Abstract
Abiotic and biotic conditions often vary continuously across the landscape, imposing divergent selection on local populations. We used a provenance trial approach to examine microgeographic variation in local adaptation in Boechera stricta (Brassicaceae), a perennial forb native to the Rocky Mountains. In montane ecosystems, environmental conditions change considerably over short spatial scales, such that neighboring populations can be subject to different selective pressures. Using accessions from southern (Colorado) and northern (Idaho) populations, we characterized spatial variation in genetic similarity via microsatellite markers. We then transplanted genotypes from multiple local populations into common gardens in both regions. Continuous variation in local adaptation emerged for several components of fitness. In Idaho, genotypes from warmer environments (low elevation or south facing sites) were poorly adapted to the north-facing garden. In high and low elevation Colorado gardens, susceptibility to insect herbivory increased with source elevation. In the high elevation Colorado garden, germination success peaked for genotypes that evolved at similar elevations as the garden, and declined for genotypes from higher and lower elevations. We also found evidence for local maladaptation in survival and fecundity components of fitness in the low elevation Colorado garden. This approach is a necessary first step in predicting how global change could affect evolutionary dynamics.
Keywords: environmental gradient, fecundity, fitness, germination success, microsatellite, common garden experiment, subalpine meadow
Introduction
Natural landscapes are highly heterogeneous. This environmental variation can impose divergent selection on natural populations, and result in the evolution of local adaptation (Alberto et al. 2013; Bennington et al. 2012; Hereford 2009; Kalske et al. 2012; Kremer et al. 2012; Savolainen et al. 2007; Vergeer and Kunin 2013; Wang et al. 2010). Reciprocal transplant experiments provide powerful tests of local adaptation to discrete habitats in a diversity of taxa (e.g., Hendry et al. 2002; Hereford 2009; Hereford and Winn 2008; Kawecki and Ebert 2004; Kim and Donohue 2013; Leimu and Fischer 2008; Lowry et al. 2008; Via 1991). Yet, most species encounter continuous gradients of environmental conditions across their ranges, and do not simply inhabit two contrasting habitat types. We know little about clinal variation in fitness and local adaptation across complex gradients that occur over short spatial scales (Gould et al. 2014; Richardson et al. 2014; Wilczek et al. 2014).
Natural populations often adapt to local climates across latitudes (e.g., Ågren et al. 2013; Etterson and Shaw 2001). Climatic conditions change only gradually along latitudinal gradients, and gene flow is likely minimal among distant populations at different latitudes. In contrast, elevation gradients present superb opportunities for investigating spatial variation in adaptation and the balance between migration and selection. In montane and alpine systems, climatic conditions vary predictably with elevation, producing similar climatic gradients as those present across latitudes but over shorter geographic distances (Dunne et al. 2003; Kelly and Goulden 2008; Knowles et al. 2006; Wang et al. 2012). These environmental differences can result in consistent patterns of local adaptation to elevation (e.g., Byars et al. 2007; Clausen et al. 1940; Kim and Donohue 2013; Storz et al. 2007). Gene flow likely connects populations at different elevations that occur in close geographically proximity, but experience disparate selective regimes. Is divergent selection at fine spatial scales strong enough to counteract the homogenizing forces of gene flow (Richardson et al. 2014)?
Evolutionary consequences of migration-selection balance depend on spatial variation in natural selection and the extent of gene flow across neighboring populations (Lenormand 2002; Sexton et al. 2014; Slatkin 1987). If gene flow declines with geographic distance (isolation by distance), but genotypes migrate across environments, then adjacent populations in different environments could experience continual influx of maladapted alleles, reducing adaptive population differentiation. In contrast, migrants may not successfully establish in environments that differ in environmental conditions from their natal habitats, limiting gene flow across environmental gradients (isolation by environment). In that case, populations from similar climates would be most likely to exchange genes, which could increase genetic variation in local populations (Sexton et al. 2014).
To assess the microgeographical scale of local adaptation, we adopted a common garden approach similar to provenance trials from the forestry literature. In provenance trials, researchers transplant families from multiple populations across the range of a species into several test gardens to quantify landscape-level patterns of local adaptation and complex traits (Aitken et al. 2008; Kremer et al. 2012; Wang et al. 2010). Foresters originally designed provenance trials to quantify genetic differences among populations and identify appropriate seed sources for reforestation (Wang et al. 2010). Recently, data from provenance trials have been used to demonstrate that adaptation will likely lag behind climate change in trees (Wang et al. 2010; Wilczek et al. 2014). This approach has two primary advantages that increase our understanding of clinal variation in local adaptation. First, in contrast with the traditional home vs. away reciprocal transplant experiment, provenance trials include a greater number of families that evolved under different environmental conditions in populations distributed across gradients. Second, these trials use an increased number of test environments, which improves our ability to discern genotype by environment interactions for fitness. Using this approach, researchers have documented local adaptation to climate in Pinus contorta and other trees (Aitken et al. 2008; Kremer et al. 2012; Wang et al. 2010) and the model organism Arabidopsis thaliana (Wilczek et al. 2014). The accuracy of predictions about fitness improves with the number of populations and test gardens included in the study (Wang et al. 2010). The provenance trial approach can also reveal instances of maladaptation, such as when foreign genotypes outperform local genotypes (from the “local vs. foreign” criterion of local adaptation, Kawecki and Ebert 2004).
Here, we test whether divergent selection across landscapes with continuous variation in environmental conditions could be strong enough to counteract ongoing gene flow (Richardson et al. 2014). The objectives of our current study were to (1) examine clinal variation in fitness and foliar damage from herbivores and local adaptation of natural accessions under realistic field conditions across years, and (2) characterize isolation by distance and estimate spatial genetic structure in populations from known geographic locations using microsatellite loci. We transplanted experimental Boechera stricta (Brassicaceae) genotypes into three common gardens to determine whether the fitness of immigrants is related to the physical distance or elevation change from their source populations. Herbivores impose strong selection on plant populations (e.g., Carmona and Fornoni 2013; Kalske et al. 2012; Prasad et al. 2012; Wise and Rausher 2013), and interactions between plants and their insect herbivores can vary across elevation gradients (Rasmann et al. 2014). In addition to characterizing fitness clines, we examined local adaptation in relation to the insect herbivore community by quantifying foliar damage from herbivores.
We performed complementary studies in Colorado and Idaho to test whether the balance between migration and selection operates similarly in different portions of the range, producing consistent fitness clines. Meta-analysis has shown that local adaptation may be more apparent for fecundity than viability components of selection (Hereford 2009). In contrast, we previously documented reciprocal polygenic local adaptation to latitude (Colorado vs. Montana) in viability, but not fecundity, components of fitness for B. stricta (Anderson et al. 2014). We examined multiple fitness components from germination success of seeds, to juvenile survival and fecundity of transplanted seedlings to test the prediction that the local environment more severely filters inappropriate genotypes early in life history. Finally, we test the contributions of geographic distance (isolation by distance) and elevation (isolation by environment) to genetic structure in both regions (Sexton et al. 2014).
Materials and Methods
Focal species and study sites
Boechera stricta (Brassicaceae) is a short-lived perennial herbaceous plant native to the U.S. Rocky Mountains. This species spans a large gradient in elevation (from 700 m – 3900 m) and latitude (from Arizona to Alaska), where populations evolve under different environmental and climatic regimes (Al-Shehbaz and Windham 2010; Rushworth et al. 2011; Song et al. 2009; Song et al. 2006). As with 10-15% of plant species (Goodwillie et al. 2005), B. stricta primarily self-pollinates (FIS: 0.74-0.97, Song et al. 2006). Previous studies have documented local adaptation to latitude (Anderson et al. 2014) and differential water availability (Lee and Mitchell-Olds 2013). Yet, we do not know whether populations are locally adapted to climatic gradients that occur over short distances within a geographic region. We conducted complementary studies in two portions of the range: the southern Rocky Mountains (Gunnison National Forest, Gunnison County, Colorado), and the northern Rocky Mountains (Lost River Range, Challis National Forest, Custer County, Idaho). The center of genetic diversity for this species occurs in the Colorado Plateau and central Rocky Mountains (Kiefer et al. 2009). Phylogeographic data suggest long-term stable populations of B. stricta in that portion of the range, with rapid expansion into northern sites after the retreat of the glaciers (Kiefer et al., 2009). Our design allows us to examine whether patterns of clinal variation in local adaptation are congruent in the two regions.
Population genetic study
As a first step in characterizing genetic isolation, we scored 16 microsatellites markers for 127 individuals from Idaho populations (elevation range: 2316-2910 m), and 97 individuals from Colorado populations (elevation range: 2842-3690m; see Online Appendix 1 for oligonucleotide sequences for primers, and Online Appendix 2 for coordinates of all populations). In the Colorado populations, the minimum distance between the two closest populations was 0.028 km, the maximum distance between the two farthest populations was 9.6 km, and the average distance (± S.D.) was 3.46 ± 2.09 km. For the Idaho populations, the minimum distance was 0.017 km, the maximum was 6.2 km, and the average (± S.D.) was 1.56 ± 0.96 km. Of the 16 microsatellines, 10 were polymorphic in the Idaho samples and 13 were polymorphic in the Colorado samples (Online Appendix 1). We genotyped one individual per population, as B. stricta is a selfing species that maintains low within population genetic variation (Song et al. 2009; Song et al. 2006). This protocol allowed us to gather data from a greater number of populations.
We ground leaf samples in liquid N2, and extracted DNA with Qiagen plant Dneasy mini kits (Qiagen Inc., Valencia, CA). Polymerase chain reaction (PCR) conditions are in Online Appendix 1. Forward primers were either pre-labeled with M13, or the M13 label was added during PCR, to produce dye-labeled amplified alleles. We resolved genotypes via electrophoresis on the ABI 3730 of the Duke University Genome Sequencing Resource Core. We scored data with Genemarker (ver. 2.20), verifying each allele score manually.
We first tested for deviation from Hardy-Weinberg equilibrium using GenAlEx 6.5 (Peakall and Smouse 2006). We then calculated genetic identity as the proportion of identical alleles between samples across loci. We analyzed genetic similarity as a function of geographic distance (isolation by distance) and elevational distance (isolation by environment) between all pairs of samples within each region. We performed Mantel matrix permutation tests to assess statistical significance, shuffling rows (and columns) of one matrix, calculating the slope of genetic identity as a function of (horizontal or vertical) distance in meters, and comparing the observed slope to the permuted distribution under the null hypothesis that identity and distance are independent. This Python program is available from Mitchell-Olds on request.
Common garden experiments
To test for adaptive differentiation among populations that are likely connected by gene flow, we conducted field experiments in Colorado and Idaho between 2012 and 2014. Near the Rocky Mountain Biological Laboratory in Colorado we established gardens at two elevations: 2891 m (38°57.086” N and 106°59.4645”W) and 3133 m (39°02.346”N and 107°03.818”W). In Idaho, we installed one common garden (elevation: 2535 m; 44°10.906” N and 113°44.363”W). Researchers maximize power to detect local adaptation by transplanting few genotypes from many populations into experimental gardens rather than many genotypes from only a few populations (Blanquart et al. 2013; Goudet and Buchi 2006). For that reason, we selected selfed full siblings of one genotype (one maternal family) from each of N=24 populations local to the Colorado sites and N=26 populations local to the Idaho garden. Prior to transplanting, we grew field-collected seeds for one generation in the greenhouse to reduce maternal effects.
Elevations of origin ranged from 2869-3682 m for the Colorado genotypes and 2414-2884 m for the Idaho genotypes. These local families were collected from populations near each garden, with geographic distance ranging from: 0.55 – 3.4 km away from the Idaho garden, 0.20-8.3 km away from the low elevation Colorado garden, and 3.2-11.7 km away from the high elevation Colorado garden (Online Appendix 3 lists coordinates of all families). We transplanted full siblings from Idaho genotypes into the Idaho garden and Colorado genotypes into the Colorado gardens. We did not plant Idaho genotypes in the Colorado gardens or Colorado genotypes in the Idaho garden, as we have already documented polygenic local adaptation to Montana vs. Colorado environments in previous studies (Anderson et al. 2014; Anderson et al. 2013). Furthermore, there is negligible gene flow between Colorado and Idaho populations (Song et al. 2009; Song et al. 2006), and the focus of the current study was to examine local adaptation in the context of gene flow.
In our study systems, temperatures decline, soil moisture increases, and the length of the growing season decreases with elevation (Anderson and Gezon 2015; Dunne et al. 2003). For example, in our Colorado study sites, mean annual temperature declines by 0.0023°C to 0.0046°C for every 1m gain in elevation, and total annual precipitation increases by 0.042 cm / 1 m gain of elevation (Anderson and Gezon 2015). Furthermore, growing season begins later in high elevation sites; for example, in 2013 and 2014, snow melted 11 days later at our high elevation Colorado garden (elevation: 3133m) than our low elevation Colorado garden (elevation: 2891m). Thus, elevation can serve as a proxy for climate. We predict that genotypes that evolved at elevations similar to that of each common garden will show the highest fitness there, and those from higher or lower elevations will have reduced fitness. Furthermore, as environmental conditions vary spatially, we predict that fitness will decline as a function of geographic distance between the source location and the site of the common garden. Other environmental and geological factors, such as bedrock, might not covary with geographic distance and elevation; thus, geographic distance and elevation are imperfect proxies for environmental variation.
In September 2011, we transplanted N=2293 juvenile rosettes (~95.5 individuals per family, N=24 families) into the lower elevation Colorado garden, and N=1150 rosettes into the Idaho garden (50 individuals per family, N=23 families). We had not yet established the higher elevation Colorado garden in 2011. In September 2012, we planted another set of rosettes into high and low elevation Colorado gardens (N=1096 individuals in the lower garden and N=1121 individuals in the higher garden from the same 24 families as in 2011). We also planted a second cohort into the Idaho garden (N=672 individuals, N=24 families). In the Idaho garden, two families planted in 2011 were not included in the 2012 cohort due to limited seed availability. We added three new families to the Idaho 2012 cohort. In sum, we exposed 24 maternal families to the Colorado gardens and 26 families to the Idaho garden. In both years and sites, we transplanted 2 siblings per genotype into blocks of 48 individuals. We disrupted natural vegetation minimally during planting.
To quantify germination success, we planted seeds in the Colorado gardens in September 2012. Seeds were only available for a subset of families. We used garden staples to affix 1cm tall seed planting grids to the ground. We then filled all 1 cm × 1 cm cells in the grid with potting soil derived from Colorado compost. We did not use native soil at each site to reduce possible sources of contamination by B. stricta seeds that may have been in the seed bank. After randomizing the planting order of genotypes, we placed one seed per cell into a ~2mm hole and then covered the seed with potting soil. We deliberately left blank control cells (potting soil, but no seed) in each grid to check for contamination from natural seed sources, and we planted late in the season, after local B. stricta completed fruiting and seed dispersal. Seedlings did not recruit into these control cells. We established 4 seed grids in each Colorado garden, with 9 seeds per genotype planted into each grid (36 seeds/genotype/garden; low elevation: 17 genotypes, N=612 seeds total; high elevation: 16 genotypes, N=571 seeds total because we only had 31 seeds for one genotype). We monitored germination success in 2013 and 2014.
We visited Colorado gardens 3-4 times/week from May - August 2012-2014, and the Idaho garden 1-2 times/month during the growing seasons of 2012 and 2013. We recorded survival, flowering success, and numbers of fruits. In 2012 and 2013, we quantified foliar damage from herbivores (% of leaf tissue removed by herbivores) as: [(number of leaves with herbivory damage) ×(average % damage on those leaves)]/total number of leaves on a plant. Foliar damage is inversely proportional to resistance to herbivores, and is subject to strong selection in our system (Prasad et al. 2012).
Over the winter of 2013-2014, gophers breached the underground fence at the high elevation Colorado site and damaged the experiment through tunneling activities (not herbivory). In spring 2014, we attributed mortality of plants at that site directly to gophers in instances where tunnels were present at the site of the plant, and the plant was missing (N=517 plants died from gopher activity, leaving N=534 individuals alive at that point); this process was unambiguous. Death from gopher tunneling is indiscriminate and unrelated to a plant's climatic tolerance or extent of local adaptation. To calculate lifetime fitness (fecundity) through September 2014, we excluded plants whose deaths were due to gophers.
Statistical analyses
We modeled data separately by region because we transplanted different maternal families into gardens in Colorado vs. Idaho. To quantify clinal variation in local adaptation, we analyzed fitness components and herbivore resistance as a function of source elevation of transplanted genotypes and geographic distance between the garden and the source population in a multiple regression framework including both predictors. We modeled quadratic effects if residuals showed curvature. For both regions, we included fixed effects for cohort, and interactions between cohort and elevation, and cohort and distance. For the Colorado analyses, we also incorporated garden as a fixed effect, and interactions between garden and (linear and quadratic effects of) elevation, and garden and geographic distance.
We monitored plants for different durations in each garden and cohort based on planting date. For example, lifetime fitness was assessed over three growing seasons (2012-2014) for the 2011 cohort planted into the low elevation CO garden, but only two growing seasons (2013- 2014) for the 2012 cohort planted into both CO gardens. Results are qualitatively similar when we model each garden and cohort separately (Online Appendix 4). We conducted a complementary repeated measures analysis to test whether clinal variation in fitness changed across the three years of the study of the 2011 cohort in the low elevation CO garden (Proc Mixed, SAS ver. 9.3, Online Appendix 5). Finally first year fitness is correlated with lifetime fecundity in both cohorts of the low elevation Colorado garden (2011 cohort: F1,22=21.3, p=0.0001, R2=0.49, β=3.15 ± 0.68 lifetime fruits/ first season fruits; 2012 cohort: F1,22=12.7, p=0.0017, R2=0.37, β=1.12 ± 0.31 lifetime fruits/first season fruits), and in the high elevation CO garden (F1,22=66.34, p<0.0001, R2=0.75, β=1.05 ± 0.13 lifetime fruits/first season fruits), suggesting that first year fitness can serve as a proxy for lifetime fitness.
We first calculated lifetime fitness for every individual plant, as total numbers of fruits produced over the course of the study (Idaho 2011 cohort: 2012 + 2013 growing seasons; Idaho 2012 cohort: 2013; Colorado 2011 cohort: 2012-2014; Colorado 2012 cohort: 2013 + 2014). To summarize overall levels of genetic variation in these regional pools of populations, we computed heritabilities in lifetime fitness and foliar damage from herbivores. In selfing species like B. stricta, selection acts on total genetic variance, not just additive genetic variance (Roughgarden 1979). We estimated genetic variance (VG) and broad-sense heritability (H2= VG/VP, where VP=family variance + block variance + error variance) of lifetime fitness via the restricted maximum likelihood (REML) method (Proc Mixed, SAS ver. 9.3).
Fitness components included: germination success (Colorado gardens only), survival to the end of the study, and lifetime fitness (total numbers of fruits). We conducted these analyses using family-level means. For logistic regressions using binary fitness components (Proc Logistic), the numerator of the response variable was the total number of successful individuals (#germinated, or # survived) and the denominator was the number of individuals planted or the number of individuals that survived the first winter for each family. To generate family-level LSMEANS for lifetime fecundity, we regressed fitness on genotype, with initial plant size as a covariate and block as a random effect; plants that died or otherwise failed to fruit were given values of 0 for fitness (Proc Mixed). We used the family averages (LSMEANS) in all subsequent analyses, and visualized the relationships between predictor variables (source elevation or geographic distance) and fitness components using the R package ggplot2 (ver. 0.9.3.1). For the 2011 cohorts planted into Colorado and Idaho, we have data on foliar damage from herbivores in two growing seasons (2012 and 2013). We conducted repeated measures analyses to test for clinal variation in herbivory in those cases. For the 2012 cohort, we only have foliar damage data from 2013.
Owing to the limited sample size of genotypes included in the seed germination study (N=16 families in the high elevation Colorado garden and 17 families in the low elevation Colorado garden), we only considered models with fixed effects of garden, linear and quadratic effects of source elevation, and interactions between garden and elevation, excluding geographic distance.
The Colorado 2011 low elevation cohort suffered 49.8% mortality the first winter after planting (winter 2011-2012), leaving 1150 individuals alive on the first census date of 1 June 2012. We found no evidence for clinal variation in overwintering success in this cohort (Online Appendix 6). Sources of this high mortality include inappropriate climatic tolerance as well as indiscriminate mortality caused by strong runoff from early spring snowmelt, which disproportionately affected blocks in on the northern side of the garden. Dead plants are typically easy to identify because dead biomass remains where the plant once was. In the case of this first winter in the northern part of the garden, plants were entirely missing and plants tags were in disarray, lying on the top of the soil facing in the same direction. In contrast, in other parts of the garden, plants were alive and tags were in place. Mortality over the first winter was much lower for the 2012 Colorado cohort (low elevation garden: 5%; high elevation garden: 1%). Furthermore, subsequent overwinter mortality was low in the 2011 Colorado cohort (9.6% mortality from September 2012- June 2013, and 15.4% from September 2013- June 2014). Therefore, we excluded plants that died in the initial winter from fitness analyses in the main text, but we show analyses with those plants in Online Appendix 6. Fitness clines are qualitatively similar whether we include or exclude these plants.
To complement our analyses of family means, we ran zero-inflated negative binomial analyses of the individual level lifetime fitness data in the R package glmmADMB (ver. 0.8.0). These analyses included random effects for genotype and block and a covariate for initial plant size. Models followed a zero-inflated distribution, as many individuals died or failed to fruit before the end of the study, resulting in values of 0 for lifetime fitness. Negative binomial distributions fit better than Poisson distributions. Results of these models were highly consistent with those of the family-level analyses (Online Appendix 7).
Results
Population genetic study
We found significant evidence for isolation by distance and isolation by elevation in both regions from our individual-based analyses. In the Colorado populations, the average genetic identity of pairs of individuals declined by 1.47% for every 1 km change in geographic distance (p=0.0002) and by 2.32% per 100 meters of elevation difference (p=0.0002). Similarly, in Idaho, average genetic similarity decreased by 1.71% for every 1 km change in geographic distance (p=0.0004) and by 1.14% per 100 meters of elevation difference (p=0.0178). Additionally, we found high levels of genetic similarity among nearby individuals (the genetic similarity of individuals 0m distant from each other, estimated from the y-intercept of the models of genetic similarity vs. geographic distance). The intercepts of these models suggest that adjacent individuals would share 39.0% of alleles in the Colorado populations and 46.4% in the Idaho populations.
Common garden experiments
Heritability
We found significant heritability in lifetime fitness in all gardens for at least one of the cohorts, and for foliar damage from insect herbivores in the high and low elevation Colorado gardens (Table 1).
Table 1.
Broad sense heritability for lifetime fitness and foliar damage from herbivores in three gardens and two cohorts. There is no high elevation 2011 cohort in Colorado (NA=Not applicable). P-values in bold are significant after Bonferroni correction for multiple tests (α=0.005), and the italicized p-value is marginally significant at that level.
| Idaho garden | Low elevation Colorado garden | High elevation Colorado garden | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | H2 | χ 2 | p-value | H2 | χ 2 | p-value | H2 | χ 2 | p-value |
| Lifetime fitness, 2011 cohort | 0.176 ± 0.048 | 194.8 | <0.0001 | 0.031 ± 0.012 | 38.8 | <0.0001 | NA | ||
| Lifetime fitness, 2012 cohort | 0.038 ± 0.078 | 0.3 | 0.58 | 0.022 ± 0.012 | 7.7 | 0.0055 | 0.23 ± 0.06 | 102 | <0.0001 |
| Foliar damage from herbivores, 2011 cohort in 2012 growing season | 0.015 ± 0.01 | 2.8 | 0.094 | 0.067 ± 0.026 | 30.1 | <0.0001 | NA | ||
| Foliar damage from herbivores, 2012 cohort | 0.163 ± 0.10 | 4.2 | 0.04 | 0.001 ± 0.009 | 0 | 1 | 0.026 ± 0.014 | 9.3 | 0.0023 |
Resistance against insect herbivores
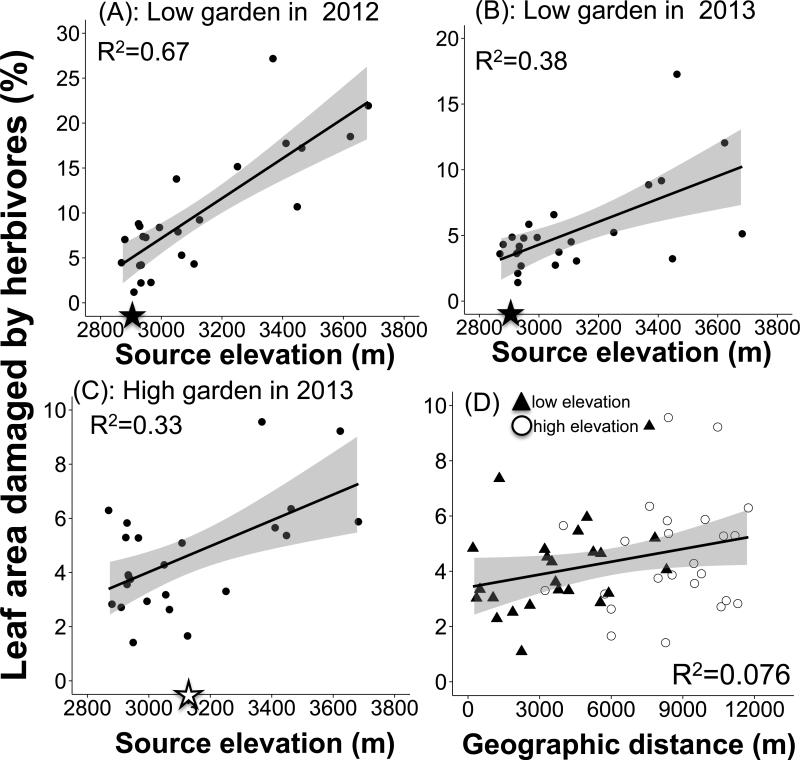
In the Colorado gardens, foliar damage by insect herbivores increased with source elevation for the low elevation 2011 cohort (measured in 2012 and 2013), and for the 2012 cohort in the high elevation garden (Fig. 1). Repeated measures analysis of the 2011 cohort revealed significant effects of source elevation (F1,21=32.6, p<0.0001), year (F1,21=6.8, p=0.016), and an interaction between year and elevation (F1,21=7.31, p=0.013), but no effect of geographic distance (F1,21=0.74, p=0.40) or year by distance (F1,21=0, p=0.96). The source elevation by year interaction resulted from a stronger positive slope in 2012 (β=0.024±0.004, t-value=6.11, p<0.0001, Fig. 1A) than in 2013 (β=0.010±0.004, t-value=2.63, p=0.016, Fig. 1B). For the 2012 cohort, we found a significant effect of garden (F1,20=5.98, p=0.024), source elevation (F1,20=7.37, p=0.013), elevation by garden interaction (F1,20=4.72, p=0.042), geographic distance (F1,21=5.97, p=0.024), but no distance by garden interaction (F1,20=0.27, p=0.61). The interaction between elevation and garden was caused by a significant positive relationship between foliar damage and elevation in the high elevation garden (β=0.0057±0.001, t-value=4.01, p=0.0007), but no relationship in the low elevation garden (β=0.00097±0.002, t-value=053, p=0.61). These results indicate that high elevation genotypes are less resistant to herbivory than low elevation families, irrespective of the location of the common garden. We found no evidence for clinal variation in leaf damage from herbivores in the Idaho genotypes, which span smaller geographic and elevational gradients than the Colorado accessions, in repeated measures analysis of the 2011 cohort (elevation, p=0.82; year, p=0.59; elevation by year, p=0.84; distance, p=0.79, distance by year, p=0.88) or the 2012 cohort (2012: elevation: p=0.22, distance: p =0.41),
Fig. 1.
Variation in foliar damage from herbivores in Colorado environments. In low (panels A + B) and high (panel C) elevation gardens, leaf damage was positively correlated with source elevation, indicating that high elevation populations are more susceptible to damage by insect herbivores across test sites. The elevation of the garden is indicated on the X-axis with closed star at 2891m for low elevation, and an open star at 3133m for high elevation (panels A-C). In addition (D), foliar damage varied with geographic distance for the 2012 cohort across both gardens; data points from the low elevation garden are closed triangles and data points from the high elevation garden are open circles. Shading around predicted regression lines indicates 95% Confidence Intervals.
Clinal variation in fitness: Colorado
We predicted that fitness would decline with provenance elevation and geographic distance in the low elevation garden (2891 m). In contrast, for the high elevation garden, we predicted that negative quadratic fitness functions would emerge, such that families from elevations similar to that of the garden (3133 m) would have optimal fitness.
Germination success
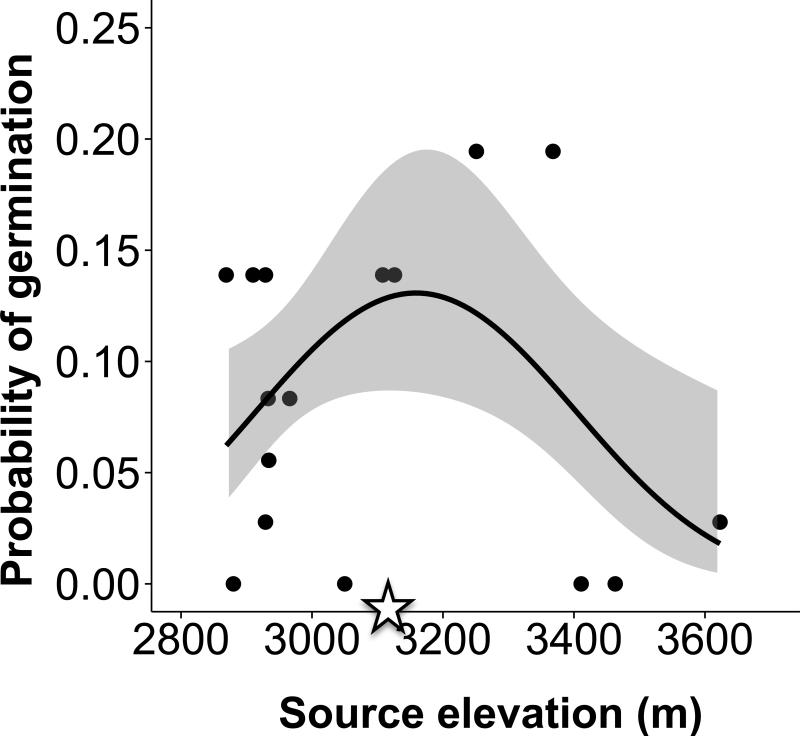
In the low elevation garden, 67 seeds germinated of the 612 seeds planted (10.9%) in 2013. In the high elevation garden, only 8.6% of seeds germinated during the spring of 2013 (49 germinants of 571seeds planted). No seeds germinated after June 2013 in either garden, and we monitored the seed grids until August 2014. As a point of comparison, germination success of these genotypes under controlled laboratory conditions is >90%. Germination success differed between gardens (χ2=4.8, p=0.029), but did not vary with linear (χ2=2.09, p=0.15) or quadratic (χ2=2.2, p=0.14) effects of elevation. Nevertheless, we found significant interactions between garden and linear (χ2=4.71, p=0.03) and quadratic (χ2=4.6, p=0.03) effects of provenance elevation. To examine interactions terms, we modeled germination success separately for each garden. For the low elevation garden, we found no evidence for clinal variation in germination success as a function of source elevation (χ2=2.50, p=0.11). In contrast, for the high elevation garden, we found the predicted quadratic relationship with elevation, such that germination success peaked for genotypes from source elevations similar to the garden (Fig. 2; linear effect of source elevation: χ2=5.08, p=0.024; quadratic effect of source elevation: χ2=5.11, p=0.024).
Figure 2.
In the high elevation Colorado garden, germination success followed the predicted quadratic fitness function, peaking for genotypes from similar elevations as the common garden (3133 m). Shading around predicted regression lines indicates 95% Confidence Intervals. The open star on the X-axis indicates the elevation of the garden.
Juvenile survival
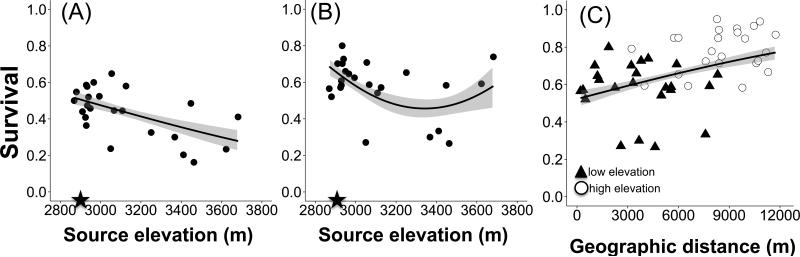
Analyses of both cohorts and gardens together indicate a significant effect of cohort, and interactions between cohort and (linear and quadratic effects of) source elevation on the probability of survival (Table 2). To interpret these interaction terms, we analyzed each cohort and garden separately (Appendix 4). For the 2011 cohort, the odds of survival from initial overwintering success to September 2014 declined by 11.4% for every 100m increase in source elevation (odds ratio: 0.886; 95% confidence interval: 0.829,0.946; p=0.0003 Appendix 4; Figure 3A), consistent with expectations. For the 2012 cohort in the low elevation garden, linear and quadratic effects of source elevation on survival revealed curvature to the fitness function: Survival decreased with elevation to a minimum at ~3300m, then increased owing to high performance of three high elevation families (linear source elevation: χ2=25.3, p<0.0001, quadratic effect of source elevation: χ2=18.2, p<0.0001; Appendix 4; Fig. 3B). In addition, the significant distance by cohort interaction (Table 4) was caused by an unexpected positive relationship between geographic distance and survival across gardens in the 2012 cohort (t=3.05, p=0.0041, Fig. 3C). We found no evidence for clinal variation in fitness in the high elevation Colorado garden (Appendix 4).
Table 2.
Clinal variation in survival and fecundity components of fitness in two Colorado gardens. We included genotype as a random effect in these models. The low elevation CO garden included two cohorts of experimental transplants from the same 24 maternal families, whereas there was only one cohort planted into the high elevation garden. For that reason, we could not model garden by cohort interactions. Family means for the high elevation CO garden excluded plants that died because of gopher activity for the CO 2012 high elevation garden. Family means for the low elevation CO 2011 cohort excluded plants that died over the first winter (2011-2012) because excessive mortality was driven by strong runoff from snowmelt. Including those early overwinter deaths does not fundamentally alter fitness clines (Online Appendix 6).
| Probability of survival | Fecindity | |||
|---|---|---|---|---|
| F1,39 | p-value | F1,39 | p-value | |
| Garden | 3.92 | 0.055 | 0.69 | 0.41 |
| Cohort | 8.45 | 0.0060 | 5.57 | 0.023 |
| Source elevation | 0.07 | 0.80 | 2.85 | 0.099 |
| Source elevation × Garden | 3.77 | 0.06 | 0.73 | 0.40 |
| Source elevation × Cohort | 8.41 | 0.0061 | 5.39 | 0.025 |
| (Source elevation)2 | 0.01 | 0.92 | 0.69 | 0.41 |
| (Source elevation) × Garden | 3.42 | 0.072 | 0.81 | 0.37 |
| (Source elevation)2 × Cohort | 8.44 | 0.006 | 5.34 | 0.026 |
| Geographic distance | 1.25 | 0.27 | 0.10 | 0.75 |
| Distance × Garden | 0.16 | 0.69 | 0.17 | 0.69 |
| Distance × Cohort | 6.59 | 0.014 | 0.00 | 0.99 |
| Genotype | χ2=28.0 | <0.0001 | χ2=16.4 | <0.0001 |
Figure 3.
Clinal variation in survival in the Colorado gardens as a function of: (A) source elevation for the 2011 cohort in the low elevation garden (2891m), (B) source elevation for the 2012 cohort in the low elevation garden (2891m), and (C) geographic distance for the 2012 cohort across gardens. Closed stars on the X-axis indicate the elevation of the garden. In panel C, data points from the low elevation garden are closed triangles and data points from the high elevation garden are open circles. Shading around predicted regression lines indicates 95% Confidence Intervals.
Lifetime fitness
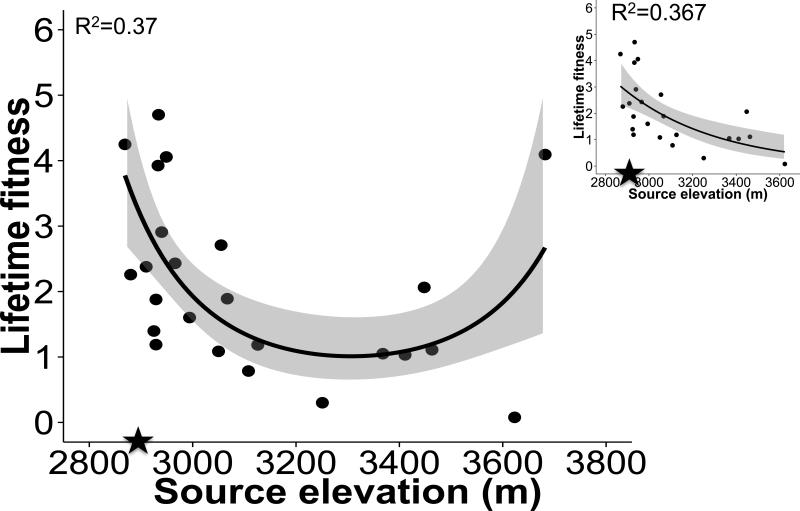
Consistent with analyses of survival, we found a significant effect of cohort, and interactions between cohort and (linear and quadratic) effect of elevation on lifetime fecundity (Table 2). For the 2011 CO cohort, lifetime fitness followed a positive quadratic relationship with source elevation, declining across most populations, but increasing at source elevations above 3300m (linear source elevation: F1,19=8.31, p=0.0095, quadratic effect of source elevation: F1,19=8.24, p=0.0098; Appendix 4; Fig. 4). The individual level zero-inflated negative binomial model showed quantitatively similar patterns (Online Appendix 7). These results are consistent with survival patterns in this garden. There was no evidence for clinal variation in lifetime fitness in the 2012 cohort in either garden.
Figure 4.
In the CO low elevation garden (2011 cohort), lifetime fitness varied with linear and quadratic effects of source elevation. The elevation of the garden (2891m) is indicated by a closed star on the X-axis. Fitness declined to a minimum for genotypes from mid-elevation populations and then increased to an unexpected peak in genotypes from high source elevation. The highest elevation family (3682m) is an influential outlier; when removed (small panel) fitness declines with source elevation, as expected. These results suggest local maladaptation, with a foreign high elevation family achieving or exceeding fitness levels of the lowest elevation families. Shading around predicted regression lines indicates 95% Confidence Intervals.
Repeated measures analysis of the 2011 Colorado cohort
Repeated measures analysis of fitness across three years of data for the low elevation Colorado cohort revealed significant effects of source elevation on survival and fecundity, but no significant year effect or year by elevation, nor any effect of geographic distance (Online Appendix 5). Consistent with predictions and with results from the overall model (Table 2, Fig. 3A), survival declined by 10.4% for every 100m increase in source elevation when analyzed across years (odds ratio: 0.896; 95% confidence interval: 0.83,0.97; F1,21=8.28, p=0.0009, Appendix 5). As with lifetime fitness, repeated measures analysis of annual fecundity indicated curvilinear relationships with source elevation (linear effect of elevation: F1,19=5.43, p=0.031; quadratic effect of elevation: F1,19=8.97, p=0.0074, Online Appendix 5). The relationship between annual fecundity and elevation was qualitatively identical to the relationship between lifetime fecundity and elevation: a negative quadratic function indicating high fitness for low and high elevation genotypes, with minimum fitness for mid-elevation genotypes (Fig. 4 and Online Appendix 5). Thus, at least at that site, fitness clines appear to be stable across three growing seasons.
Clinal variation in fitness: Idaho
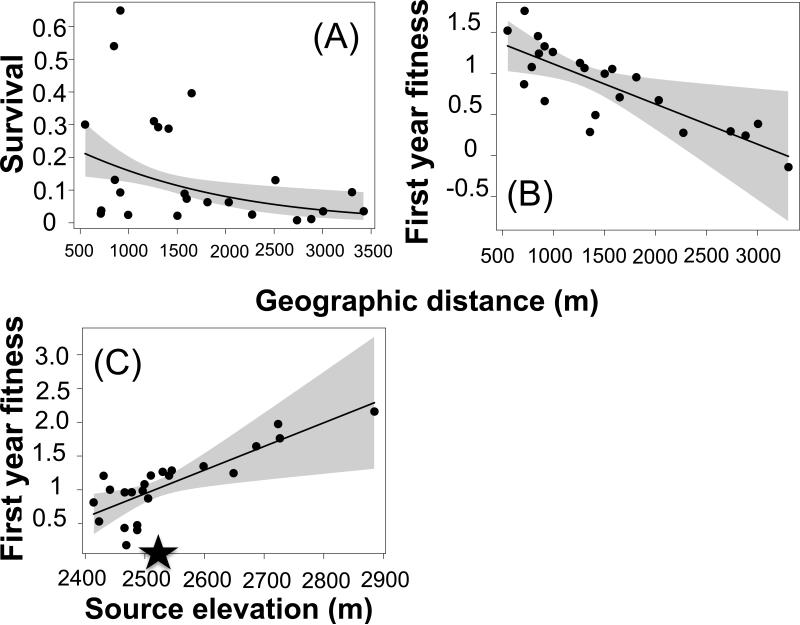
We planted two cohorts into the Idaho garden (Idaho 2011 cohort and Idaho 2012 cohort). Survival decreased with geographic distance for the 2012 cohort (survival × cohort interaction, Table 3, Appendix 4, Fig. 5A), but was not related to distance for the 2011 cohort or elevation for either cohort. We found no evidence for clinal variation in lifetime fitness (Table 3); however, models of fitness in the first growing season revealed a positive relationship with source elevation and a negative relationship with geographic distance for the 2011 cohort (Figs. 5B+C, Table 4 and Appendix 4: slope estimate for elevation in 2011 cohort: β=0.012 ± 0.004 fruits/m elevation, t=3.1, p=0.0062; slope estimate for distance in 2011 cohort: β= -0.00174 ± 0.0006 fruits/m elevation, t=-2.98, p=0.008).
Table 3.
Clinal variation in fitness for the two cohorts planted into the Idaho garden. As we found no evidence for clines in lifetime fitness, we also analyzed fitness in the first growing season (Year 1 fitness). We included genotype as a random effect in these models. There was no evidence for curvilinear fitness clines in this dataset.
| Probability of survival | Year 1 fitness | Lifetime fitness | ||||
|---|---|---|---|---|---|---|
| F1,18 | p-value | F1,18 | p-value | F1,18 | p-value | |
| Cohort | 5.31 | 0.033 | 4.98 | 0.039 | 1.58 | 0.22 |
| Source elevation | 1.18 | 0.29 | 3.90 | 0.064 | 1.33 | 0.26 |
| Source elevation × Cohort | 3.39 | 0.082 | 6.48 | 0.020 | 2.56 | 0.13 |
| Geographic distance | 0.67 | 0.42 | 3.68 | 0.07 | 1.48 | 0.24 |
| Distance × Cohort | 5.78 | 0.027 | 6.45 | 0.0205 | 2.86 | 0.11 |
| Genotype | χ2=4.31 | 0.019 | χ2=0.3 | 0.58 | χ2=0 | 1 |
Figure 5.
Clinal variation in fitness in the Idaho garden (elevation: 2535). Fitness declined with geographic distance for (A) survival in the 2012 cohort, and (B) first year fecundity in the 2011 cohort. Additionally, (C) first year fecundity of the 2011 cohort increased with source elevation. Shading around predicted lines represents 95% CI, and the star on the X-axis of panel C shows the elevation of the garden (2535m).
Discussion
We found clinal variation in components of fitness in the northern and southern Rocky Mountains, consistent with fine-scale local adaptation to environmental gradients that span short geographic distances; our study also documented several instances of local maladaptation in the low elevation Colorado garden. These results are compatible with migration-selection balance as an important cause of genetic variation within populations. Our field experiments show genetic variation in lifetime fitness and susceptibility to insect herbivory, suggesting that populations could evolve higher levels of fitness through ongoing natural selection in contemporary landscapes (Shaw and Shaw 2014).
In our system, gene flow is likely spatially restricted because of limited pollination and seed dispersal movements in this primarily self-pollinating plant (isolation by distance). Changing environmental conditions across elevations could also restrict gene flow (isolation by environment) (Sexton et al. 2014). Elevation and geographic distance are correlated in our samples in both regions; therefore, it is difficult to disentangle the effects of environment (elevation) from those of distance on genetic structure. Future studies using loci with lower mutations rates will be needed to achieve more accurate estimates of the rate of decline in genetic similarity across space. High levels of genetic similarity across space indicate that populations in close geographic proximity likely exchange genes. Yet, gene flow is not sufficient to counteract selection that varies continuously across environmental gradients.
Microgeographic variation in local (mal)adapatation
By transplanting genotypes from multiple populations across the range of a species into several common gardens, researchers can examine patterns of adaptation to environmental gradients (Fournier-Level et al. 2011; Wang et al. 2010; Wilczek et al. 2014). In our study, we documented microgeographic local adaptation in three distinct gardens. In Idaho, genotypes from nearby populations maintained high fitness in the mid-elevation north-facing common garden, whereas transplants from warmer sites (low elevation or south-facing populations) had depressed fitness. The strongest patterns appeared when we examine clinal variation in fitness jointly across both cohorts in the two years of this study: Survival declined with geographic distance in the 2012 cohort, and first year fecundity increased with elevation and decreased with distance in the 2011 cohort. These results highlight the need to replicate studies to test for general patterns.
In the high elevation Colorado garden, a quadratic fitness function indicated that genotypes from elevations similar to the garden had a fitness advantage for germination success, but we found no clinal variation in survival or fecundity of transplanted rosettes there. In contrast, a meta-analysis of reciprocal transplant studies found stronger patterns of local adaptation in fecundity components of fitness than for survival (Hereford 2009). Yet, the studies available for that meta-analysis likely did not assess variation in fitness at very early life history stages. Indeed, few reciprocal transplant studies have investigated the seed to seedling transition; therefore, we lack a comprehensive understanding of local adaptation in germination success (Donohue et al. 2010; Geber and Eckhart 2005; Kim and Donohue 2013; Wilczek et al. 2014). Germination is a critical life history transition, as seeds that fail to germinate have no future fecundity. Natural selection could filter out inappropriate genotypes early in the life cycle, but that pattern would not be apparent in studies that only transplant juveniles and do not plant seeds. In some environments, such as this common garden, immigrant genotypes could have low germination rates, but the few seedlings that manage to establish may not differ from local genotypes in subsequent survival and fecundity. We do not yet have data on B. stricta seed dormancy rates in field conditions. From our laboratory studies, seed dormancy appears minimal; we can achieve >70% germination success within 1-3 weeks simply by planting seeds onto moist filter paper in petri dishes or directly into the soil with no cold treatment or stratification. Seeds appear to require only a short period of after-ripening (~1 month of dry storage) and moisture to enable germination. Seeds that did not germinate in the field study could have been nonviable or dormant; alternatively, they could have germinated and died prior to monitoring. Dormant seeds that failed to germinate during the 2 year monitoring effort (2013 and 2014) would likely have reduced genetic contribution to future generations compared with seeds that germinated in 2013 and reproduced in 2014. The extent of local adaptation likely changes across life history stages, and could be most evident early in life history.
In addition to documenting local adaptation, our study also revealed evidence for local maladaptation, which was particularly prominent in the low elevation garden in Colorado. For this garden we predicted that fitness would be negatively correlated with source elevation and geographic distance, as the elevation of the garden (2891m) nearly matches the source elevation of the lowest family (2869m). Indeed, the probability of survival for the 2011 cohort conformed to expectations. However, we found evidence of local maladaptation in lifetime fitness of the 2011 cohort, and survival of the 2012 cohort. In these cases, fitness declined to a minimum at mid-source elevations, but then increased to a second peak at high source elevation. This second, unexpected, fitness peak resulted from high survival and fecundity of one or two high elevation genotypes transplanted into this low elevation garden. It is possible that these successful families evolved in populations that share non-climatic conditions with the low elevation common garden. Elevation serves as a reliable proxy for climate, but other environmental variables (e.g., bedrock) vary spatially, but do not necessarily covary with elevation. Alternatively, some genotypes in this regional pool of populations could occur in environments where they have sub optimal fitness. Spatially restricted gene flow could prevent these genotypes from rapidly colonizing more appropriate sites, and could prevent more successful genotypes from colonizing sites dominated by suboptimal genotypes. Maladaptation could also potentially arise from genetic drift and limited genetic diversity in local populations. Future studies that include more local gardens, especially at higher elevations, will quantify the extent of maladaptation across this landscape and examine the mechanism underlying local maladaptation.
Clinal variation in susceptibility to insect herbivory
We examined a key trait that is subject to strong selection in plant populations: leaf damage by insect herbivores (e.g., Carmona and Fornoni 2013; Kalske et al. 2012; Prasad et al. 2012; Wise and Rausher 2013). In both Colorado gardens, foliar herbivory increases with source elevation, indicating that across test sites, high elevation populations are more susceptible to damage by insect herbivores. Interestingly, we also found that foliar damage increased with geographic distance in the 2012 cohort (for both Colorado gardens), suggesting that there could be some adaptation to local herbivore populations.
We hypothesize that short growing seasons and cool temperatures at high elevations limit herbivore populations, reduce selection for resistance, and mediate selection for rapid development. The Colorado populations show clear genetic clines in flowering phenology and size at flowering: In both common gardens, high elevation gentoypes flower rapidly at small sizes and for a shorter duration than low elevation genotypes (Anderson and Gezon 2015). We suggest that foliar traits that enable rapid development, such as high foliar nitrogen content and thin leaves (high specific leaf area), increase susceptibility of leaves to insect herbivores (Anderson and Gezon 2015). Rapid responses of herbivores to climate change (Rasmann et al. 2014) could disproportionately diminish the future fitness of plants from high elevation populations if B. stricta does not keep pace with climate change via migration, adaptation and plasticity.
Climate change and local adaptation
Climate change has already induced local maladaptation in the model organism Arabidopsis thaliana (Wilczek et al. 2014) and will likely alter the locations where tree genotypes have optimal performance (Wang et al. 2010). Transplanted genotypes from historically warmer populations outperformed local genotypes in 4 common gardens in the native range of A. thaliana (Wilczek et al. 2014). The demographic consequences of local maladaptation mediated by climate change include reduced population growth rates (λ) and local population extinction (Wilczek et al. 2014). An additional consequence of reduced λ that has not been highlighted in the literature is that fewer individuals will emigrate from contracting populations, which would decrease the rate at which populations migrate to track favorable climatic conditions.
We found evidence for clinal variation in local adaptation in all gardens, and maladaptation in a subset of the datasets. Maladaptation may become more apparent as climate change proceeds and genotypes from warmer, lower elevation populations gain a fitness advantage over local genotypes. In addition, the poor performance of mid-elevation families in the low elevation Colorado garden foreshadows maladaptation in their home sites in the near future as temperatures continue to increase. Future manipulative field studies are needed to examine the evolutionary consequences of climate change in this landscape, and these studies should examine fitness across life history stages especially at the seed-seedling transition. Seeds rely on climatic cues such as temperature and water to initiate dormancy and to germinate; therefore, germination success could be highly susceptible to climate change (Walck et al. 2011). Finally, genetic variation in fitness suggests that these regional pools of populations have the capacity to evolve higher fitness in contemporary environments, but changing conditions could alter this evolutionary potential. Not only do we need more estimates of genetic variation in components of fitness in nature (Shaw and Shaw 2014), but we need additional estimates in treatments that mimic future conditions to test the potential of global change to disrupt adaptive evolution.
Conclusions
Our field and population genetic studies documented microgeographic variation in fitness and local adaptation, as well as high levels of genetic similarity among individuals in different populations, and moderate isolation by distance. Our approach revealed evidence for local adaptation and maladaptation that would not have been apparent in traditional home vs. away reciprocal transplant designs. In Colorado, fitness clines were consistent with local adaptation for early life history events (germination in the high elevation garden and survival in the low elevation garden), but local maladaptation became apparent for fecundity owing to the unexpectedly high fitness of one high elevation genotype. In the Idaho garden, fitness clines were generally consistent across survival and fecundity, and show that genotypes from warm environments (low elevation or south facing sites) have reduced success in the cool north-facing garden. This approach is key for detecting adaptation to continuous environmental variation along gradients, and can reveal the evolutionary consequences of rapid climate change (Wang et al. 2010; Wilczek et al. 2014).
Supplementary Material
Acknowledgements
We are grateful to Caroline Daws, Anna Battiata, Cassidy Way, Tatiana Vasquez, and Natalie Lowell for field and labwork at the Rocky Mountain Biological Laboratory, to Maggie Wagner, Cathy Rushworth, Cheng-Ruei Lee, and Rose Keith for fieldwork in Idaho, and to Kathy Ghattas for assistance at Duke. We thank Dr. Mike Whitlock and three anonymous reviewers for comments on a previous draft. Funding was provided by the University of South Carolina and University of Georgia (to J.T.A.), the National Institutes of Health (R01 GM086496 to T.M.O.) and the National Science Foundation (EF-0723447 to T.M.O.).
Footnotes
Online Appendices
Online Appendix 1: Oligonucleotide sequences for primers of the microsatellite loci scored in this study.
Online Appendix 2: Coordinates of accessions genotyped to quantify genetic similarity.
Online Appendix 3: Coordinates for gardens and maternal families in the Colorado and Idaho common garden experiments.
Online Appendix 4: Lifetime fitness components (survival and fecundity) analyzed separately for each garden and cohort.
Online Appendix 5: Repeated measures analysis of fitness components in the three years of the study of the low elevation Colorado garden (2012, 2013, and 2014), documenting that the fitness clines are stable through time.
Online Appendix 6: Fitness analyses including plants that died over the first winter for the Colorado 2011 low elevation cohort.
Online Appendix 7: Clinal variation in fitness, modeled using individual level lifetime fitness data with a zero-inflated negative binomial distribution (R package glmmADMB ver. 0.8.0).
Literature cited
- Ågren J, Oakley CG, McKay JK, Lovell JT, Schemske DW. Genetic mapping of adaptation reveals fitness trade-offs in Arabidopsis thaliana. Proceedings of the National Academy of Sciences. 2013;110:21077–21087. doi: 10.1073/pnas.1316773110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken SN, Yeaman S, Holliday JA, Wang T, Curtis-McLane S. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evolutionary Applications. 2008;1:95–111. doi: 10.1111/j.1752-4571.2007.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shehbaz IA, Windham MD. In: Boechera. F. o. N. A. E. Committee, editor. Oxford University Press; Floral of North America North of Mexico. New York and Oxford: 2010. pp. 348–412. [Google Scholar]
- Alberto F, Aitken SN, Alía R, González-Martínez SC, Hänninen H, Kremer A, Lefèvre F, et al. Potential for evolutionary responess to climate change - evidence from tree populations. Global Change Biology. 2013;19:1645–1661. doi: 10.1111/gcb.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Gezon Z. Plasticity in functional traits in the context of climate change: A case study of the subalpine forb Boechera stricta (Brassicaceae). Global Change Biology. 2015 doi: 10.1111/gcb.12770. [DOI] [PubMed] [Google Scholar]
- Anderson J, Lee C-R, Mitchell-Olds T. Strong selection genome-wide enhances fitness tradeoffs across environments and episodes of selection. Evolution. 2014;68:16–31. doi: 10.1111/evo.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Lee C-R, Rushworth C, Colautti R, Mitchell-Olds T. Genetic tradeoffs and conditional neutrality contribute to local adaptation. Molecular Ecology. 2013;22:699–708. doi: 10.1111/j.1365-294X.2012.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennington C, Fetcher N, Vavrek M, Shaver G, Cummings K, McGraw J. Home site advantage in two long-lived arctic plant species: results from two 30-year reciprocal transplant studies. Journal of Ecology. 2012;100:841–851. [Google Scholar]
- Blanquart F, Kaltz O, Nuismer SL, Gandon S. A practical guide to measuring local adaptation. Ecology Letters. 2013;16:1195–1205. doi: 10.1111/ele.12150. [DOI] [PubMed] [Google Scholar]
- Byars S, Papst W, Hoffmann AA. Local adaptation and cogradient selection in the alpine plant, Poa hiemata, along a narrow altitudinal gradient. Evolution. 2007;61:2925–2941. doi: 10.1111/j.1558-5646.2007.00248.x. [DOI] [PubMed] [Google Scholar]
- Carmona D, Fornoni J. Herbivores can select for mixed defensive strategies in plants. New Phytologist. 2013;197:576–585. doi: 10.1111/nph.12023. [DOI] [PubMed] [Google Scholar]
- Clausen J, Keck D, Hiesey W. Effects of varied environments on Western North American Plants. Carnegie Institution of Washington; Washington, D.C., U.S.A.: 1940. Experimental studies on the nature of species. I. [Google Scholar]
- Donohue K, Rubio de Casas R, Burghardt LT, Kovach K, Willis CG. Germination, postgermination adaptation, and species ecological ranges. Annual Review of Ecology and Systematics. 2010;41:293–319. [Google Scholar]
- Dunne JA, Harte J, Taylor KJ. Subalpine meadow flowering phenology responses to climate change: Integrating experimental and gradient methods. Ecological Monographs. 2003;73:69–86. [Google Scholar]
- Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334:86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- Geber M, Eckhart V. Experimental studies of adaptation in Clarkia xantiana. II Fitness variation across a subspecies border. Evolution. 2005;59:521–531. [PubMed] [Google Scholar]
- Goodwillie C, Kalisz S, Eckert CG. The evolutionary enigma of mixed mating systems in plants: Occurrence, theoretical explanations, and empirical evidence. Annual Review Of Ecology Evolution And Systematics. 2005;36:47–79. [Google Scholar]
- Goudet J, Buchi L. The effects of dominance, regular inbreeding and sampling design on QST, an estimator of population differentiation for quantitative traits. Genetics. 2006;172:1337–1347. doi: 10.1534/genetics.105.050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould B, Moeller DA, Eckhart V, Tiffin P, Fabio ES, Geber M. Local adaptation and range boundary formation in response to complex environmental gradients across the geographical range of Clarkia xantiana ssp. xantiana. Journal of Ecology. 2014;102:95–107. [Google Scholar]
- Hendry AP, Taylor EB, McPhail JD. Adaptive divergence and the balance between selection and gene flow: Lake and stream stickleback in the misty system. Evolution. 2002;56:1199–1216. doi: 10.1111/j.0014-3820.2002.tb01432.x. [DOI] [PubMed] [Google Scholar]
- Hereford J. A quantitative survey of local adaptation and fitness trade-offs. American Naturalist. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Hereford J, Winn AA. Limits to local adaptation in six populations of the annual plant Diodia teres. New Phytologist. 2008;178:888–896. doi: 10.1111/j.1469-8137.2008.02405.x. [DOI] [PubMed] [Google Scholar]
- Kalske A, Muola A, Laukkanen L, Mutikainen P, Leimu R. Variation and constraints of local adaptation of a long-lived plant, its pollinators and specialist herbivores. Journal of Ecology. 2012;100:1359–1372. [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Kelly AE, Goulden ML. Rapid shifts in plant distribution with recent climate change. Proceedings of the National Academy of Sciences. 2008;105:11823–11826. doi: 10.1073/pnas.0802891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C, Dobeš C, Sharbel T, Koch M. Phylogeographic structure of the chloroplast DNA gene pool in North American Boechera – A genus and continental-wide perspective. Molecular Phylogenetics and Evolution. 2009;52:303–311. doi: 10.1016/j.ympev.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Kim E, Donohue K. Local adaptation and plasticity of Erysimum capitatum to altitude: its implications for responses to climate change. Journal of Ecology. 2013;101:796–805. [Google Scholar]
- Knowles N, Dettinger MD, Cayan DR. Trends in snowfall versus rainfall in the Western United States. Journal of Climate. 2006;19:4545–4559. [Google Scholar]
- Kremer A, Ronce O, Robledo-Arnuncio J, Guillaume F, Bohrer G, Nathan R, Bridle J, et al. Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecology Letters. 2012;15:378–392. doi: 10.1111/j.1461-0248.2012.01746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-R, Mitchell-Olds T. Complex trait divergence contributes to environmental niche differentiation in ecological speciation of Boechera stricta. Molecular Ecology. 2013;22:2204–2217. doi: 10.1111/mec.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimu R, Fischer M. A meta-analysis of local adaptation in plants. Public Library of Science One. 2008;3:e4010. doi: 10.1371/journal.pone.0004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T. Gene flow and the limits to natural selection. Trends in Ecology & Evolution. 2002;17:183–189. [Google Scholar]
- Lowry DB, Rockwood RC, Willis JH. Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution. 2008;62:2196–2214. doi: 10.1111/j.1558-5646.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Song B-H, Olson-Manning C, Anderson J, Lee C-R, Schranz M, Windsor A, et al. A gain of function polymorphism controlling complex traits and fitness in nature. Science. 2012;336:1081–1084. doi: 10.1126/science.1221636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmann S, Pellissier L, Defossez E, Jactel H, Kunstler G. Climate-driven change in plant-insect interactions along elevation gradients. Functional Ecology. 2014;28:46–54. [Google Scholar]
- Richardson JL, Urban MC, Bolnick DI, Skelly DK. Microgeographic adaptation and the spatial scale of evolution. Trends in Ecology & Evolution. 2014;29:165–176. doi: 10.1016/j.tree.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Roughgarden J. Theory of population genetics and evolutionary ecology: an introduction. Macmillan; New York: 1979. [Google Scholar]
- Rushworth C, Song B-H, Lee C-R, Mitchell-Olds T. Boechera, a model system for ecological genomics. Molecular Ecology. 2011;20:4843–4857. doi: 10.1111/j.1365-294X.2011.05340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen O, Pyhajarvi T, Knurr T. Gene flow and local adaptation in trees. Annual Review of Ecology Evolution and Systematics. 2007;38:595–619. [Google Scholar]
- Sexton JP, Hangartner S, Hoffmann A. Genetic isolation by environment of distance: Which pattern of gene flow is most common? Evolution. 2014;68:1–15. doi: 10.1111/evo.12258. [DOI] [PubMed] [Google Scholar]
- Shaw R, Shaw F. Quantitative genetic study of the adaptive process. Heredity. 2014;112:13–20. doi: 10.1038/hdy.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Song B-H, Windsor AJ, Schmid KJ, Ramos-Onsins S, Schranz ME, Heidel A, Mitchell-Olds T. Multilocus pattern of nucleotide diversity, population structure and linkage disequilibrium in Boechera stricta, a wild relative of Arabidopsis. Genetics. 2009;181:1021–1033. doi: 10.1534/genetics.108.095364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BH, Clauss M, Pepper A, Mitchell-Olds T. Geographic patterns of microsatellite variation in Boechera stricta, a close relative of Arabidopsis. Molecular Ecology. 2006;15:357–369. doi: 10.1111/j.1365-294X.2005.02817.x. [DOI] [PubMed] [Google Scholar]
- Storz J, Sabatino S, Hoffmann F, Gering E, Moriyama H, Ferrand N, Monteiro B, et al. The molecular basis of high-altitude adaptation in deer mice. PLoS Genetics. 2007;3:e45. doi: 10.1371/journal.pgen.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergeer P, Kunin WE. Adaptation at range margins: Common garden trials and the performance of Arabidopsis lyrata across its northwestern European range. New Phytologist. 2013;197:989–1001. doi: 10.1111/nph.12060. [DOI] [PubMed] [Google Scholar]
- Via S. The genetic structure of host plant adaptation in a spatial patchwork: demographic variability among reciprocally transplanted pea aphid clones. Evolution. 1991;45:827–852. doi: 10.1111/j.1558-5646.1991.tb04353.x. [DOI] [PubMed] [Google Scholar]
- Walck J, Hidayati S, Dixon KW, Thompson K, Poschlod P. Climate change and plant regeneration from seed. Global Change Biology. 2011;17:2145–2161. [Google Scholar]
- Wang T, Hamann A, Spittlehouse D, Murdock T. Climate WNA-High-resolution spaital climate data for Western North America. Journal of Applied Meteorology and Climatology. 2012;51:16–29. [Google Scholar]
- Wang T, O'Neill GA, Aitken SN. Integrating environmental and genetic effects to predict responses of tree populations to climate. Ecological Applications. 2010;20:153–163. doi: 10.1890/08-2257.1. [DOI] [PubMed] [Google Scholar]
- Wilczek AM, Cooper MD, Korves TM, Schmitt J. Lagging adaptation to warming climate in Arabidopsis thaliana. Proceedings of the National Academy of Sciences. 2014;111:7906–7913. doi: 10.1073/pnas.1406314111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ, Rausher MD. Evolution of resistance to a multiple-herbivore community: Genetic correlaions, diffuse coevolution, and constraints on the plant's response to selection. Evolution. 2013;67:1767–1779. doi: 10.1111/evo.12061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.