed by monitoring cell morphology changes, reproducible induction of known lineage markers, and similarity of the end-point transcriptome to differentiated cells from the steady-state samples of FANTOM5 (14) (auxiliary data table S1).
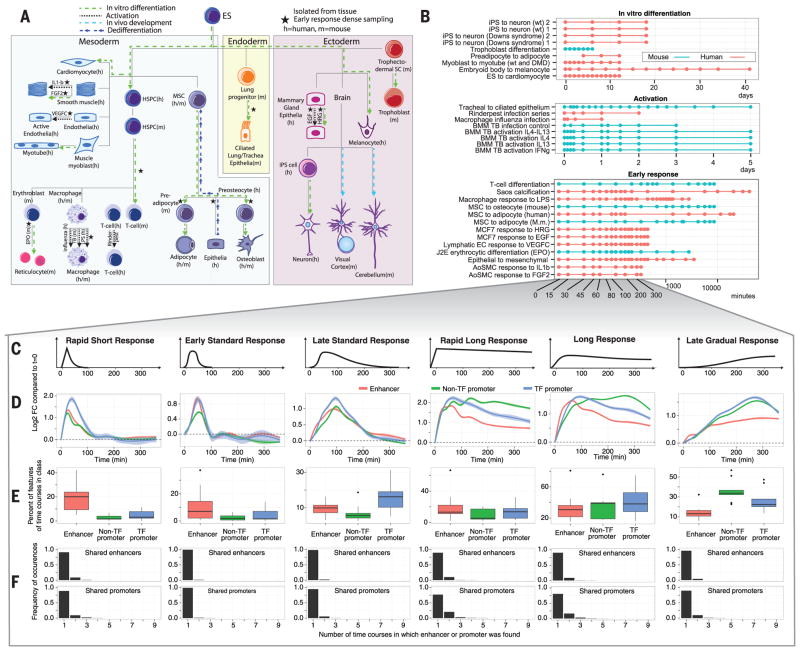
Fig. 1. Time course design and definition of response classes.
(A) Schematic illustration of the time course experiments included in the study, arranged according to a development tree. Germ layers are shown as boxes. Black stars indicate time series sampled with high resolution. (B) Overview of time courses according to sampling strategy. The x axis indicates time after induction. Each dot indicates CAGE sampling, typically done in biological triplicates. (C) Stylistic representation of each of the major up-regulated response patterns (classes) identified as described in the main text. The y axis shows log2 fold change versus time 0; the x axis shows time in minutes. (D) Mean expression log2 fold change across time courses for enhancers and promoters classified into each response pattern [as in (C)]. The 95% confidence intervals of means are shown. (E) Boxplots of fractions showing the preference for enhancers, TF promoters, and other promoters for respective response class. (F) Overlap between time courses in terms of enhancers and promoters in respective class. Barplots show the frequency (y axis) of the number of time courses (out of 9) sharing a specific feature (x axis).
The current data expand the set of known human and mouse core promoters from the FANTOM5 body-wide steady-state atlas (14) to 201,802 and 158,966, and the set of transcribed enhancers to 65,423 and 44,459. Of all identified core promoters in human and mouse, 51% and 61% varied significantly in expression in at least one time course. Out of the 103,355 differentially expressed human promoters, 80,152 were within genes on the same strand. Of these, 55,626 are potential alternative promoters (see supplementary methods), overlapping a total of 13,138 genes. We found 65 human genes that had a dynamic switch between alternative promoters within a time course, leading to exclusion of exons encoding protein domains (table S4).
Of all enhancers identified in FANTOM5, 42,274 human (65%) and 34,338 mouse (77%) enhancers were expressed in at least one CAGE library in the current study. Of these, 5371 (13%) human and 6824 (20%) mouse enhancers changed expression significantly over time in at least one time course. Most of these enhancer changes were time-course specific (56% in human, 67% in mouse). In contrast, the fraction of promoters regulated in only a single time course was smaller (29% in human, 33% in mouse).
We profiled 13 cellular systems with high temporal resolution within the first hours of cellular induction (Fig. 1B). We focused on the first 6 hours in nine of these time courses (five human and four mouse having sufficient numbers of dynamic promoters and enhancers; table S1).
Based on unsupervised clustering, we identified a set of distinct response pattern classes, shared by multiple time courses, by analyzing expression fold changes versus time 0 in each time course. For each response class, we defined specific expression rules (fig. S1), enabling consistent response class labeling of any dynamically transcribed enhancer or TSS in a time-course–specific fashion (figs. S2 to S4). Transcription factor (TF) promoters were analyzed as a distinct group. Because most enhancers and promoters that were dynamically changing in this set were up-regulated over time (fig. S5), we focused on the six up-regulated response classes (Fig. 1C).
Multiple enhancers, TF promoters, and non–TF promoters were found in all response classes (Fig. 1D, fig. S6, and auxiliary data table S3), but with different preferences. Enhancers were more common in the early peaking classes (“rapid short,” “early standard,” and “rapid long” responses). TF promoters were generally induced after enhancers (preferring the “late standard” response and “long response” classes) and non–TF promoters were most common in the “late gradual response” class that increased gradually with time (Fig. 1E), suggesting that many of these genes were the direct or indirect targets of the induced transcription factors. Simulation studies, as well as gene-specific RNA half-life data (15), showed that differential degradation rates of RNA species (11) could not explain the observed class preferences (supplementary text and figs. S7 and S8). Although these patterns were evident across cell types and species, few promoters (mean 8.5% across classes) and even fewer enhancers (mean 5.1% across classes) were assigned to the same response class in two or more time courses (Fig. 1F).
We looked further at a literature-curated set of 232 immediate early response (IER) genes (table S5). Although 65% of the IER genes had at least one promoter that was up-regulated within the first 2 hours in at least two time courses, no consistent pattern of IER expression was obvious between time courses (fig. S9). For example, only 42 promoters were induced early in five or more human time courses (fig. S10A). Even fewer enhancers shared an early response: Only 11 were induced in three or more time courses (fig. S10B), and of these, half neighbored a known IER gene. Thus, the IER pattern is generalizable across different cell states, but the cohort of IER genes are not.
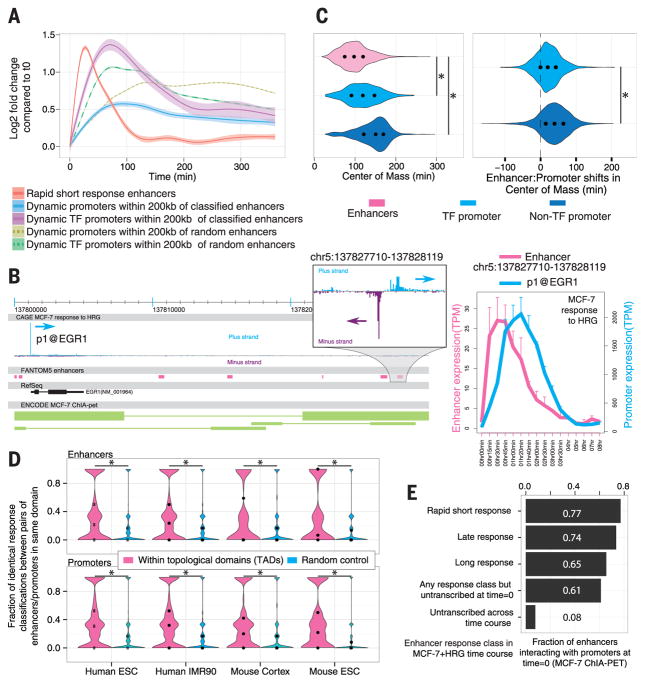
In general, up-regulated enhancers in the rapid short response class were transcribed earlier than their proximal (±200 kb) promoters (Fig. 2, A and B, and fig. S11). Proximal TF promoters were, in turn, more highly and more rapidly activated than proximal non–TF promoters. To compare the responses over time, we used the “center of mass” (CM) statistic identifying the time point by which 50% of the expression change in the enhancer or promoter had occurred. Enhancers changed most rapidly, followed by TF promoters, then non–TF promoters (Fig. 2C). The temporal differences were highlighted further when enhancers were compared to their proximal promoters (within ±200 kb) (Fig. 2C). For 85.8% of enhancer–non–TF promoter pairs and 74.6% of enhancer–TF promoter pairs, the CM occurred earlier for the enhancer (P < 1.0 × 10−106, Wilcoxon signed rank test). We hypothesized that these results might reflect larger chromatin structures; indeed, enhancer-promoter pairs defined by topological domains (TADs) (16) gave similar results (figs. S11 and S12), and moreover, enhancers (or promoters) within the same TAD were more likely to be in the same response class (Fig. 2D). Similarly, groups of enhancer-promoter pairs (defined either by genomic proximity or TAD boundaries) were more similar in terms of CM shifts than expected by chance (fig. S13, P < 1.0 × 10−14, Mann-Whitney U test).
Fig. 2. Temporal shifts between enhancer and promoter activity.
(A) Smoothed mean expression over time for all enhancers classified into the rapid short response group and all differentially expressed proximal (±200 kb) promoters, split by gene type. Controls for class specificity (dotted lines) constitute promoters proximal to randomly sampled enhancers from other classes. Shaded areas indicate 95% confidence intervals. (B) Example of expression timing in an enhancer-promoter pair (EGR1), showing activation of enhancers before promoter activation. MCF-7 ChIA-PET interaction data are visualized at the bottom as green lines; each line represents a cluster of ChIA-PET paired tags consisting of at least three pairs, where line end thickness is proportional to the number of paired tags in the cluster. Right panel shows the expression level of promoter and enhancer in MCF-7 cells after induction with HRG. Error bars indicate SD. (C) Left: Distribution of center of mass (CM) of expression changes (see main text) for enhancers, TF promoters, and promoters of other genes. Right: difference in CM (“shift”) between enhancers-promoter pairs linked by proximity (±200 kb) split by gene type. Black dots indicate 25th, 50th, and 75th percentiles. Asterisks indicate significance (P < 1.0 × 10−106, Mann-Whitney U test). (D) The similarity of enhancer or promoter response classification (Fig. 1C) within each TAD was analyzed by calculating the frequency of identically classified enhancers or promoters in all pairwise comparisons. Frequency distributions are shown as violin plots. Controls are made by randomly sampling the same number of enhancers or promoters and calculating the classification similarity as above (repeated 100 times for each TAD). Asterisks indicate significance (P < 0.01, Mann-Whitney U test); dots represent percentiles, as in (C). (E) Fraction of enhancers that interact (by RNAPII-ChIA-PET) with promoters in unstimulated MCF-7 cells, split by enhancer response class in the MCF-7+HRG time course.
We used ENCODE (13) data to demonstrate that enhancers dynamically expressed in the MCF-7+HRG time course were more likely to be marked with high deoxyribonuclease (DNAse) sensitivity and enriched in H3K27ac and RNA polymerase II (RNAPII) chromatin immuno-precipitation signal in steady-state MCF-7 cells than enhancers that were not active throughout the time course (fig. S14A). Indeed, chromatin interaction analysis with paired-end-tag sequencing (ChIA-PET) data from steady-state MCF-7 cells (17) showed that these dynamic enhancers interacted with promoters to a much larger extent than nonactive enhancers, but the fraction of promoter-interacting enhancers was high regardless of response class (Fig. 2E), suggesting that many dynamically changing enhancers are proximal to their promoter target(s) and primed beforehand in terms of chromatin state. However, chromatin patterns in the unstimulated state were not sufficient to distinguish between temporal enhancer classes (fig. S14B).
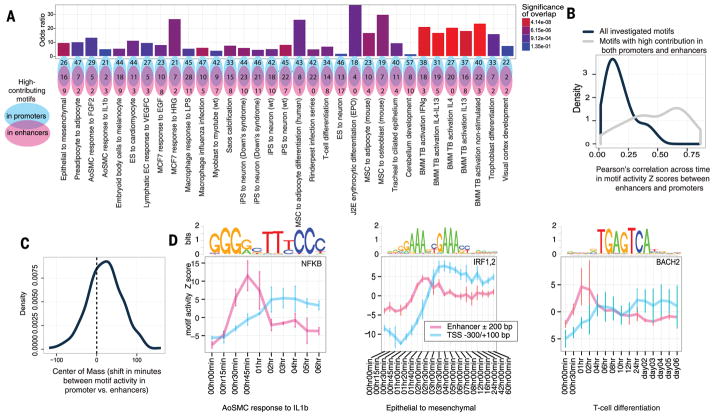
Transcription factor binding sites implicated in regulating enhancer and promoter expression were assessed by inferring motif activities (18)—a statistic that describes the ability of a DNA motif to explain observed expression changes across a given set of samples—based on motif occurrence in the regions −300 to +100 base pairs (bp) from the major TSSs of each promoter and ±200 bp from the center of each enhancer, resulting in a derived activity profile across time for each DNA binding motif and time course. Motif sets with high predictive power in enhancers and promoters overlapped significantly (false discovery rate < 0.05, Fisher’s exact test) in 29 out of 33 time courses (Fig. 3A). Many of these highly contributing motifs described binding sites for known lineage-specific regulators in specific time courses, such as FOS in MCF-7 cells stimulated by HRG, GATA6 in cardiomyocyte differentiation, and nuclear factor κB (NF-κB) in macrophages. On average, motif activity scores correlated positively across time between enhancers and promoters, with significantly higher correlation for motifs identified as significantly active (supplementary text) in both enhancers and promoters (P < 6.9 × 10−8, Mann-Whitney test) (Fig. 3B); however, in general, motif activity reached a maximum in enhancers earlier than in promoters (P < 1.8 × 10−14, Wilcoxon signed rank test; Fig. 3, C and D). Thus, the general observation of enhancer transcription waves preceding those of promoters identified above was supported by motif activity.
Fig. 3. Motif analysis of linked enhancers and promoters over time.
(A) Overlap of motifs classified as significant for driving expression in enhancers and promoters. Top row: bar plot of motif overlap odds ratios, colored by significance. Bottom row: Venn diagrams of motif set overlap. (B) Distributions of average Pearson correlation coefficient between motif activities in enhancers and promoters in all motifs investigated (black) and motifs significantly active in both enhancers and promoters (gray). (C) Distribution of shift (minutes) in motif activity center of mass (see Fig. 2D) in promoters compared to enhancers. (D) Examples of motif activity in enhancers preceding that of promoters. Motif activity is plotted as the average of activity Z scores per time point. Error bars indicate the SD.
In summary, by using a large-scale comparative analysis across many different tissues and time courses, and simultaneously sampling expression at gene promoters and enhancers, we reveal that enhancer transcription is the most common rapid transcription change occurring when cells initiate a state change. Enhancer RNA concentration peaked as early as 15 min after the transition trigger was applied in some time courses. Although earlier studies of single time courses have reported enhancer activity before gene activation in a small set of enhancer-gene pairs (8, 9, 19), we can now establish this phenomenon as a general feature of mammalian transcriptional regulation, across a multitude of biological systems. This challenges previous models that suggested that linked enhancers and promoters are coexpressed over time [e.g., (8, 15, 19, 20)]. Indeed, even in the case of late response classes, candidate enhancers appear to be activated in advance of promoters in their vicinity (fig. S11). The rapid burst of eRNA activity at 15 min was frequently followed by a rapid return to baseline (Fig. 1D). In these cases, it may be that once the target promoter has been activated, enhancer activity is no longer required. Other enhancers were rapidly activated and then continuously expressed. These eRNAs may have additional functional roles, such as the recently suggested role in promoting elongation (15).
Supplementary Material
Acknowledgments
For a full list of acknowledgements and contributions, see supplementary text. FANTOM5 was made possible by a Research Grant for RIKEN Omics Science Center from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) to Y. Hayashizaki. It was also supported by Research Grants for RIKEN Preventive Medicine and Diagnosis Innovation Program to Y. Hayashizaki and RIKEN Centre for Life Science Technologies, Division of Genomic Technologies (from the MEXT, Japan). Additional funding is listed in the supplementary text. All CAGE data needed to reproduce the study have been deposited at the DNA Data Bank of Japan (DDBJ) under accession numbers DRA000991, DRA002711, DRA002747, and DRA002748. Additional visualizations of the data are available at http://fantom.gsc.riken.jp/5/. The human induced pluripotent stem cell lines that were subjected to cortical neuronal differentiation can be made available after completion of a materials transfer agreement with the Australian Institute for Bioengineering and Nanotechnology of The University of Queensland.
Footnotes
The list of author affiliations is available in the Supplementary Text.
www.sciencemag.org/content/347/6225/1010/suppl/DC1
Materials and Methods
Auxiliary data tables S1 to S3
References (21–32)
REFERENCES AND NOTES
- 1.Levine M, Cattoglio C, Tjian R. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonn S, et al. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 3.Nord AS, et al. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stergachis AB, et al. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim TK, et al. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, et al. PLOS Genet. 2014;10:e1004610. doi: 10.1371/journal.pgen.1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rönnerblad M, et al. Blood. 2014;123:e79–e89. doi: 10.1182/blood-2013-02-482893. [DOI] [PubMed] [Google Scholar]
- 8.Kaikkonen MU, et al. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, et al. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersson R, et al. Nat Commun. 2014;5:5336. doi: 10.1038/ncomms6336. [DOI] [PubMed] [Google Scholar]
- 12.Andersson R, et al. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ENCODE Project Consortium. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FANTOM Consortium and the RIKEN PMI and CLST (DGT) Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaukowitch K, et al. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon JR, et al. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G, et al. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.FANTOM Consortium et al. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh C-L, et al. Proc Natl Acad Sci USA. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.IIott NE, et al. Nat Commun. 2014;5:3979–3979. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.