SUMMARY
Despite the remarkable clinical response of melanoma harboring BRAF mutations to BRAF inhibitors (BRAFi), most tumors become resistant. Here, we identified the downregulation of the ubiquitin ligase RNF125 in BRAFi-resistant melanomas and demonstrated its role in intrinsic and adaptive resistance to BRAFi in cultures as well as its association with resistance in tumor specimens. Sox10/MITF expression correlated with and contributed to RNF125 transcription. Reduced RNF125 was associated with elevated expression of receptor tyrosine kinases (RTKs), including EGFR. Notably, RNF125 altered RTK expression through JAK1, which we identified as an RNF125 substrate. RNF125 bound to and ubiquitinated JAK1, prompting its degradation and suppressing RTK expression. Inhibition of JAK1 and EGFR signaling overcame BRAFi resistance in melanoma with reduced RNF125 expression, as shown in culture and in in vivo xenografts. Our findings suggest that combination therapies targeting both JAK1 and EGFR could be effective against BRAFi-resistant tumors with de novo low RNF125 expression.
Graphical Abstract

INTRODUCTION
A missense mutation (V600E) in the activation loop of serine-threonine protein kinase B-RAF (BRAFV600E) is the most prevalent coding region mutation in melanoma and is seen in >50% of melanoma tumors (Davies et al., 2002). Tumors harboring constitutively active BRAFV600E exhibit highly active mitogen-activated protein kinase (MAPK) signaling, which is implicated in their transformation (Lopez-Bergami, 2011).
Success in targeting oncogenic kinase activity has encouraged the development of therapies targeting the BRAF mutation, an approach that has produced a growing number of BRAF inhibitors (BRAFi), including vemurafenib and dabrafenib. These reagents represent significant advances in the clinical management of melanoma relative to the previous first-line therapy, dacarbazine (Chapman et al., 2011; Flaherty et al., 2010; Hauschild et al., 2012; Sosman et al., 2012). Nonetheless, some tumors treated with BRAFi exhibit intrinsic drug resistance, while others develop adaptive resistance over time. This remains a major obstacle in the long-term effectiveness of BRAFi-based therapy (Ribas and Flaherty, 2011) and thus is the subject of intense study.
Numerous pathways reportedly underlie BRAFi resistance, including reactivation of MAPK signaling through NRAS or MEK1 mutations, BRAF splicing or gene amplification, and upregulation of receptor tyrosine kinases (RTKs) or growth factors (Abel et al., 2013; Nazarian et al., 2010; Poulikakos et al., 2011; Shi et al., 2012; Wagle et al., 2011; Wilson et al., 2012). In addition, altered signaling pathways, such as PI3K/AKT/mTOR and MITF/PGC1alpha, are implicated in BRAFi resistance (Haq et al., 2013; Shi et al., 2011; Villanueva et al., 2010). However, it is currently not possible to predict which tumors will exhibit chemoresistance. These hurdles have stimulated interest in novel combination therapies, including BRAFi, but it remains challenging to identify which patients should undergo such regimens (Sullivan and Flaherty, 2013). Defining the mechanisms that underlie intrinsic/primary resistance or adaptive resistance and detecting them prior to initiating treatment could accelerate the development of rational combination therapies aimed at overcoming BRAFi resistance.
Given the importance of ubiquitin proteasome system (UPS) components in tumor development, progression, and resistance mechanisms (Hoeller and Dikic, 2009; Qi et al., 2008, 2010, 2013), we sought to determine whether UPS components may also contribute to BRAFi resistance of melanoma. To identify components of the UPS that potentially drive BRAFi resistance, we performed functional screening of a small interfering RNA (siRNA) library against UPS-related genes. We then assessed positive hits for differentially expressed genes in data sets of BRAFi-resistant melanomas. The combined analyses led us to identify the E3 ubiquitin ligase RNF125, which is downregulated in resistant melanomas, as a component of intrinsic resistance to BRAFi. We demonstrate the role of RNF125 in regulating JAK1 and EGFR expression, and establish the importance of this regulation for chemoresistance of melanoma to BRAFi.
RESULTS
Identification of RNF125 in BRAFi-Resistant Melanomas
To define mechanisms underlying melanoma cell resistance to BRAFi, we evaluated the potential deregulation of UPS factors in BRAFi-resistant melanoma. To this end, we performed an unbiased screen of a siRNA library, including 1,173 genes encoding most of the UPS-associated proteins. We performed the screen using melanoma cell lines (Lu1205 parental, sensitive [Lu1205S]), which became resistant in the presence of increasing concentrations (up to 5 μM) of the BRAFi PLX4032 (Lu1205 resistant [Lu1205R]; Figures 1A and S1A). As previously reported, resistant cultures exhibited a high ERK activation correlated with BRAFi resistance, with an overall IC50 increase of 20- to 400-fold (Greger et al., 2012; Su et al., 2012; Figure S1A). Potential changes in viability of the parental and BRAFi-resistant Lu1205 cultures were monitored following transfection of cells with three siRNAs targeting each of the 1,173 UPS-related genes (Figure 1A). An initial screen of the parental line identified 18 genes for which inhibition conferred a growth advantage in the presence of BRAFi (1 μM; Figures 1B and 1C). Among these genes, inhibition of CUL3, RBX1, or WDR24 conferred the most potent net growth advantage (Figures 1B and 1C). In agreement with this finding, an independent study reported that downregulation of Cul3 and Rbx1 confers a growth advantage in melanoma cells treated with BRAFi (Shalem et al., 2014). None of the UPS-related genes identified in the Lu1205S cells were able to alter the growth of Lu1205R cells (Figure S1B). Overall, we selected 18 genes for re-analysis in a second BRAFi-resistant melanoma cell line (A375 resistant [A375R]), again using three different siRNAs. Depletion of nine of the original 18 genes significantly increased BRAFi resistance in both the Lu1205 and A375 lines (Figure 1D).
Figure 1. Identification of RNF125 in BRAFi-Resistant Melanoma Based on siRNA Screening and Gene Expression Analysis.
(A) Experimental scheme of siRNA screening.
(B) Fold change in the growth of Lu1205 cells transfected with siRNAs and cultured in BRAFi (PLX4032, 1 μM) or DMSO (vehicle) for 72 hr. Red lines indicate >60% viability and a >1.25-fold growth increase relative to siRNA controls in cells cultured in BRAFi.
(C) Genes selected in the first screening.
(D) Fold change in the growth of Lu1205 and A375 cells transfected with siRNAs targeting genes selected in the first screening. The red line indicates a 1.25-fold growth increase; arrowhead indicates RNF125.
(E) Fold change in UPS-related gene expression in a data set of BRAFI-resistant melanoma (Nazarian et al., 2010). Red lines indicate a 1.25-fold difference. Selected genes were tested (see Figure S1C).
(F) Top: fold change in RNF125 expression in established cultures of resistant (Res) lines relative to parental (Par) cells. Bottom: immunoblot showing the protein levels of RNF125 and its substrate DDX58/RIG-I.
Specific RNF125 bands are indicated with an arrow. Upper non-specific (n.s.) band is marked. Data are presented as mean ± SD. See also Figure S1.
To narrow the list of UPS-related candidate genes, we assessed gene expression data sets obtained from melanoma tumor-derived cultures resistant to BRAFi (GEO: GSE24862) (Nazarian et al., 2010). Of 740 UPS-related genes, only 12 were differentially expressed in the two resistant lines (eight downregulated and four upregulated). When compared with the nine genes identified in our original siRNA screen, the ubiquitin ligase RNF125 emerged as the primary candidate in both analyses. RNF125 expression was significantly lower in resistant cells than in parental cells, and RNF125 depletion in parental cells conferred a growth advantage in the presence of BRAFi (Figures 1D, 1E, and S1C). In agreement with microarray data sets, RNF125 transcript and protein levels were markedly decreased in the two resistant cultures (A375R and Lu1205R) compared with parental cultures (Figures 1F and S1C). RNF125 is implicated in T cell activation via regulation of the T cell receptor and in the innate immune response to viral infection via regulation of DDX58/RIG-I (Arimoto et al., 2007; Giannini et al., 2008; Shoji-Kawata et al., 2007; Zhao et al., 2005). Correspondingly, the expression levels of the known RNF125 substrate DDX58/RIG-I were increased in BRAFi-resistant cells (Figure 1F).
Altered RNF125 Expression Impacts Melanoma Resistance to BRAFi
Next, we asked whether RNF125 expression is associated with intrinsic BRAFi resistance. We conducted independent analyses of melanoma cell lines (10 and 12 melanoma cell lines in two respective experiments) and found an inverse correlation between BRAFi resistance and RNF125 expression levels (r = −0.75, p = 0.0051) (Figures 2A, 2B, and S2A; Table S1). Notably, RNF125 knockdown in melanoma cultures increased the BRAFi IC50 (p < 0.0001; Figures 2C and S2B–S2E). Interestingly, RNF125 depletion in parental cells did not confer a degree of resistance comparable to that seen in resistant cells, implying that RNF125 may play a role in growth adaptation or fitness phenotypes of BRAFi-resistant cells.
Figure 2. RNF125’s Function in BRAFi Resistance.
(A) IC50 of the BRAFi (PLX4032) in melanoma cell lines (Table S1).
(B) Correlation of PLX4032 IC50 with RNF125 expression (based on qPCR; Table S1).
(C) BRAFi-induced growth inhibition of UACC91 cells expressing scrambled shRNA or two shRNAs targeting RNF125. Shown are the IC50 changes and corresponding protein expression following shRNA treatment. Effectiveness of shRNF125 measured by qPCR and by western blot, which depicts level of RNF125 and its reported substrate DDX58. Specific RNF125 band is pointed by arrow. The p values (paired, two-tailed Student’s t test) in the analyses using two shRNAs were <0.0001.
(D) Fold difference in the growth of A375 cells transfected with empty vector (EV), wild-type RNF125 (WT), or the RING mutant (RM) and cultured for 72 hr in media with or without BRAFi (DMSO in parental and BRAFi [5 μM] in resistant cells). The growth was assessed using ATPlite (see Experimental Procedures). The fold difference was calculated to set the value in EV to one.
(E) Growth of A375R cells in soft agar. A375R cells expressing the indicated levels of RNF125 were plated and cultured in the presence of BRAFi (5 μM). Colony number was quantified using ImageJ.
(F) UACC1113 cell growth upon expression of the indicated plasmids for 72 hr. The fold difference in growth was assessed as described above.
Data are presented as mean ± SD (A and C–F). See also Figure S2 and Table S1.
To confirm that lower RNF125 expression is associated with BRAFi resistance of melanoma cells, we carried out RNF125 gain-of-function experiments to monitor potential changes in the resistance of these cultures. Notably, overexpression of wild-type (WT) RNF125 slowed the growth of resistant, but not parental, cells, as seen in short-term 2D and long-term 3D cultures (Figures 2D and 2E). Moreover, this effect required E3 ligase activity (Figures 2D and 2E), as overexpression of the RNF125 RING mutant had no effect. Similarly, expression of WT protein, but not the RING mutant, inhibited growth in UACC113 melanoma cells expressing low levels of RNF125 (Figure 2F). These data establish the importance of RNF125 depletion in intrinsic BRAFi resistance and adaptive resistance, both of which play key, albeit distinct, roles in the propensity of melanoma to resist therapy (Kugel and Aplin, 2014).
Identification of JAK1 as an RNF125 Substrate
To identify RNF125 substrates that mediate these effects, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis in BRAFi-resistant melanoma cultures using the RING mutant form of RNF125, which is more stable. Among 21 putative RNF125 substrates was JAK1, a key regulator of immune cell activation and interferon responses (Table S2), consistent with RNF125’s reported role in immune responses (Arimoto et al., 2007; Zhao et al., 2005). When we over-expressed tagged forms of JAK1 and the RNF125 RING mutant in HEK293T cells, we found that they interacted (Figure 3A). In the presence of MG132, we detected interaction of endogenous RNF125 and JAK1 using two different JAK1 antibodies (Figure 3B). Next, we assessed whether RNF125 regulated JAK1 stability. Following ectopic expression of RNF125 and JAK1, we found that RNF125 expression decreased JAK1 protein steady-state levels in a manner dependent on RNF125 E3 ligase activity (Figures 3C and 3D). Accordingly, RNF125 depletion by two different small hairpin RNAs (shRNAs) increased the steady-state levels of JAK1 protein, but not those of other JAK family members (Figures S2B, S3E, and 3E). Notably, MG132 treatment increased JAK1 steady-state levels, masking the effect of RNF125 depletion and partially blocking deregulated JAK1 expression by ectopically expressed RNF125 (Figure 3F). Accordingly, the WT, but not the RING mutant form of RNF125, decreased the JAK1 half-life by 50% (from 16.3 hr to 7.2 hr, based on linear regression analysis; Figure 3G). Collectively, these findings identify JAK1 as a previously undisclosed RNF125 substrate.
Figure 3. JAK1 Is a Novel RNF125 Substrate.
(A) The RNF125-JAK1 interaction was analyzed in HEK293T cells ectopically expressing FLAG-tagged RNF125 and V5-tagged JAK1. IgLC, IgG light chain. Arrows point to the position of RNF125 protein; n.s. indicates non-specific band.
(B) Interaction of endogenous RNF125 with JAK1 analyzed in A375 cells pre-treated with MG132 (5 μM) for 10 hr. Two independent experiments were conducted using two different JAK1 antibodies. Asterisks indicate RNF125.
(C) JAK1 deregulation by RNF125 in HEK293T cells ectopically expressing the indicated amounts of plasmids. GFP served as a transfection control.
(D) JAK1 expression analyzed in Lu1205 cells transfected with the indicated RNF125 plasmids (empty vector [EV], wild-type [WT], or RING mutant [RM]).
(E) JAK1 expression was analyzed in A375 cells expressing scrambled shRNA (Scr) or two shRNAs (#1 and #2) targeting RNF125.
(F) JAK1 expression was analyzed in A375 cells expressing scrambled shRNA or shRNF125 (#2) and treated with MG132 (5 μM, 10 hr), and in Lu1205 cells expressing WT RNF125 and treated with MG132.
(G) A375 cells expressing scrambled shRNA or shRNF125 (#2) were subjected to cycloheximide chase (50 μg/ml for indicated times). Band intensity was quantified using LICOR.
(H) Increased expression of JAK1 protein in established UACC91-resistant (UACC91R) cells in the absence of changes in JAK1 transcript levels. Specific RNF125 band is pointed by arrow. Upper non-specific (n.s.) bands are marked. Data are presented as mean ± SD.
(I) JAK1 expression in A375R and Lu1205R cells was analyzed following transfection with WT RNF125 plasmid.
(J) Cycloheximide chase analysis was performed in Lu1205 parental and resistant cells. Band intensity was quantified as above.
Regulation of JAK1 by RNF125 in BRAFi-Resistant Cells
Given the regulation of JAK1 stability by RNF125, we asked whether JAK1 expression is altered in BRAFi-resistant cells. We detected increased levels of JAK1 protein, but not mRNA, in BRAFi-resistant cells (Figure 3H). To confirm that RNF125 deregulation increases JAK1 protein levels, we re-expressed WT RNF125 and observed reduced JAK1 protein levels (Figure 3I). Cycloheximide chase experiments revealed an increased JAK1 protein half-life in resistant cells (Figure 3J), which correlated with decreased RNF125 expression (Figure 3H).
The SOX10/MITF Axis Is an Upstream Regulator of RNF125 Expression
To identify genes that are potentially affected (positively or negatively) by RNF125/JAK1 signaling, we used two different data sets for resistant (Nazarian et al., 2010) or primary (Krauthammer et al., 2012) melanoma cultures. We further analyzed genes that were co-regulated with RNF125 expression (Pearson’s r > 0.7 or r < −0.7) using Ingenuity Pathway Analysis (IPA). Among the top five co-regulated genes that we considered as putative upstream components were HNF4A, MITF, and SOX10. Among these, SOX10 expression levels were positively correlated with RNF125. Thus, we set out to determine whether SOX10 and its downstream target, MITF, may be upstream regulators of RNF125. SOX10 or MITF depletion significantly decreased RNF125 expression, whereas HNF4a depletion did not (Figures 4A and S3A). By assessing independent data sets (GEO: GSE37059, GSE50535) for genes regulated by SOX10, we confirmed a positive correlation with RNF125 expression (Shakhova et al., 2012; Sun et al., 2014). These observations suggest that the SOX10/MITF axis is an upstream regulator of RNF125, which carries corresponding SOX10-binding motifs and M box elements in its promoter regions (not shown).
Figure 4. EGFR Post-transcriptional Regulation by RNF125.
(A) RNF125 expression in A375 cells expressing indicated shRNAs.
(B) Left: EGFR expression in A375 and WM793 cells expressing control or shRNF125. Upper non-specific (n.s.) bands are marked. Right: corresponding changes in EGFR mRNA levels. Specific RNF125 bands are indicated with an arrow.
(C) Analysis of EGFR and JAK1 protein (upper) and transcript (lower) levels in UACC lines. Specific RNF125 bands are indicated with an arrow. Upper non-specific (n.s.) bands are marked.
(D) Left: UACC1113 cells expressing JAK1 shRNAs were analyzed for EGFR, JAK1, and SOX10 expression. Right: corresponding transcript levels were analyzed.
(E) EGFR expression in HeLa cells transfected with indicated plasmids.
(F) A375 cells were transduced with the indicated shRNAs and assessed for expression of EGFR, JAK1, and SOX10.
(G) Corresponding changes in transcript levels (EGFR, JAK1, and SOX10).
(H) Top: A375R (M) cells, which are BRAFi-resistant cells generated in vivo, were treated with the indicated concentrations of a JAKi for 24 hr and assessed for protein expression. Bottom: corresponding changes in transcript levels.
*p < 0.01 and **p < 0.05 unless otherwise specified. Data are presented as mean ± SD (A–D, G, and H).
RNF125/JAK1 Affects EGFR Expression
A recent report links EGFR and SOX10 expression in BRAFi-resistant melanomas (Sun et al., 2014). Notably, our gene clustering analysis for genes that are co-expressed in melanoma identified a link between RNF125 and EGFR expression (Figures S3B and S3C). Thus, we assessed the potential effect of RNF125 expression on EGFR expression. RNF125 depletion increased EGFR protein, but not mRNA, levels in A375, WM793, and Lu1205 (Figures 4B and S4A) cells; however, further analysis indicated that RNF125 does not interact with EGFR directly, thus excluding EGFR as an RNF125 substrate (not shown). Furthermore, RNF125 expression in additional melanoma lines was also positively correlated with SOX10 expression (r = 0.75) and inversely correlated with EGFR transcript (r = −0.65) and protein expression (Figures 4C and S4B), suggesting transcriptional and post-translational regulation of EGFR by SOX10/RNF125.
As anticipated, JAK1 protein levels were higher in melanoma cells that expressed low levels of RNF125 (Figure 4C). Given the reported crosstalk between EGFR and JAK1, we asked whether RNF125 regulates EGFR in a JAK1-dependent manner (Cordero et al., 2012; Yamauchi et al., 1997). Notably, JAK1 depletion in melanoma cells expressing higher levels of EGFR significantly lowered EGFR protein levels without altering EGFR transcription, supporting this hypothesis (Figure 4D). Conversely, JAK1 overexpression increased endogenous EGFR protein levels (Figure 4E). Notably, SOX10 depletion decreased RNF125 expression and increased EGFR transcript levels, consistent with our own results (Figure S4C) and independent studies (Sun et al., 2014). To assess a potential causative role for SOX10 in the RNF125/JAK1/EGFR pathway, we assessed the potential association of JAK1 levels with SOX10 expression. However, we found that JAK1 depletion did not alter SOX10 expression, suggesting that JAK1 does not regulate EGFR via SOX10 (Figure 4D). Transient SOX10 depletion in A375 cells also led to reduced JAK1 mRNA and protein levels, but increased those of EGFR (Sun et al., 2014) despite reduced RNF125 expression (Figures S4C and S4D). This suggests that there is an alternate route for control of JAK1 under certain conditions that is independent of RNF125. Notably, unperturbed melanoma cells, which exhibit reduced SOX10, also exhibited reduced RNF125 expression and elevated JAK1 and EGFR expression (Figure 4C), pointing to the primary role of RNF125 in controlling JAK1 expression concomitantly with the effect of EGFR expression. Importantly, JAK1 knockdown in SOX10-depleted melanoma cells completely abrogated SOX10-mediated EGFR protein upregulation without altering EGFR transcript levels. These observations suggest that in addition to the SOX10/EGFR regulatory axis, the RNF125/JAK1 pathway constitutes an independent regulatory module that controls EGFR expression, as seen in melanoma harboring low SOX10 and high EGFR expression (Figures 4F and 4G).
To substantiate the link between JAK1 and EGFR expression, we assessed pharmacological inhibitors of JAK, which are known to affect different members of the JAK family. Notably, inhibition of JAK activity decreased EGFR expression (Figures 4H and S4E). Interestingly, downregulation of EGFR following treatment with JAK inhibitors (JAKi) occurred at transcriptional and post-translational levels depending on the inhibitor used to attenuate JAK activity (Figure 4H). Other mechanisms that could account for the effect of JAK1 on EGFR expression may involve the adaptor protein GRB2 and the ubiquitin ligase CBL, both of which interact with JAK1 and EGFR and modulate EGFR expression (Giorgetti-Peraldi et al., 1995; Meisner et al., 1995). The reduced EGFR expression seen in JAK1-depleted cells was unchanged by the presence of proteasome inhibitors, indicating that this effect is independent of the UPS (Figure S4F).
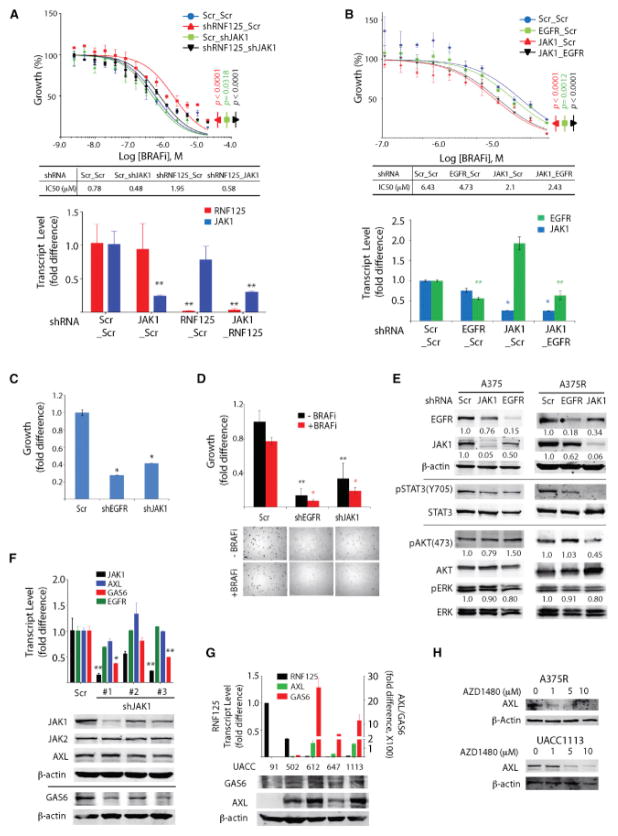
Inhibition of JAK1 Resolves Melanoma Resistance to BRAFi
We next asked whether the RNF125/JAK1/EGFR axis contributes to BRAFi resistance. First, we determined whether JAK1 depletion altered BRAFi resistance in an RNF125-dependent manner. RNF125 depletion increased the BRAFi IC50 from 0.78 to 1.95 μM, and decreased it to 0.58 μM upon additional depletion of JAK1 (Figure 5A). These observations suggest that the BRAFi resistance seen in RNF125-depleted cells is primarily due to perturbations in JAK1 expression/activity. Next, we tested whether depletion of JAK1 and EGFR altered the resistance of cells showing a high BRAFi IC50. Whereas EGFR depletion partially attenuated BRAFi resistance (6.43–4.73 μM), JAK1 depletion alone or together with EGFR depletion had a more pronounced effect (2.1 and 2.43 μM, respectively; Figure 5B).
Figure 5. JAK1 and EGFR Signaling Maintains the Growth of Resistant Cells.
(A) UACC91 cells were subjected to double knockdown with the indicated combinations of shRNAs and analyzed for cell growth. Bottom: RNF125/JAK1 knockdown efficiency was evaluated by qPCR.
(B) Analysis of BRAFi-mediated growth inhibition of UACC1113 cells expressing the indicated shRNAs. Bottom: knockdown efficiency was evaluated by qPCR.
(C) Analysis of growth of A375R cells transiently infected with lentivirus harboring the indicated shRNAs after 72 hr of culture.
(D) Growth of the A375R cells in (C) analyzed in soft agar.
(E) Cell lysates from parental (A375) and resistant (A375R) cells expressing the indicated shRNAs were analyzed using the indicated antibodies. EGFR and JAK1 band intensities were normalized to β-actin and quantified. Phospho-AKT and phospho-ERK were normalized to the respective total proteins and quantified.
(F) Expression of the indicated transcripts (upper) and proteins (lower) in UACC1113 cells depleted of JAK1.
(G) Expression of GAS6 and AXL transcripts and protein in lysates of UACC lines expressing low (612, 647, and 1,113) and high (91 and 502) RNF125 levels.
(H) Changes in AXL expression following treatment of mouse-derived A375R (M) cells and UACC1113, which represent melanoma with intrinsic resistance (lower) with various concentrations of JAKi (AZD1480) for 24 hr.
*p < 0.01 and **p < 0.05 unless otherwise specified. Data are presented as mean ± SD (A–D, F, and G). See also Figure S4.
Given the robust effect of RNF125 depletion on BRAFi-resistant cell growth, we asked whether altered JAK1 or EGFR expression phenocopied altered RNF125 expression. Similar to the effects of RNF125 on BRAFi-resistant cells, depletion of JAK1 or EGFR significantly inhibited the growth of BRAFi-resistant cells in 2D culture (to 42% of controls following JAK1 knockdown and 28% following EGFR knockdown; Figure 5C). Similarly, depletion of JAK1 or EGFR attenuated the 3D growth of resistant melanoma cells in soft agar (Figure 5D). Analyses of MAPK/ERK and PI3K/AKT pathways in parental and BRAFi-resistant cells depleted of JAK1 or EGFR also revealed a markedly reduced AKT activity in resistant, but not parental, cells (Figure 5E), pointing to a MAPK/ERK-independent mechanism underlying BRAFi resistance.
Unlike JAK1 depletion, EGFR depletion did not rescue drug sensitivity in BRAFi-resistant cells. Thus, we asked whether JAK1 signaling regulates other RTKs or their ligands that we identified in our bioinformatics analysis (Figure S3) or have been shown independently to be associated with BRAFi resistance (Konieczkowski et al., 2014; Müller et al., 2014; Nazarian et al., 2010; Sos et al., 2014). The transcript levels for four of the five genes encoding RTKs or their ligands (GAS6, IL6, KIT, and PDGFR, but not AXL) decreased following JAK1 knockdown (Figures 5F and S4G). Notably, levels of AXL protein, an RTK linked to BRAFi resistance (Konieczkowski et al., 2014; Müller et al., 2014), were reduced following JAK1 depletion by two of the three shRNAs used, or treatment with a JAK1 inhibitor (Figures 5F and 5H). The latter is consistent with the inverse correlation we observed for GAS6/AXL and RNF125 expression, and is similar to that seen for EGFR and RNF125 (Figure 5G). In all, our data suggest that JAK1 regulates the expression of several factors implicated in BRAFi resistance, including EGFR, GAS6/AXL, IL6, KIT, and PDGFR.
To confirm the genetic data, we assessed whether treatment with pharmacological inhibitors of EGFR (gefitinib) or JAK1 (pyridone 6 or AZD1480) would alter the growth of BRAFi-resistant A375R or Lu1205R cells. Gefitinib, either alone or in combination with a BRAFi, did not block the growth of either line in soft agar; however, a combination of a BRAFi and the JAKi pyridone 6 or a triple combination of BRAFi plus pyridone 6 and gefitinib significantly and dose-dependently attenuated the growth of BRAFi-resistant melanomas (Figure 6A). Similarly, combined treatment with a BRAFi and the JAKi AZD1480 blocked the growth of Lu1205R cells (Figure S5A), suggesting that increased expression of JAK1 and EGFR is essential to maintain BRAFi-resistant cell growth.
Figure 6. Inhibition of JAK1 and EGFR Blocks the Growth of Resistant Cells.
(A) A375R cells were grown for 2 weeks in soft agar in the presence of BRAFi (PLX4032), EGFRi (gefitinib), and JAKi (pyridone 6).
(B) BRAFi IC50 in parental and in vivo-generated resistant cells. IC50 = 0.003 and 3.18 μM in parental and resistant cells, respectively.
(C) Levels of the indicated transcripts and proteins in parental and resistant cells. *p < 0.05.
(D) Tumor growth changes in athymic nude mice injected with A375R (M) were monitored in two groups (BRAFi only and BRAFi+JAKi+EGFRi). Tumor volume is shown as the fold difference relative to tumor volume at day 0. Inset: images of three representative tumors at the end of the experiment. Because some tumors developed slower, their treatment begun at later time points, and their collectionband respective pictures were delayed (indicated by dotted line). See also the Experimental Procedures.
(E) Levels of the indicated proteins in the tumors shown in (D).
Data are presented as mean ± SD (A–C). See also Figure S5.
To validate these findings, we employed a xenograft model in mice using BRAFi-resistant A375 tumors that had relapsed in vivo. These mouse-derived A375R (M) cells exhibited a 100-fold higher BRAFi IC50 than their parental line and expressed low levels of RNF125 and elevated levels of JAK1 and EGFR (Figures 6B and 6C). In two independent assessments, we monitored the effect of BRAFi alone (PLX4720 fed in chow) or in combination with the JAKi AZD1480 together with the EGFR inhibitor (EGFRi) gefitinib. Significantly, the growth of the BRAFi-resistant A375 tumors was attenuated (60%) when mice were administered the combination treatment compared with BRAFi alone (n = 9 per group, p = 0.01147; Figure 6D). As observed in cultured cells, expression of both EGFR and AXL also decreased in the BRAFi+JAKi+EGFRi-treated group (Figure 6E).
RNF125 Expression Inversely Correlates with BRAFi Resistance in Melanoma Specimens
To confirm the cell culture and xenograft analyses, we assessed tumor specimens obtained from patient-derived xenografts (PDXs). PDXs generated from BRAFi-resistant tumors showed reduced RNF125 expression in nine of 17 tumors (a marked reduction in three and a more modest but significant reduction in six; Figure S5B). Interestingly, three of the 17 tumors exhibited a marked reduction in RNF125 expression that was associated with BRAFi resistance (Figure S5B), indicating that the RNF125/JAK1 axis would be relevant in a fraction of BRAFi-resistant tumors. To further assess changes associated with JAK1 expression, we monitored STAT3 activation, a reliable surrogate for JAK1 activity, which is also implicated in BRAFi resistance (Figure S5C; Girotti et al., 2013; Liu et al., 2013; Sos et al., 2014). Increased STAT3 phosphorylation (pSTAT) was seen in specimens exhibiting reduced RNF125 expression but not in tumors with unaltered RNF125 expression (Figures 7A and S5D). Moreover, analyses of three melanoma transcriptome data sets (GEO: GSE24862, GSE31534, GSE36139) identified a five-gene signature of STAT3-regulated genes, which coincided with reduced RNF125 and enhanced BRAFi resistance (Figures S6A–S6F; Barretina et al., 2012; Nazarian et al., 2010; Wang et al., 2012). Notably, that signature was highly correlated with pSTAT3 staining (Figures 7A and 7B). Likewise, in sections from BRAFi-resistant, PDX-derived specimens, pSTAT3 staining was inversely correlated with RNF125 staining (Figure 7C). Notably, expression of the STAT3-signature genes was significantly higher in tissue sections derived from patients with progressed forms of the disease (Figure 7D).
Figure 7. Correlation of RNF125 Expression and BRAFi Resistance in Melanoma Patients.
(A) RNF125 and pSTAT3 expression levels were assessed in tissue sections from PDX tumors treated with or without BRAFi. The upper two sections (3936-1 and 3936-2) and the lower section (4070-2) represent responders (in which RNF125 levels are reduced by BRAFi) and non-responders (no changes in RNF125 following BRAFi treatment), respectively (Figure S5B). Scale bar indicates 100 μm.
(B) Analysis of five STAT3 signature genes in three responders and two non-responders (Figures S5B and S6).
(C) RNF125 and pSTAT3 expression was assessed in tissue sections from patients before treatment (PRE) and after disease progression (PRO). Size bar indicates 500 (4×) and 100 (20×) μm, respectively.
(D) Analysis of five STAT3-signature genes in 11 PRE (black) and 14 PRO (red) melanoma patients. Following normalization to 18S RNA, the fold difference for the expression of each gene was calculated relative to the average value of the PRE samples (set to one). The statistical significance (p value) was determined by the Mann-Whitney U test. The number of samples analyzed for each gene is indicated. Bars indicate mean ± SEM
(E) The expression of RNF125 transcript was analyzed in melanoma tissue collected before BRAFi treatment (PRE), during treatment (ON), and after the development of progressive disease (PRO). “n” indicates the number of patients.
(F) Kaplan-Meier analysis of melanoma patients exhibiting a low or high increase in RNF125 expression upon treatment with BRAFi.
(G) RNF125 protein expression in melanoma tissues collected from patients before treatment (PRE) and after disease progression (PRO).
(H) RNF125 expression (before treatment) normalized to GAPDH was analyzed in terms of duration of response to BRAFi (time to progression). Shown is RNF125 expression versus time to progression in 22 patients. A correlation coefficient was calculated from 16 patients (filled circles), excluding six outliers (blank circles).
(I) RNF125 expression was analyzed in terms of the best RECIST score. As in Figure 7H, Pearson’s correlation coefficient (r) and p values were calculated from 16 (filled circles) of 22 (outliers, blank circles) patients.
See also Figures S5 and S6 and Table S3.
We next validated these findings using tumor specimens derived from melanoma patients. RNF125 expression in samples from patients treated with BRAFi was significantly higher in the initial phase of treatment (ON, response phase) but decreased during the progression phase (PRO, resistant phase) (Figure 7E; Table S3). Notably, Kaplan-Meier analysis suggested that patients whose tumors showed higher RNF125 levels at the response phase (ON treatment) had a longer survival time than those whose tumors exhibited lower levels (n = 17, p = 0.02; Figure 7F). A detailed comparison of RNF125 expression in six matched samples revealed that RNF125 expression was reduced in about half of the cases, indicating that our findings are relevant to a significant subset of resistant tumors (Figure S5E). As seen in resistant cells, RNF125 protein expression during the progression/resistance phases decreased relative to levels seen in the pre-treatment phase in patient tissues (Figures 7C and 7G). Further analysis of the correlation between RNF125 expression and time to progression, or best response evaluation criteria in solid tumors (RECIST) score, showed that 16 of 22 patients exhibited a significant correlation (r = 0.70, p = 0.003; Figure 7H). Of interest, the level of RNF125 expression during treatment (ON phase) could be associated with survival, even in the outlier cases (Figures 7F and S5F). Correspondingly, analysis of the RECIST scores also suggested that patients expressing higher RNF125 levels showed a better response to BRAFi (Figure 7I).
DISCUSSION
The need to identify mechanisms underlying melanoma cell resistance to existing therapies has fueled intensive investigation. Most studies have focused on pathways that alter MAPK signaling, either through mutation or differential splicing of BRAF or through upregulation of related receptors, including PDGFR and EGFR. Here, we identify a previously undisclosed mechanism that increases EGFR expression through deregulation of the ubiquitin ligase RNF125. We find that RNF125 deregulation alters EGFR expression indirectly by controlling JAK1, a newly described RNF125 substrate. In addition to its effect on EGFR, JAK1 expression was also found to regulate other genes, including GAS6/AXL, IL6, KIT, and PDGFR, all of which are implicated in BRAFi resistance (Konieczkowski et al., 2014; Müller et al., 2014; Nazarian et al., 2010; Sos et al., 2014). As previously demonstrated, targeting multiple pathways is likely to be more efficacious for eradicating tumor cells than targeting EGFR alone (Müller et al., 2014). Thus, the effect of JAKis on the regulation of multiple RTKs linked to BRAFi resistance may offer an efficient therapeutic modality.
We observed a link between RNF125 activity and melanoma resistance in cultures of melanoma cells obtained from resistant tumors, in melanoma cells that developed BRAFi resistance in culture, in xenografts and PDX from BRAFi-resistant tumors, and in tumor samples obtained at the time of resistance onset. Each assessment supports a causal role for RNF125 and JAK in BRAFi-resistant melanoma. Significantly, the growth of BRAFi-resistant xenografts was effectively attenuated using a combination of BRAFi, JAKi, and EGFRi. Our bioinformatics analysis indicated an effect of RNF125/JAK1 on several RTKs implicated in BRAFi resistance and confirmed them as part of the greater JAK1 network (Figures S3B–S3D). Given that multiple mechanisms underlie BRAFi resistance, including NRAS and MEK mutations and alternate BRAF splicing, the mechanism identified here should be relevant to the subset of BRAFi-resistant melanoma tumors. Our initial assessment of three independent sets of patient tumors resistant to BRAFi suggests that the RNF125/JAK1 regulatory axis described here is relevant to a sizeable fraction of these tumors, which is a prediction that should be confirmed using larger cohorts.
Interestingly, RNF125 levels are also notably lower in other tumor types (such as colorectal cancer) that exhibit a poor response to BRAFi (GEO: GSE36139) (Barretina et al., 2012; Prahallad et al., 2012). Thus, it will be interesting to assess RNF125’s function in conferring resistance to BRAFi or other MAPK inhibitors in these cancer types.
Our findings also suggest that SOX10 regulates EGFR both transcriptionally (directly) and post-translationally (via RNF125 destabilization of JAK1), indicating that SOX10 constitutes a common regulatory hub underlying the control of resistance phenotypes. Consistent with this observation, SOX10 regulation of MITF expression and increased signaling via NF-κB and the receptor tyrosine kinase AXL have been linked to primary resistance to BRAFi in a subset of melanomas (Kim and Ronai, 2015; Konieczkowski et al., 2014; Müller et al., 2014). In all cases, reduced levels of SOX10, MITF, or RNF125 were shown to confer resistance. Importantly, our observations suggest that RNF125 downregulation maintains resistance phenotypes, consistent with our current understanding of adaptive resistance and its importance for cell growth and survival (Kugel and Aplin, 2014). The lower RNF125 expression seen in cells with intrinsic or adaptive resistance confers fitness benefits by upregulating JAK and EGFR signaling. A critical role for the RNF125 regulatory axis is also reflected in our network analysis, which demonstrates that RNF125 control of JAK1 is linked to the expression of many genes that are deregulated in association with BRAFi resistance (Figure S3D) confirming, the importance of this axis in melanoma resistance. Taken together, our data suggest that combined inhibition of JAK and EGFR offers a significant advantage for attenuating the growth of BRAFi-resistant melanoma and could constitute a therapeutic modality to overcome BRAFi-resistant tumors.
EXPERIMENTAL PROCEDURES
Cell Lines and Establishment of Resistant Cells
Melanoma (A375, Lu1205, WM9, WM35, WM793, SK-Mel-28, and SK-Mel-29), HeLa, and HEK293T cells were cultured in DMEM (Life Technology) supplemented with 10% fetal bovine serum and penicillin/streptomycin. Melanoma UACC cell lines were maintained in RPMI1640 medium (Life Technology) supplemented with 10% fetal bovine serum and penicillin/streptomycin. Resistant cells were established in culture by increasing PLX4032 levels at each passage and maintaining them in media containing PLX4032 (A375 [5 μM], Lu1205 [5 μM], and UACC91 [1 μM]). A375R (M) cells (in vivo-generated BRAFi-resistant cells) were isolated from relapsed tumors and expanded in media without BRAFi for xenograft analysis and characterization.
Reagents and Antibodies
The BRAFi PLX4032, the EGFRi gefitinib/ZD1839, and the JAKi AZD1480 were purchased from Selleckchem. The JAKi pyridone 6 was purchased from Millipore. The following antibodies were used: FLAG-tag from Sigma-Aldrich; V5-tag from Life Technology or Bethyl Laboratories; Stat3, β-actin, GFP, and control IgG from Santa Cruz; TYK2 from BD Biosciences; JAK2, JAK3, EGFR, AXL, pSTAT3, and DDX58/RIG-I from Cell Signaling; GAS6 from R&D Systems; RNF125 from Life Technology, Bethyl Laboratories, AbCam, or Sigma-Aldrich; and JAK1 from Santa Cruz, BD Biosciences, Cell Signaling, or EMD Millipore.
Screen for BRAFi-Resistance-Associated Genes
Screening was performed using siRNAs targeting 1,173 UPS-related genes (consisting of ubiquitin ligases, ubiquitin-like proteins, and deubiquitination enzymes). Each gene was represented by three different sets of siRNAs. siRNA transfection was performed according to the reverse transfection method in 384-well format using RNAimax (Life Technology). Using 48 nonspecific siRNAs and three killer siRNAs (targeting genes essential for viability), we tested the transfection efficiency and calculated the z-factor of viability using two different BRAFi concentrations (1 and 5 μM). At both concentrations, the latter value ranged from 0.8 to 0.9 in Lu1205 parental and resistant cells. For reverse transfection of siRNAs, 1 μl of each siRNA (0.5 μM), including control siRNAs (nonspecific and killer), was spotted per well. After a 30-min reaction in the presence of RNAimax (0.1 μl in 10 μl Opti-MEM; Life Technology), parental or BRAFi-resistant cells (3,500 cells/40 μl of medium) were added. Twenty-four hours later, 10 μl of BRAFi (final concentration: 1 μM for parental cells and 5 μM for resistant cells) was added. Cell growth and viability were determined 72 hr later using ATPlite (Perkin Elmer) according to the manufacturer’s recommendation. Fold differences in cell growth were calculated by dividing values from vehicle (DMSO)-treated wells by values from BRAFi-treated wells. The fold differences from each siRNA set were further analyzed. Secondary screening was repeated using A375 and Lu1205 lines under the same conditions.
Cell Viability Assay in 2D Culture
Cell growth was assayed using ATPlite (Perkin Elmer). Briefly, melanoma cells (5,000 cells/well) were cultured in 96-well plates with a transparent bottom. Twenty-four hours after seeding, inhibitors were added for 72 or 120 hr. Cell growth or viability was measured using ATPlite (Perkin Elmer) and quantified by monitoring luminescence intensity.
Soft Agar Assay
Soft agar culture was performed using standard methods. Briefly, 50,000 melanoma cells in top agar (0.35% in 1× DMEM) were plated over a second layer of agar (0.7% in 1× DMEM). After 24 hr, medium containing various compounds was added over the top agar and changed every 5 days. Colonies were visualized by crystal violet staining (0.005% crystal violet in 4% paraformaldehyde/PBS). Images were acquired with a microscope (×40) or scanned (HP ScanJet G4010). Colony number and size were analyzed using ImageJ software (NIH).
Patient Samples
Patients with metastatic melanoma containing a BRAFV600E mutation (confirmed by genotyping) were enrolled in clinical trials of a BRAFi or combined BRAF+MEK inhibitors (Table S3). Consent was obtained for tissue acquisition in accordance with a protocol approved by the Institutional Review Board and the conventions of the Helsinki Declaration on Human Rights. Tumor biopsies were collected pre-treatment (on day 0), after 10–14 days of treatment, and/or at the time of progression, if applicable. Formalin-fixed tissue was analyzed with H&E staining to confirm the presence of a viable tumor. Additional tissue was snap-frozen and stored in liquid nitrogen or was processed immediately for RNA purification.
PDXs were established by grafting melanoma tumor biopsies from patients subcutaneously into immunodeficient NSG mice with Matrigel (BD Biosciences). The animals were monitored weekly for tumor growth and live tissue was banked or serially transplanted when the tumors reached maximal volume. In PDXs established from BRAFi therapy-relapsed patients, tumors were grown under a PLX4720 200 ppm diet. All studies were conducted in accordance with the Wistar Institutional Animal Care and Use Committee.
Immunohistochemistry
Tumor biopsies were stained with primary antibodies for RNF125 (HPA041514; Sigma-Aldrich) followed by a secondary antibody conjugated to horseradish peroxidase and treated with 3,3′-diaminobenzidine (DAB) chromogen. Stained slides were interpreted by a dedicated dermatopathologist. Only positive signals with clear melanoma morphology were evaluated. For immunofluorescence, the primary antibodies were RNF125 and pSTAT3 (both diluted 1:1,000; Cell Signaling), and signals were amplified using the TSA-Biotin Plus System (Perkin Elmer) according to the manufacturer’s instructions. The same slides were counterstained with S-100 (1:400; Dako). Immunofluorescence-stained slides were visualized using a fluorescence microscope with a Slidebook or Aperio slide scanner.
DNA Constructs, Transfection, and Transduction
FLAG-tagged RNF125 vectors were constructed by subcloning PCR products from MEF cDNA into pcDNA3.0-C-FLAG. The FLAG-tagged fragment was then subcloned into the pBabe retroviral vector. RNA125 RING domain mutations (C37 and 40A) were introduced by site-directed mutagenesis (Agilent Technology). A JAK1 expression vector was constructed in pLX304-V5 Gateway vectors from pDONR223-JAK1 (Addgene #23932).
Cells were transiently transfected using Jet-Prime (Polyplus Transfection). For viral transduction, viral particles were harvested after calcium phosphate transfection of HEK293T cells with plasmid vectors and appropriate packaging plasmids. Target cells were infected with virus particles by spinoculation (1,600 × g for 30 min at room temperature) in the presence of 4 μg/ml polybrene (Sigma-Aldrich). Stable clones were established by culture in media containing puromycin (1 μg/ml; InvivoGen) or blasticidin (5 μg/ml; Sigma-Aldrich).
Cycloheximide Chase Assay
Cycloheximide chase was performed as described previously (Kim et al., 2014). Briefly, cycloheximide (50 μg/ml) was added to cells at the indicated times. Cell lysates were prepared as described previously (Kim et al., 2014) and analyzed with the indicated antibodies.
Immunoprecipitation and Immunoblotting
To detect protein interactions, cell lysates were prepared using 1% Tritonlysis buffer (50 mM Tris-HCl [pH 7.4] and 150 mM NaCl) spiked with protease and phosphatase inhibitors (Thermo Scientific). Lysates pre-cleared with beads (Protein A/G agarose bead; Santa Cruz) were incubated with the appropriate antibodies overnight at 4°C, and then protein A/G agarose beads were added and the incubation continued for an additional 2 hr at 4°C. Beads were washed with lysis buffer, boiled in Laemmli buffer, and subjected to SDS-PAGE. To detect endogenous JAK1-RNF125 interactions, A375 cells were pretreated with MG132 (10 μM; Selleckchem) for 5 hr before lysis and immunoprecipitation with an anti-JAK1 antibody (BD Bioscience and EMD Millipore). For immunoblotting, cell or tumor lysates were prepared using RIPA buffer (50 mM Tris-HCl [pH 7.4], 1% [v/v] NP-40, 0.1% [w/v] sodium de-oxycholate, 0.1% [w/v] SDS, 150 mM NaCl, 1 mM EDTA, a protease inhibitor cocktail [Roche], and PhoStop [Roche]). Imaging of immunoblots was performed with the aid of LICOR system using respective fluorescence antibodies. A horizontal line indicates cases where the membrane was split to enable reactions with multiple antibodies. In cases of overexpression, RNF125 is seen as a single band. When endogenous protein was followed, a doublet was identified. The identity of the specific RNF125 band was confirmed by qPCR where a corresponding change were also seen at the transcript levels. Specific RNF125 band is pointed by arrow. Non-specific (n.s.) band is noted.
qPCR
Total RNA was obtained with GenElute (Sigma-Aldrich) and subjected to reverse transcription using high-capacity cDNA synthesis kits (Applied Bio-systems). For qPCR, cDNAs were analyzed with CFX Connect (BioRad) using Faststart Universal Cyber Green Master Mix (Roche) according to the manufacturer’s directions. Total RNA from patient biopsies was obtained using an RNeasy kit on a QiaCube apparatus (QIAGEN) and served as the template (250 ng) to generate cDNA (Superscript VILO cDNA Synthesis Kit; Invitrogen). Real-time qPCR was carried out on a LightCycler (Roche) using Essential Green Master Mix (Roche). Transcript level differences were analyzed according to the ΔΔct method. Primer sequences are listed in Table S4. Imaging of immunoblots was performed with the aid of LICOR system using respective fluorescence antibodies. A horizontal line indicates cases where the membrane was split to enable reactions with multiple antibodies. In cases of overexpression, RNF125 is seen as a single band. When endogenous protein was followed, a doublet was identified. The identity of the specific RNF125 band was confirmed by qPCR where corresponding change were also seen at the transcript levels. Specific RNF125 bands are indicated with an arrow. Non-specific (n.s.) bands are noted.
Mass Spectrometry
Melanoma A375 cells were transfected with control plasmid (pcDNA3.0 with a FLAG-tagged C terminus) or plasmid encoding FLAG-tagged RNF125 (WT or mutant RNF125RM form). Cells were lysed in 1% Triton-lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% [v/v] Triton X-100, a protease inhibitor cocktail [Roche], and PhoStop [Roche]) and pre-cleared with protein A/G agarose beads (Santa Cruz). Immunoprecipitation was performed using FLAG-M2-agarose beads (Sigma-Aldrich). After washing with lysis buffer, bound proteins were eluted with FLAG-peptide (Sigma-Aldrich) according to the manufacturer’s directions.
Samples for LC-MS/MS analysis were prepared according to standard procedures (Kim et al., 2014). In brief, protein samples were reduced, alkylated, trypsin digested, and then desalted. Desalted samples were resuspended in 125 μl 0.1% TFA/0.2% acetonitrile in glass sample vials and then analyzed using a Michrom MDLC Paradigm MS4 HPLC system, HTC-PAL autosampler, and LTQ OrbitrapXL mass spectrometer.
In Vivo Xenograft
In vivo-generated BRAFi-resistant cells were injected (1.0 × 106, s.c.) into the lower left flank of 6-week-old female athymic nude mice. Both the BRAFi-only and BRAFi+JAKi+EGFRi groups were first fed BRAFi chow (PLX4720, 417 ppm in chow; Research Diets) on the day of injection. Once the tumors reached ~50–100 mm3, the experimental group also received JAKi (AZD1480, 50 mg/kg, oral gavage, once daily) and EGFRi (gefitinib, 100 mg/kg, oral gavage, once daily), and the BRAFi-only control group received vehicle only. Tumor size was monitored every 3–4 days until the mice were euthanized. Tumors were collected from euthanized mice and processed for further analyses. Because tumor growth rate varied among the individual tumors, indicated treatment was initiated at different time points (based on tumor size rather than time from injection). Accordingly, tumor collection took place at two time points, respective of treatment initialization. The Institutional Animal Care and Use Committee (IACUC) of Sanford-Burnham Medical Research Institute approved our study protocols.
Gene Silencing
pLKO.1 vectors targeting RNF125, JAK1, EGFR, HNF4a, SOX10, and MITF were purchased from Thermo Scientific. After transduction of shRNA lentivirus, silencing efficiency was validated from cell lysates of stable clones using the appropriate antibodies or by qPCR analysis of mRNA using the primers shown in Table S4.
Statistical Analysis
Unless specified, data are presented as means ± SD and the significance of differences was analyzed using a two-tailed, unpaired t test. Differences with p values < 0.05 were considered significant. For analyses of a linear correlation between two groups, Pearson’s correlation was calculated using Excel software unless otherwise specified. IC50 values were calculated using Prism 6.0 software with non-linear regression to fit data to the log values of inhibitor concentration versus the response values (fold changes in luciferase activity). The R statistical package was used to generate box plots. The p values from Pearson correlation coefficient analysis were calculated with the following equation using Excel software: p = 1−F.dist(((n−2)*r2)/(1−r2), 1, n−2, TRUE (n, number of data pairs; r, Pearson’s coefficient).
Supplementary Material
Highlights.
RNF125 binds to, ubiquitinates, and promotes degradation of JAK1
Low RNF125 increases JAK1 and EGFR expression in BRAFi-resistant melanoma
The combination of JAK, BRAF, and EGFR inhibitors overcomes drug resistance
RNF125 expression in melanoma specimens inversely correlates with BRAFi resistance
Acknowledgments
We thank Holly Yin for providing the UACC cell lines used in this study. We also thank members of Z.A.R.’s laboratory for extensive discussions. This work was supported by grants from the NCI (CA128814), the Melanoma Research Foundation, and the Hervey Family Non-endowment Fund at The San Diego Foundation (to Z.A.R.).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.04.049.
AUTHOR CONTRIBUTIONS
H.K. designed performed experiments and analyzed data. D.T.F., Z.A.C., M.P.L., R.D., J.W., C.K., M.M.H., A.P., and K.T.F. provided and analyzed tumor specimens. Y.F. established resistant cultures and performed mouse xenograft studies. L.B. performed MS analysis. Y.S., N.K.H., T.Z., K.M.B., R.H., and A.P. provided and analyzed gene expression data in human melanoma tumor cell lines. Z.A.R. designed and analyzed experiments, and coordinated collaboration and exchange of reagents. H.K., K.T.F., and Z.A.R. wrote the manuscript.
References
- Abel EV, Basile KJ, Kugel CH, 3rd, Witkiewicz AK, Le K, Amaravadi RK, Karakousis GC, Xu X, Xu W, Schuchter LM, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. J Clin Invest. 2013;123:2155–2168. doi: 10.1172/JCI65780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci USA. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. BRIM-3 Study Group. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero JB, Stefanatos RK, Myant K, Vidal M, Sansom OJ. Non-autonomous crosstalk between the Jak/Stat and Egfr pathways mediates Apc1-driven intestinal stem cell hyperplasia in the Drosophila adult midgut. Development. 2012;139:4524–4535. doi: 10.1242/dev.078261. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O’Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini AL, Gao Y, Bijlmakers MJ. T-cell regulator RNF125/TRAC-1 belongs to a novel family of ubiquitin ligases with zinc fingers and a ubiquitin-binding domain. Biochem J. 2008;410:101–111. doi: 10.1042/BJ20070995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti-Peraldi S, Peyrade F, Baron V, Van Obberghen E. Involvement of Janus kinases in the insulin signaling pathway. Eur J Biochem. 1995;234:656–660. doi: 10.1111/j.1432-1033.1995.656_b.x. [DOI] [PubMed] [Google Scholar]
- Girotti MR, Pedersen M, Sanchez-Laorden B, Viros A, Turajlic S, Niculescu-Duvaz D, Zambon A, Sinclair J, Hayes A, Gore M, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov. 2013;3:158–167. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, Dickerson SH, Laquerre SG, Liu L, Gilmer TM. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol Cancer Ther. 2012;11:909–920. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH, Jr, Kaempgen E, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- Kim H, Ronai ZA. The UPs and DOWNs of MITF in melanoma resistance. Pigment Cell Melanoma Res. 2015;28:132–133. doi: 10.1111/pcmr.12338. [DOI] [PubMed] [Google Scholar]
- Kim H, Claps G, Möller A, Bowtell D, Lu X, Ronai ZA. Siah2 regulates tight junction integrity and cell polarity through control of ASPP2 stability. Oncogene. 2014;33:2004–2010. doi: 10.1038/onc.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, Frederick DT, Barzily-Rokni M, Straussman R, Haq R, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4:816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel CH, 3rd, Aplin AE. Adaptive resistance to RAF inhibitors in melanoma. Pigment Cell Melanoma Res. 2014;27:1032–1038. doi: 10.1111/pcmr.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Cao J, Wu J, Sullivan K, Shen J, Ryu B, Xu Z, Wei W, Cui R. Stat3-targeted therapies overcome the acquired resistance to vemurafenib in melanomas. J Invest Dermatol. 2013;133:2041–2049. doi: 10.1038/jid.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bergami P. The role of mitogen- and stress-activated protein kinase pathways in melanoma. Pigment Cell Melanoma Res. 2011;24:902–921. doi: 10.1111/j.1755-148X.2011.00908.x. [DOI] [PubMed] [Google Scholar]
- Meisner H, Conway BR, Hartley D, Czech MP. Interactions of Cbl with Grb2 and phosphatidylinositol 3′-kinase in activated Jurkat cells. Mol Cell Biol. 1995;15:3571–3578. doi: 10.1128/mcb.15.7.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Foppen MH, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Qi J, Nakayama K, Gaitonde S, Goydos JS, Krajewski S, Eroshkin A, Bar-Sagi D, Bowtell D, Ronai Z. The ubiquitin ligase Siah2 regulates tumorigenesis and metastasis by HIF-dependent and -independent pathways. Proc Natl Acad Sci USA. 2008;105:16713–16718. doi: 10.1073/pnas.0804063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Nakayama K, Cardiff RD, Borowsky AD, Kaul K, Williams R, Krajewski S, Mercola D, Carpenter PM, Bowtell D, Ronai ZA. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell. 2010;18:23–38. doi: 10.1016/j.ccr.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Tripathi M, Mishra R, Sahgal N, Fazli L, Ettinger S, Placzek WJ, Claps G, Chung LW, Bowtell D, et al. The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell. 2013;23:332–346. doi: 10.1016/j.ccr.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A, Flaherty KT. BRAF targeted therapy changes the treatment paradigm in melanoma. Nat Rev Clin Oncol. 2011;8:426–433. doi: 10.1038/nrclinonc.2011.69. [DOI] [PubMed] [Google Scholar]
- Shakhova O, Zingg D, Schaefer SM, Hari L, Civenni G, Blunschi J, Claudinot S, Okoniewski M, Beermann F, Mihic-Probst D, et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat Cell Biol. 2012;14:882–890. doi: 10.1038/ncb2535. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kong X, Ribas A, Lo RS. Combinatorial treatments that overcome PDGFRβ-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer Res. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, Ng C, Chodon T, Scolyer RA, Dahlman KB, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji-Kawata S, Zhong Q, Kameoka M, Iwabu Y, Sapsutthipas S, Luftig RB, Ikuta K. The RING finger ubiquitin ligase RNF125/TRAC-1 down-modulates HIV-1 replication in primary human peripheral blood mono-nuclear cells. Virology. 2007;368:191–204. doi: 10.1016/j.virol.2007.06.028. [DOI] [PubMed] [Google Scholar]
- Sos ML, Levin RS, Gordan JD, Oses-Prieto JA, Webber JT, Salt M, Hann B, Burlingame AL, McCormick F, Bandyopadhyay S, Shokat KM. Oncogene mimicry as a mechanism of primary resistance to BRAF inhibitors. Cell Rep. 2014;8:1037–1048. doi: 10.1016/j.celrep.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, Bradley WD, Wang Q, Yang H, Xu L, Higgins B, Kolinsky K, Packman K, Kim MJ, Trunzer K, et al. Resistance to selective BRAF inhibition can be mediated by modest upstream pathway activation. Cancer Res. 2012;72:969–978. doi: 10.1158/0008-5472.CAN-11-1875. [DOI] [PubMed] [Google Scholar]
- Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49:1297–1304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, Haanen J, Blank C, Wesseling J, Willems SM, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hurley DG, Watkins W, Araki H, Tamada Y, Muthukaruppan A, Ranjard L, Derkac E, Imoto S, Miyano S, et al. Cell cycle gene networks are associated with melanoma prognosis. PLoS ONE. 2012;7:e34247. doi: 10.1371/journal.pone.0034247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Ueki K, Tobe K, Tamemoto H, Sekine N, Wada M, Honjo M, Takahashi M, Takahashi T, Hirai H, et al. Tyrosine phosphorylation of the EGF receptor by the kinase Jak2 is induced by growth hormone. Nature. 1997;390:91–96. doi: 10.1038/36369. [DOI] [PubMed] [Google Scholar]
- Zhao H, Li CC, Pardo J, Chu PC, Liao CX, Huang J, Dong JG, Zhou X, Huang Q, Huang B, et al. A novel E3 ubiquitin ligase TRAC-1 positively regulates T cell activation. J Immunol. 2005;174:5288–5297. doi: 10.4049/jimmunol.174.9.5288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.